Screening of Mpro Protease (SARS-CoV-2) Covalent Inhibitors from an Anthocyanin-Rich Blueberry Extract Using an HRMS-Based Analytical Platform
Abstract
:1. Introduction
2. Results
2.1. Metabolomics
2.1.1. Analytical Profiling of BPE
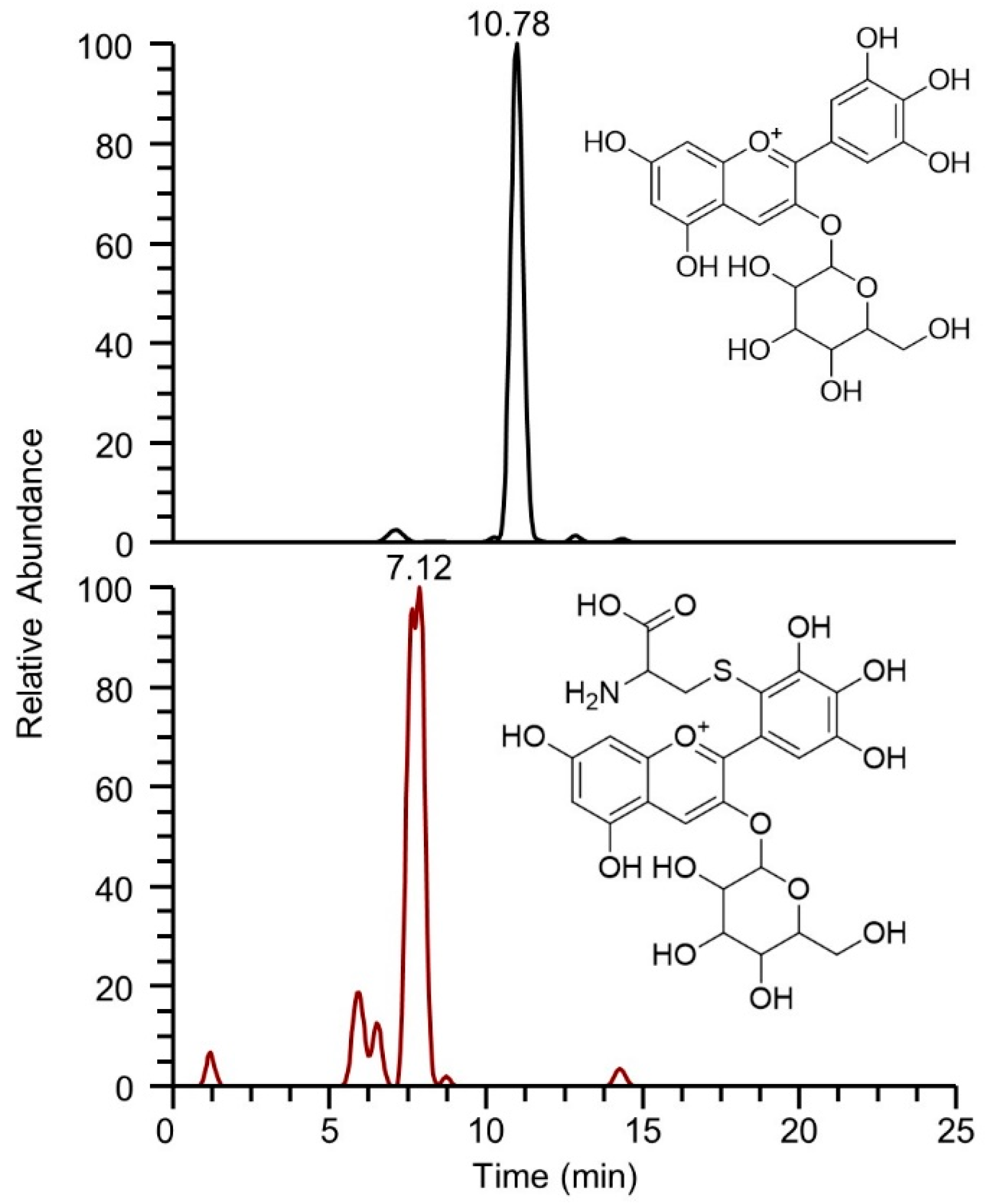
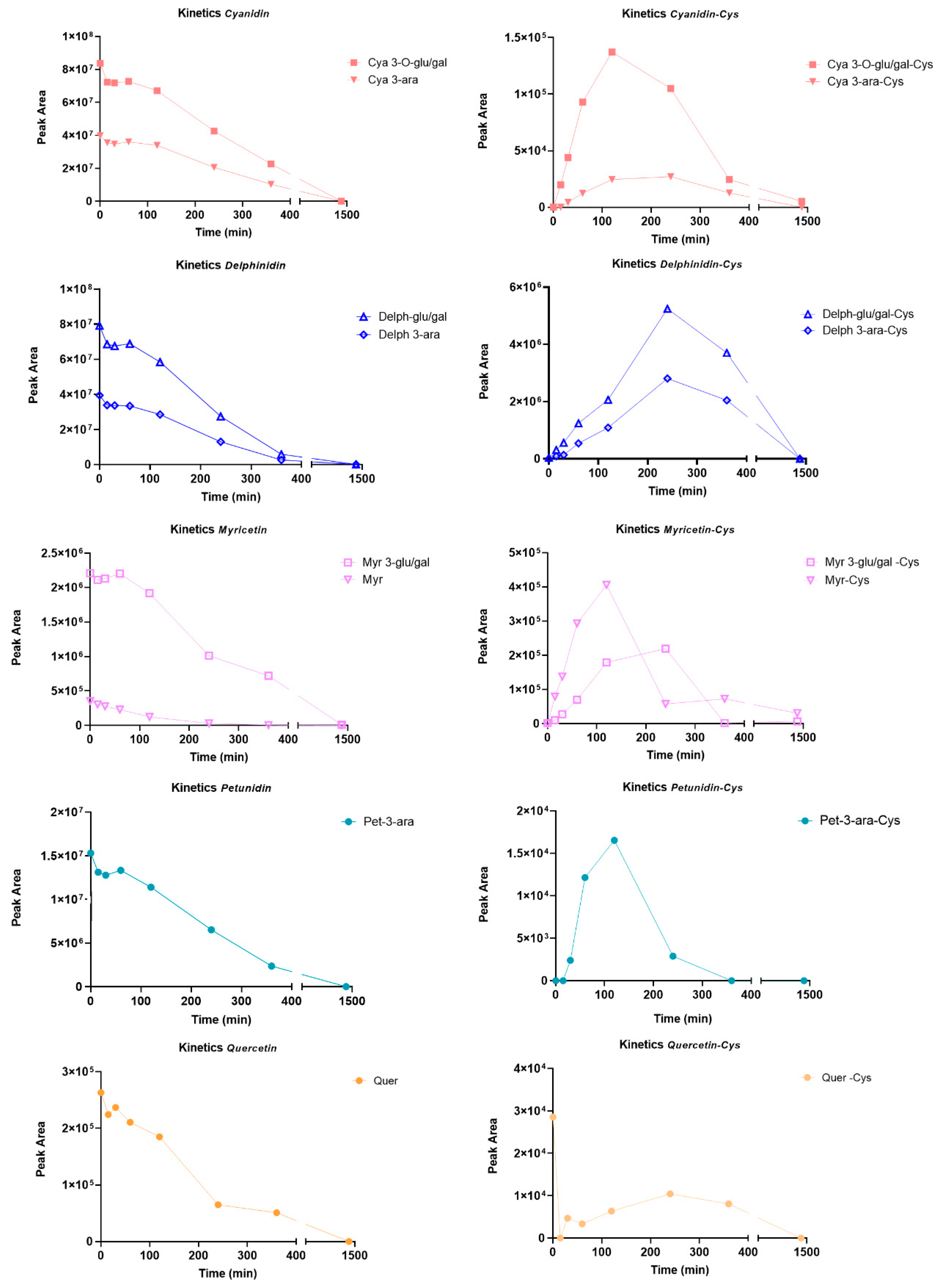
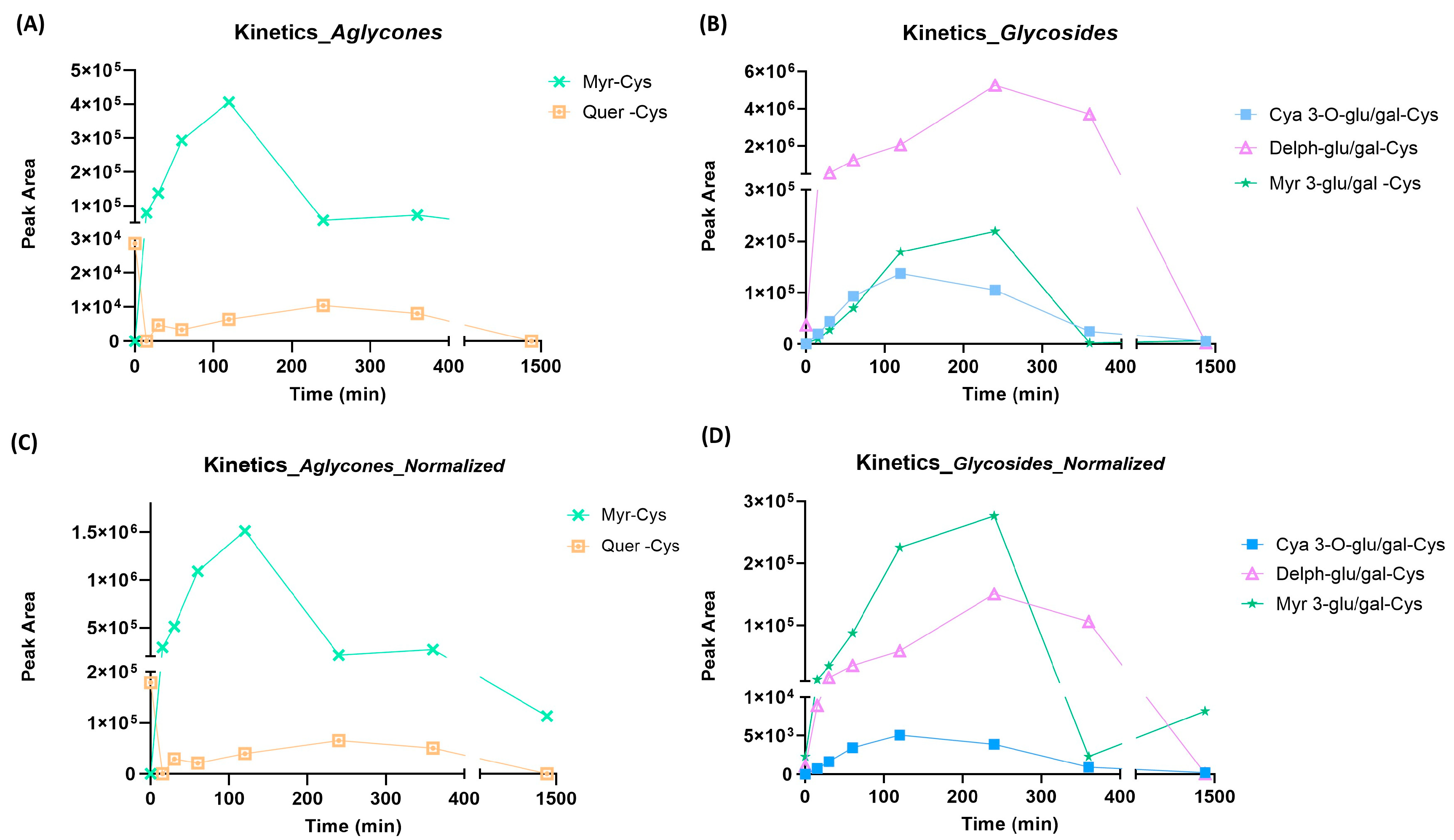
2.1.2. Identification of BPE Electrophilic Compounds and Reaction Kinetics Study
- (i)
- Among the adducts formed in the presence of the glycosidic derivatives of the target flavonoids, the delphinidin-3-glucoside/galactoside-Cys adduct is definitely the predominant one, with a significantly higher content than the adducts formed in the presence of the glycosylated forms of myricetin, cyanidin and petunidin (Delph > Myr > Cya > Pet);
- (ii)
- In general, adducts with derivatives carrying the glucose/galactose units bound are more abundant than those formed with molecules carrying arabinose units;
- (iii)
- The last plausible consideration concerns the different reactivity observed between the glycosidic forms and the corresponding aglycones; actually, the only flavonoid present in both forms is the myricetin. The glycosidic derivative has a better Tmax and a content which is twice as high as the adduct produced with the corresponding aglycone.
2.2. Protein Structure Analysis
2.2.1. Targeted Protein Structure Analysis: Characterization of Potential Covalent Binders of Mpro by nLC-HR-MS/MS Analysis of the Incubation Mixture with BPE
2.2.2. Target-Based Protein Structure Analysis of the Incubation Mixture of Mpro with Delphinidin-3-Glucoside Standard
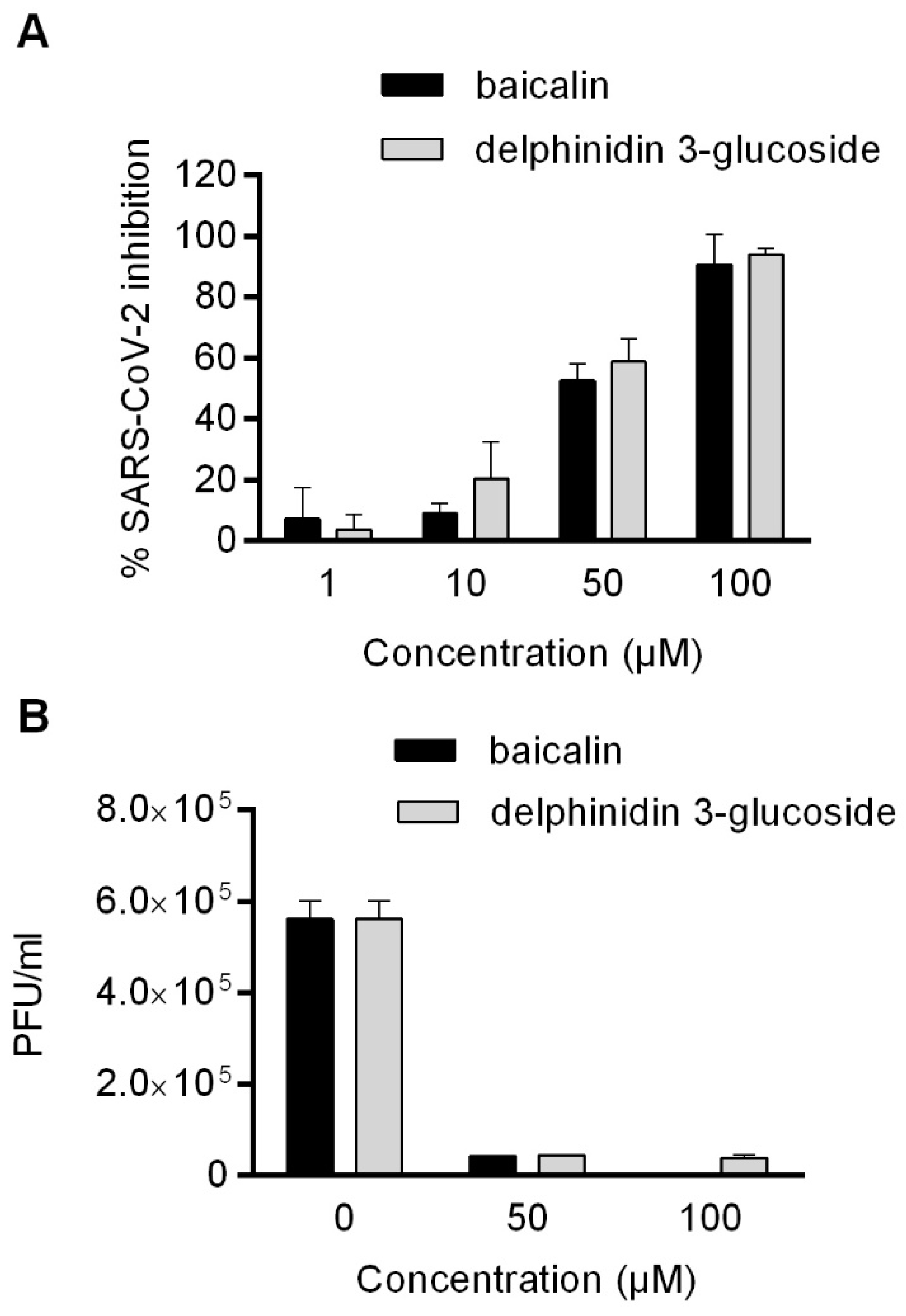
2.3. Evaluation of the Antiviral Activity
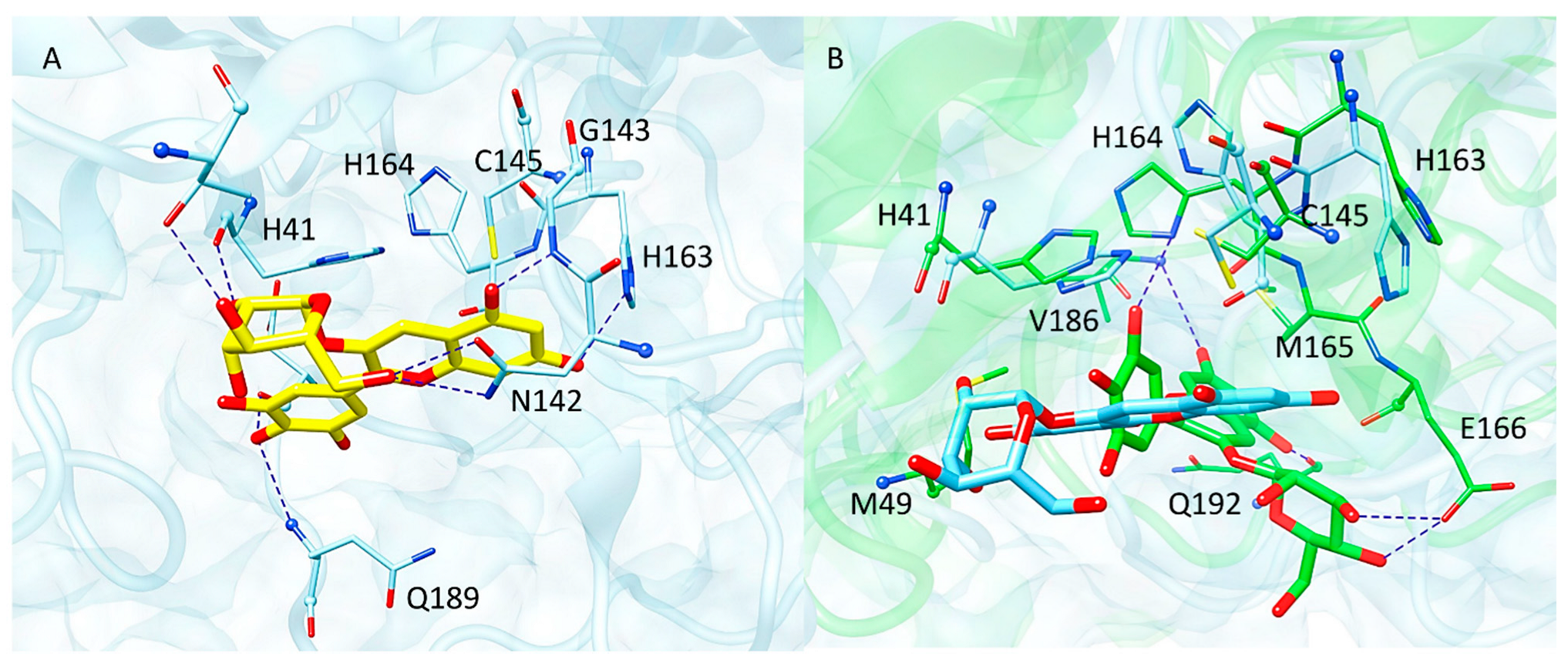
2.4. Computational Studies
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Plant Materials and Blueberry Extract Preparation
4.3. Metabolomics
4.3.1. Analytical Profiling of the Blueberry Polyphenol Extract (BPE)
4.3.2. Electrophilic Compound Identification and Reaction Kinetics Study
- Sample preparation
- Reaction kinetic study by LC-HRMS (BPE: Cysteine)
- MS data elaboration
4.4. Protein Structure Analysis
4.4.1. Characterization and Localization of Protein Adducts
- Mpro incubation with BPE
- Protein digestion (S-TRAP™ technology)
- nLC-HR-MS/MS analysis (Orbitrap Elite™ Mass Spectrometer)
- Targeted data analysis
4.4.2. Characterization of Protein Adducts Deriving from the Mpro Incubation with Delphinidin-3-Glucoside
4.5. In Vitro Evaluation of Antiviral Activity
4.5.1. Cell Culture and Virus
4.5.2. Infection, Treatment with Baicalin and Delphinidin-3-Glucoside
4.5.3. Antiviral Assays
4.5.4. Evaluation of Virucide Activity by Plaque Reduction Assay
4.5.5. Statistical Analysis
4.6. Computational Studies
4.6.1. Molecular Docking
4.6.2. Molecular Dynamics Simulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviation
| ACN | Acetonitrile |
| AUC | Area under the curve |
| BPE | Blueberry polyphenol extract |
| Cys | Cysteine |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FA | Formic acid |
| FBS | Fetal bovine serum |
| His | Histidine |
| HR-MS | High-resolution mass spectrometry |
| HTS | High Throughput Screening |
| IAA | Iodoacetamide |
| LC-HRMS | Liquid Chromatography High Resolution Mass Spectrometry |
| MD | Molecular dynamics |
| MS | Mass Spectrometry |
| Mpro | Main protease |
| Nsp | Non-structural proteins |
| PD | Proteome Discoverer |
| PSMs | Peptide spectral matches |
| TCEP | Tris(2-carboxyethyl)phosphine |
| TEAB | Tetraethylammonium bromide |
| TFA | Trifluoroacetic acid |
| TIC | Total ionic current |
References
- Available online: https://cov-lineages.org/lineage_list.html (accessed on 26 May 2023).
- Beltrán, L.A.; De La Hoz-Rodríguez, S.; Iserte, L.B.; Rodríguez, S.; Fernández-De-La-pradilla, A.; González, F.V. Advances in the Development of SARS-CoV-2 Mpro Inhibitors. Molecules 2022, 27, 2523. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Faheem; Kumar, B.K.; Sekhar, K.V.G.C.; Kunjiappan, S.; Jamalis, J.; Balaña-Fouce, R.; Tekwani, B.L.; Sankaranarayanan, M. Druggable targets of SARS-CoV-2 and treatment opportunities for COVID-19. Bioorg. Chem. 2020, 104, 104269. [Google Scholar] [CrossRef]
- Sacco, M.D.; Ma, C.; Lagarias, P.; Gao, A.; Townsend, J.A.; Meng, X.; Dube, P.; Zhang, X.; Hu, Y.; Kitamura, N.; et al. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020, 6, eabe0751. [Google Scholar] [CrossRef] [PubMed]
- El-Baba, T.J.; Lutomski, C.A.; Kantsadi, A.L.; Malla, T.R.; John, T.; Mikhailov, V.; Bolla, J.R.; Schofield, C.J.; Zitzmann, N.; Vakonakis, I.; et al. Allosteric Inhibition of the SARS-CoV-2 Main Protease: Insights from Mass Spectrometry Based Assays. Angew. Chemie Int. Ed. 2020, 59, 23544–23548. [Google Scholar] [CrossRef] [PubMed]
- Di, L.; Kerns, E.H.; Hong, Y.; Chen, H. Development and application of high throughput plasma stability assay for drug discovery. Int. J. Pharm. 2005, 297, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Sacco, M.D.; Hurst, B.; Townsend, J.A.; Hu, Y.; Szeto, T.; Zhang, X.; Tarbet, B.; Marty, M.T.; Chen, Y.; et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020, 30, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Clyde, A.; Galanie, S.; Kneller, D.W.; Ma, H.; Babuji, Y.; Blaiszik, B.; Brace, A.; Brettin, T.; Chard, K.; Chard, R.; et al. High Throughput Virtual Screening and Validation of a SARS-CoV-2 Main Protease Non-Covalent Inhibitor. bioRxiv 2021. [Google Scholar] [CrossRef]
- Stein, R.M.; Kang, H.J.; McCorvy, J.D.; Glatfelter, G.C.; Jones, A.J.; Che, T.; Slocum, S.; Huang, X.P.; Savych, O.; Moroz, Y.S.; et al. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature 2020, 579, 609–614. [Google Scholar] [CrossRef]
- Lyu, J.; Wang, S.; Balius, T.; Singh, I.; Levit, A.; Moroz, Y.S.; O’Meara, M.J.; Che, T.; Algaa, E.; Tolmachova, K.; et al. Ultra-large library docking for discovering new chemotypes. Nature 2019, 566, 224–229. [Google Scholar] [CrossRef]
- Baron, G.; Borella, S.; della Vedova, L.; Vittorio, S.; Vistoli, G.; Carini, M.; Aldini, G.; Altomare, A. An integrated metabolomic and proteomic approach for the identification of covalent inhibitors of the main protease (Mpro) of SARS-CoV-2 from crude natural extracts. Talanta 2023, 252, 123824. [Google Scholar] [CrossRef] [PubMed]
- Messaoudi, O.; Gouzi, H.; El-Hoshoudy, A.N.; Benaceur, F.; Patel, C.; Goswami, D.; Boukerouis, D.; Bendahou, M. Berries anthocyanins as potential SARS-CoV-2 inhibitors targeting the viral attachment and replication; molecular docking simulation. Egypt. J. Pet. 2021, 30, 33–43. [Google Scholar] [CrossRef]
- Silva, S.; Costa, E.M.; Veiga, M.; Morais, R.M.; Calhau, C.; Pintado, M. Health promoting properties of blueberries: A review. Crit. Rev. Food Sci. Nutr. 2020, 60, 181–200. [Google Scholar] [CrossRef]
- Sarni-Manchado, P.; Cheynier, V.; Moutounet, M. Reactions of polyphenoloxidase generated caftaric acid o-quinone with malvidin 3-O-glucoside. Phytochemistry 1997, 45, 1365–1369. [Google Scholar] [CrossRef]
- Dangles, O.; Fenger, J.A. The Chemical Reactivity of Anthocyanins and Its Consequences in Food Science and Nutrition. Molecules 2018, 23, 1970. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Yao, S.; Zhao, W.; Zhang, Y.; Liu, J.; Shao, Q.; Wang, Q.; Li, M.; Xie, H.; Shang, W.; et al. Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease. Nat. Commun. 2021, 12, 3623. [Google Scholar] [CrossRef] [PubMed]
- Joyner, P.M. Protein Adducts and Protein Oxidation as Molecular Mechanisms of Flavonoid Bioactivity. Molecules 2021, 26, 5102. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Mori, T.; Tanaka, T.; Mizuno, D.; Yamaji, R.; Kumazawa, S.; Nakayama, T.; Akagawa, M. Covalent modification of proteins by green tea polyphenol (–)-epigallocatechin-3-gallate through autoxidation. Free Radic. Biol. Med. 2008, 45, 1384–1394. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Rajamani, S.; Kaylor, J.; Han, S.; Zhou, F.; Fink, A.L. The flavonoid baicalein inhibits fibrillation of alpha-synuclein and disaggregates existing fibrils. J. Biol. Chem. 2004, 279, 26846–26857. [Google Scholar] [CrossRef]
- Kneller, D.W.; Phillips, G.; O’Neill, H.M.; Jedrzejczak, R.; Stols, L.; Langan, P.; Joachimiak, A.; Coates, L.; Kovalevsky, A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020, 11, 3202. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Hamdoun, S.; Chen, R.; Yang, L.; Ip, C.K.; Qu, Y.; Li, R.; Jiang, H.; Yang, Z.; Chung, S.K.; et al. Identification of natural compounds as SARS-CoV-2 entry inhibitors by molecular docking-based virtual screening with bio-layer interferometry. Pharmacol. Res. 2021, 172, 105820. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Cui, M.; Zheng, C.; Wang, M.; Sun, R.; Gao, D.; Bao, J.; Ren, S.; Yang, B.; Lin, J.; et al. Myricetin Inhibits SARS-CoV-2 Viral Replication by Targeting Mpro and Ameliorates Pulmonary Inflammation. Front. Pharmacol. 2021, 12, 669642. [Google Scholar] [CrossRef]
- Liu, H.; Ye, F.; Sun, Q.; Liang, H.; Li, C.; Li, S.; Lu, R.; Huang, B.; Tan, W.; Lai, L. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J. Enzyme Inhib. Med. Chem. 2021, 36, 497–503. [Google Scholar] [CrossRef]
- Pitsillou, E.; Liang, J.; Karagiannis, C.; Ververis, K.; Darmawan, K.K.; Ng, K.; Hung, A.; Karagiannis, T.C. Interaction of small molecules with the SARS-CoV-2 main protease in silico and in vitro validation of potential lead compounds using an enzyme-linked immunosorbent assay. Comput. Biol. Chem. 2020, 89, 1476–9271. [Google Scholar] [CrossRef]
- Ongkowijoyo, P.; Luna-Vital, D.A.; Gonzalez de Mejia, E. Extraction techniques and analysis of anthocyanins from food sources by mass spectrometry: An update. Food Chem. 2018, 250, 113–126. [Google Scholar] [CrossRef]
- Baron, G.; Ferrario, G.; Marinello, C.; Carini, M.; Morazzoni, P.; Aldini, G. Effect of Extraction Solvent and Temperature on Polyphenol Profiles, Antioxidant and Anti-Inflammatory Effects of Red Grape Skin By-Product. Molecules 2021, 26, 5454. [Google Scholar] [CrossRef] [PubMed]
- Gato, E.; Perez, A.; Rosalowska, A.; Celeiro, M.; Bou, G.; Lores, M. Multicomponent polyphenolic extracts from Vaccinium corymbosum at lab and pilot scale. Characterization and effectivity against nosocomial pathogens. Plants 2021, 10, 2801. [Google Scholar] [CrossRef]
- Cladis, D.P.; Debelo, H.; Lachcik, P.J.; Ferruzzi, M.G.; Weaver, C.M. Increasing Doses of Blueberry Polyphenols Alters Colonic Metabolism and Calcium Absorption in Ovariectomized Rats. Mol. Nutr. Food Res. 2020, 64, 2000031. [Google Scholar] [CrossRef]
- Müller, D.; Schantz, M.; Richling, E. High performance liquid chromatography analysis of anthocyanins in bilberries (Vaccinium myrtillus L.), blueberries (Vaccinium corymbosum L.), and corresponding juices. J. Food Sci. 2012, 77, C340–C345. [Google Scholar] [CrossRef]
- Contreras, R.A.; Köhler, H.; Pizarro, M.; Zúñiga, G.E. In Vitro Cultivars of Vaccinium corymbosum L. (Ericaceae) are a Source of Antioxidant Phenolics. Antioxidants 2015, 4, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, A.M.; Angeloni, S.; Abouelenein, D.; Acquaticci, L.; Xiao, J.; Sagratini, G.; Maggi, F.; Vittori, S.; Caprioli, G. A new HPLC-MS/MS method for the simultaneous determination of 36 polyphenols in blueberry, strawberry and their commercial products and determination of antioxidant activity. Food Chem. 2022, 367, 130743. [Google Scholar] [CrossRef] [PubMed]
- Grace, M.H.; Xiong, J.; Esposito, D.; Ehlenfeldt, M.; Lila, M.A. Simultaneous LC-MS quantification of anthocyanins and non-anthocyanin phenolics from blueberries with widely divergent profiles and biological activities. Food Chem. 2019, 277, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Mikulic-Petkovsek, M.; Slatnar, A.; Stampar, F.; Veberic, R. HPLC–MSn identification and quantification of flavonol glycosides in 28 wild and cultivated berry species. Food Chem. 2012, 135, 2138–2146. [Google Scholar] [CrossRef] [PubMed]
- Delbue, S.; D’Alessandro, S.; Signorini, L.; Dolci, M.; Pariani, E.; Bianchi, M.; Fattori, S.; Modenese, A.; Galli, C.; Eberini, I.; et al. Isolation of SARS-CoV-2 strains carrying a nucleotide mutation, leading to a stop codon in the ORF 6 protein. Emerg. Microbes Infect. 2021, 10, 252–255. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Available online: https://www.fda.gov/media/134922/download (accessed on 2 December 2022).
- Parisi, O.I.; Dattilo, M.; Patitucci, F.; Malivindi, R.; Delbue, S.; Ferrante, P.; Parapini, S.; Galeazzi, R.; Cavarelli, M.; Cilurzo, F.; et al. Design and development of plastic antibodies against SARS-CoV-2 RBD based on molecularly imprinted polymers that inhibit in vitro virus infection. Nanoscale 2021, 13, 16885–16899. [Google Scholar] [CrossRef]
- Su, H.X.; Yao, S.; Zhao, W.F.; Li, M.J.; Liu, J.; Shang, W.J.; Xie, H.; Ke, C.Q.; Hu, H.C.; Gao, M.N.; et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020, 41, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Vittorio, S.; Manelfi, C.; Gervasoni, S.; Beccari, A.R.; Pedretti, A.; Vistoli, G.; Talarico, C. Computational Insights into the Sequence-Activity Relationships of the NGF(1–14) Peptide by Molecular Dynamics Simulations. Cells 2022, 11, 2808. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J.P. Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters. J. Mol. Model. 2013, 19, 1–32. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. PLANTS: Application of ant colony optimization to structure-based drug design. In Ant Colony Optimization and Swarm Intelligence: Proceedings of the 6th International Conference, ANTS 2006, Brussels, Belgium, 4–7 September 2006; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4150, pp. 247–258. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Tubiana, T.; Carvaillo, J.C.; Boulard, Y.; Bressanelli, S. TTClust: A Versatile Molecular Simulation Trajectory Clustering Program with Graphical Summaries. J. Chem. Inf. Model. 2018, 58, 2178–2182. [Google Scholar] [CrossRef] [PubMed]
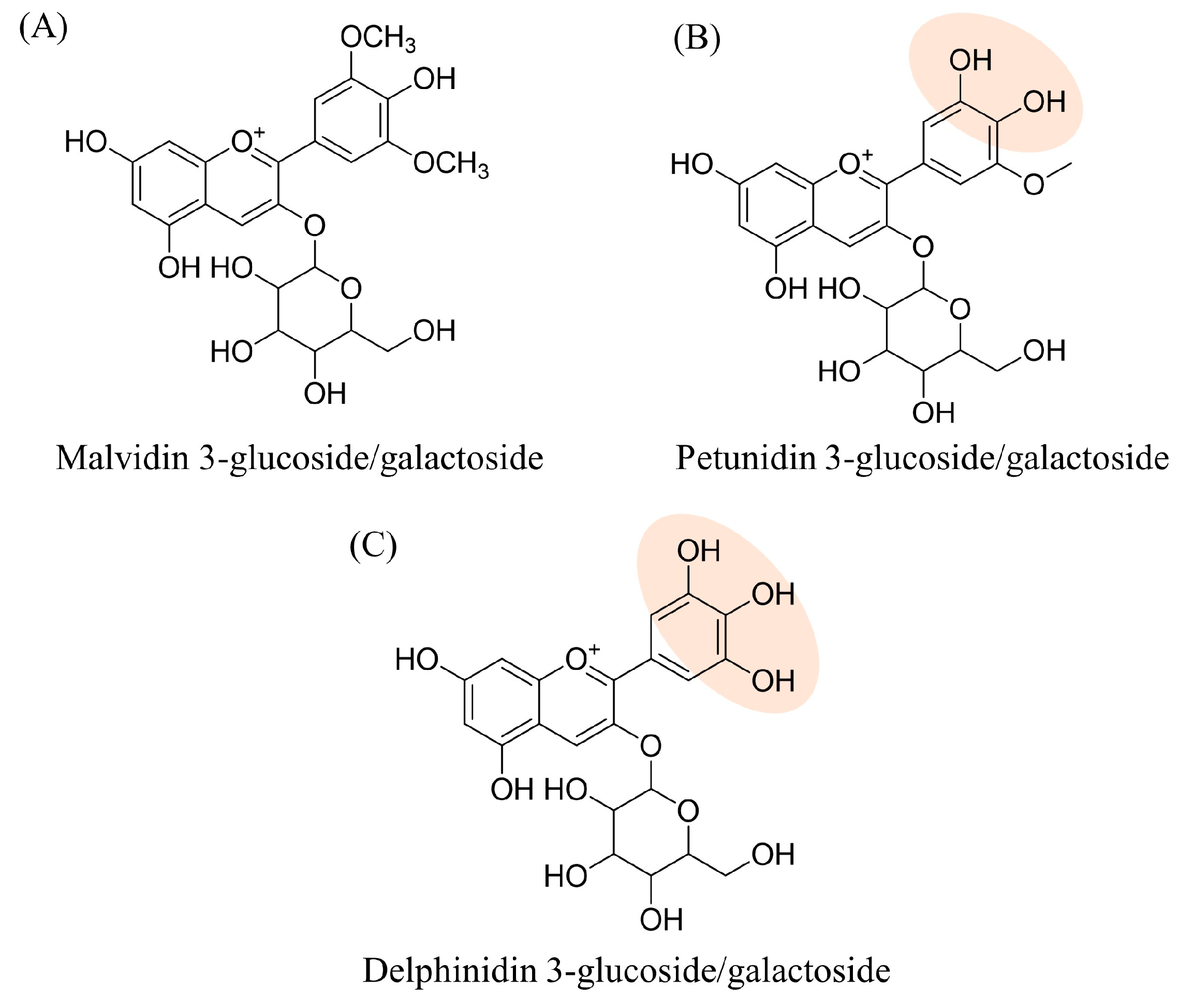
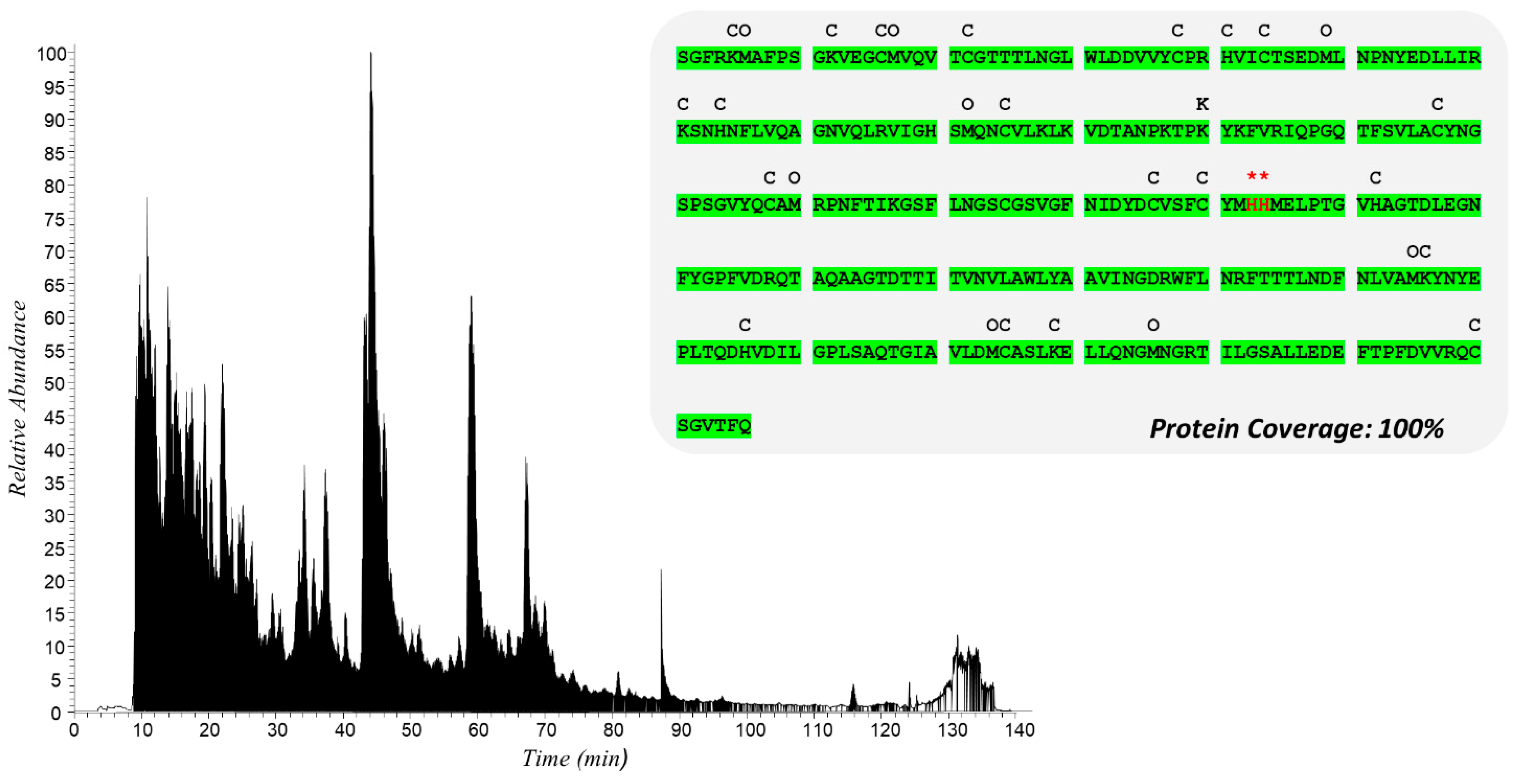
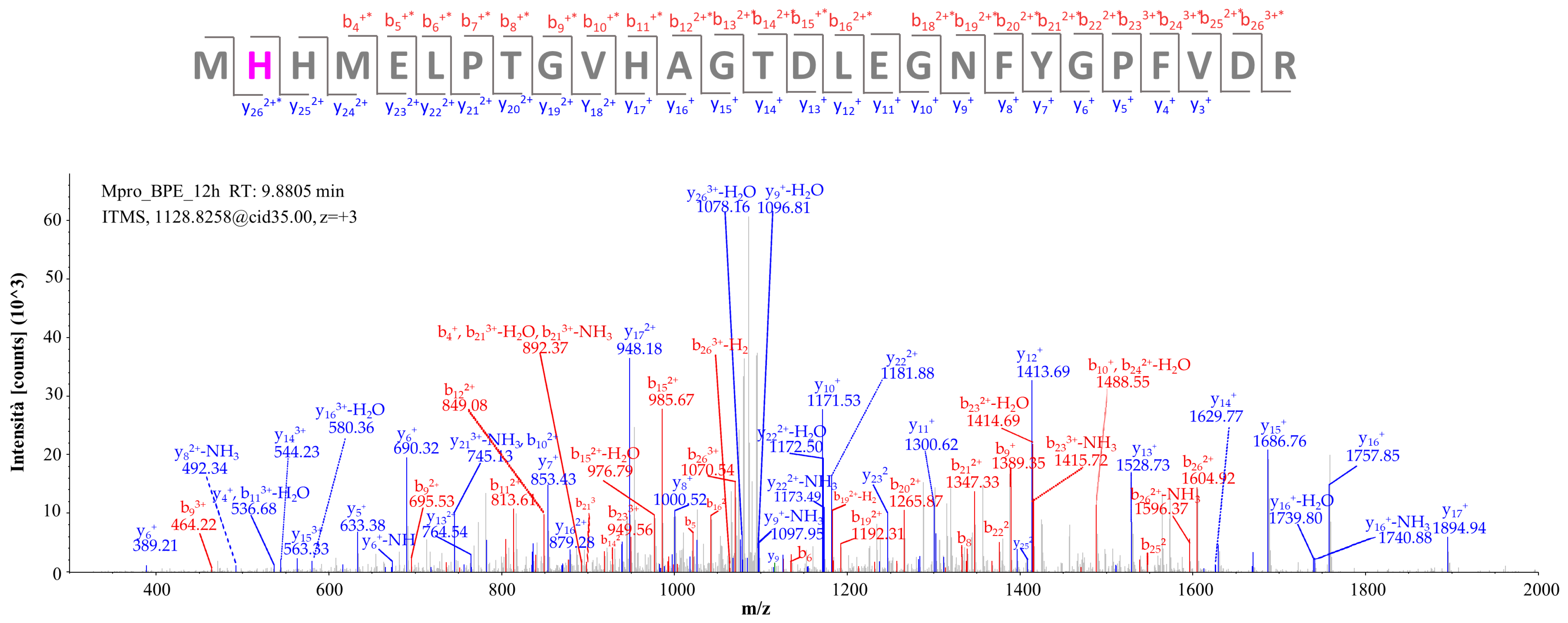
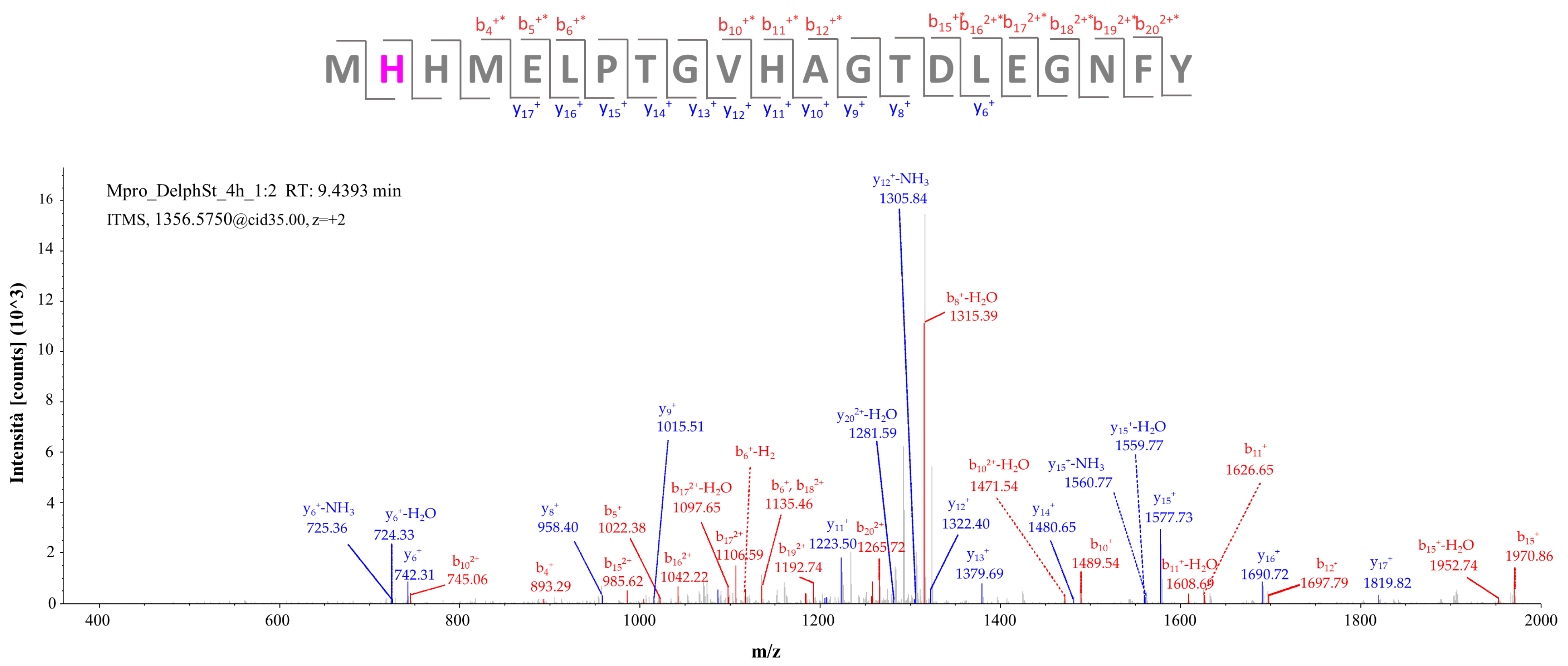
| Peak N° | Compound | Chemical Formula | Th. [M]+/[M + H]+ | Exp. [M]+/[M + H]+ | Δ ppm | MS/MS | RT (min) | Rel. Ab. (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | Delphinidin-3-glucoside/galactoside | C21H21O12+ | 465.1033 | 465.10201 | −2.77 | 303 | 3.33 | 12.11 |
| 2 | Delphinidin-3-arabinoside | C20H19O11+ | 435.09273 | 435.09188 | −1.95 | 303 | 4.31 | 6.14 |
| 3 | Cyanidin-3-glucoside/galactoside | C21H21O11+ | 449.10838 | 449.1074 | −2.18 | 287 | 4.59 | 5.33 |
| 4 | Petunidin-3-glucoside/galactoside | C22H23O12+ | 479.11895 | 479.11793 | −2.13 | 317 | 5.2 | 12.72 |
| 5 | Cyanidin-3-arabinoside | C20H19O10+ | 419.09782 | 419.09731 | −1.22 | 287 | 6.58 | 2.87 |
| 6 | Petunidin-3-arabinoside | C21H21O11+ | 449.10838 | 449.10775 | −1.4 | 317 | 7.5 | 5.83 |
| 7 | Peonidin-3-glucoside/galactoside | C22H23O11+ | 463.12403 | 463.12319 | −1.81 | 301 | 7.5 | 1.4 |
| 8 | Malvidin-3-glucoside/galactoside | C23H25O12+ | 493.1346 | 493.13362 | −1.99 | 331 | 8.67 | 22.99 |
| 9 | Peonidin-3-arabinoside | C21H21O10+ | 433.11347 | 433.11285 | −1.43 | 301 | 10.43 | 0.82 |
| 10 | Malvidin-3-arabinoside | C22H23O11+ | 463.12403 | 463.12326 | −1.66 | 331 | 11.57 | 14.2 |
| 11 | Myricetin-3-glucoside/galactoside | C21H21O13 | 481.09821 | 481.0973 | −1.9 | 319 | 13.84 | 1.1 |
| 12 | Quercetin-3-glucoside/galactoside | C21H21O12 | 465.1033 | 465.10255 | −1.61 | 303 | 20.96 | 8.64 |
| 13 | Quercetin-3-glucuronide | C21H19O13 | 479.08256 | 479.08165 | −1.9 | 303 | 21.53 | 0.53 |
| 14 | Quercetin-3-arabinoside/xyloside | C20H19O11 | 435.09273 | 435.09216 | −1.32 | 303 | 25.92 | 3.42 |
| 15 | Quercetin-3-rhamnoside | C21H21O11 | 449.10838 | 449.10793 | −1.01 | 303 | 29.77 | 1.59 |
| 16 | Isorhamnetin-3-glucoside/galactoside | C22H23O12 | 479.11895 | 479.11824 | −1.47 | 317 | 30.32 | 0.29 |
| 17 | Myricetin | C15H11O8 | 319.04539 | 319.04512 | −0.85 | 181 | 31.04 | 0.02 |
| 18 | Quercetin | C15H11O7 | 303.05048 | 303.05008 | −1.31 | 153–181 | 47.85 | 0.02 |
| Compound | Chemical Formula | Th. [M]+/[M + H]+ |
|---|---|---|
| Cyanidin-3-arabinoside | C23H24NO12S+ | 538.10192 |
| Cyanidin-3-glucoside/galactoside | C24H26NO13S+ | 568.11248 |
| Delphinidin-3-arabinoside | C23H24NO13S+ | 554.09683 |
| Delphinidin-3-glucoside/galactoside | C24H26NO14S+ | 584.10740 |
| Myricetin | C18H15NO12S | 470.03932 |
| Myricetin-3-glucoside/galactoside | C24H25NO15S | 600.10231 |
| Petunidin-3-arabinoside | C24H26NO13S+ | 568.11248 |
| Petunidin-3-glucoside/galactoside | C25H28NO14S+ | 598.12304 |
| Quercetin-3-arabinoside/xyloside | C23H23NO13S | 554.09683 |
| Quercetin-3-glucoside/galactoside | C25H27NO13S | 582.12813 |
| Quercetin-3-rhamnoside | C24H25NO13S | 568.11248 |
| Quercetin | C18H15NO9S | 422.05457 |
| Quercetin-3-glucuronide | C24H23NO15S | 598.08666 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altomare, A.; Baron, G.; Cambiaghi, G.; Ferrario, G.; Zoanni, B.; Della Vedova, L.; Fumagalli, G.M.; D’Alessandro, S.; Parapini, S.; Vittorio, S.; et al. Screening of Mpro Protease (SARS-CoV-2) Covalent Inhibitors from an Anthocyanin-Rich Blueberry Extract Using an HRMS-Based Analytical Platform. Molecules 2024, 29, 2702. https://doi.org/10.3390/molecules29112702
Altomare A, Baron G, Cambiaghi G, Ferrario G, Zoanni B, Della Vedova L, Fumagalli GM, D’Alessandro S, Parapini S, Vittorio S, et al. Screening of Mpro Protease (SARS-CoV-2) Covalent Inhibitors from an Anthocyanin-Rich Blueberry Extract Using an HRMS-Based Analytical Platform. Molecules. 2024; 29(11):2702. https://doi.org/10.3390/molecules29112702
Chicago/Turabian StyleAltomare, Alessandra, Giovanna Baron, Giulia Cambiaghi, Giulio Ferrario, Beatrice Zoanni, Larissa Della Vedova, Giulio Maria Fumagalli, Sarah D’Alessandro, Silvia Parapini, Serena Vittorio, and et al. 2024. "Screening of Mpro Protease (SARS-CoV-2) Covalent Inhibitors from an Anthocyanin-Rich Blueberry Extract Using an HRMS-Based Analytical Platform" Molecules 29, no. 11: 2702. https://doi.org/10.3390/molecules29112702










