Preliminary Screening on Antibacterial Crude Secondary Metabolites Extracted from Bacterial Symbionts and Identification of Functional Bioactive Compounds by FTIR, HPLC and Gas Chromatography–Mass Spectrometry
Abstract
:1. Introduction
2. Results
2.1. Extraction of Symbiotic Bacteria
2.2. Bacterial Identification by 16S rRNA
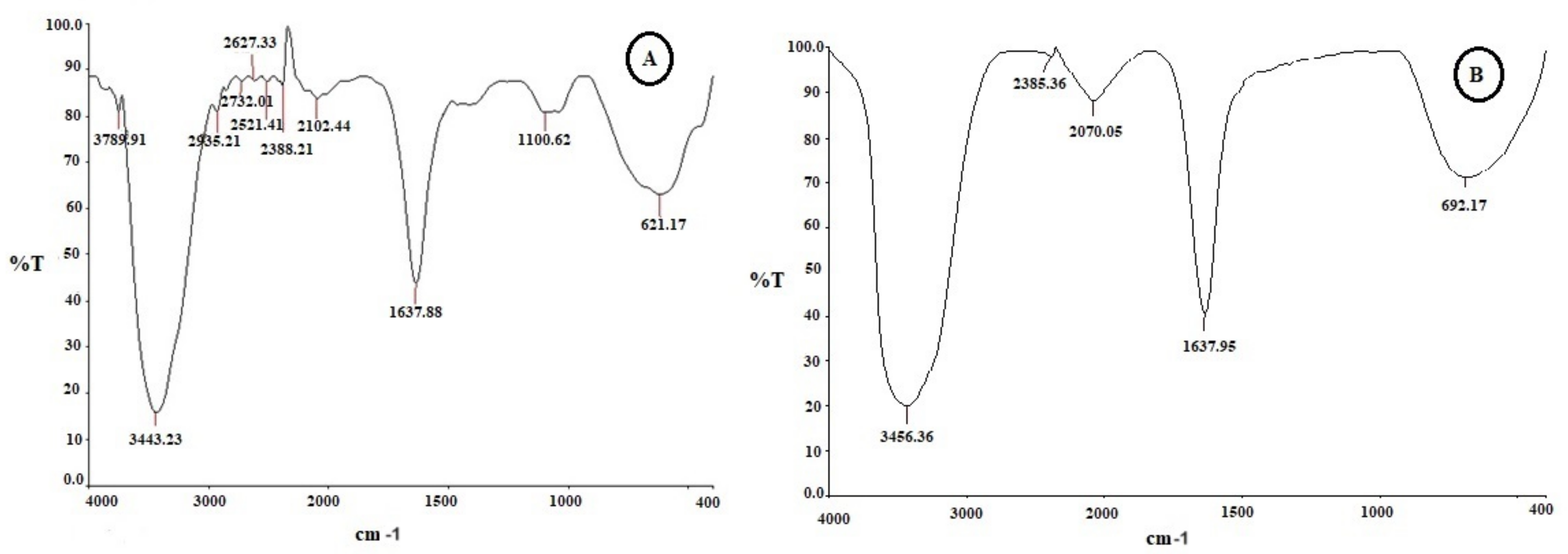
2.3. Functional Group Analysis by FTIR
2.4. Purification of Secondary Functional Compound Using HPLC and GC–MS
2.5. Antibacterial Activities of Symbiotic Bacteria
3. Discussion
4. Materials and Methods
4.1. Chemicals and Media
4.2. Identification, Extraction, and Molecular Characterization of Symbiotic Bacteria
4.3. The 16S rDNA Sequencing and Phylogenetic Analysis
4.4. Extraction of Bioactive Crude Compounds from Symbiotic Bacteria Using Ethyl Acetate
4.5. Purification of Bioactive Compound
4.6. Identification of Functional Bioactive Compounds by FTIR and GC–MS Analysis
4.7. Antibacterial Assay
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gobinath, C.; Rodríguez-Hernández, A.I.; del Rocío López-Cuellar, M.; Palma-Rodríguez, H.M.; Chavarría-Hernández, N. Bacteriocin encapsulation for food and pharmaceutical applications: Advances in the past 20 years. Biotechnol. Lett. 2019, 41, 453–469. [Google Scholar]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef]
- Ji, D.; Yi, Y.; Kang, G.H.; Choi, Y.H.; Kim, P.; Baek, N.I.; Kim, Y. Identification of an antibacterial compound, benzylideneacetone, from Xenorhabdus nematophila against major plant-pathogenic bacteria. FEMS Microbiol. Lett. 2004, 239, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B. Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 2009, 13, 224–230. [Google Scholar] [CrossRef]
- Shi, Y.M.; Hirschmann, M.; Shi, Y.N.; Ahmed, S.; Abebew, D.; Tobias, N.J.; Grün, P.; Crames, J.J.; Pöschel, L.; Kuttenlochner, W.; et al. Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria. Nat. Chem. 2022, 14, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Boemare, N. Biology, taxonomy and systematics of Photorhabdus and Xenorhabdus. In Entomopathogenic Nematology; CABI Publishing, CAB International: Wallingford, UK, 2002; pp. 35–56. [Google Scholar]
- Hazir, S.; Stackebrant, E.; Lang, E.; Ehlers, R.U.; Keskin, N. Two new subspecies of Photorhabdus luminescens, isolated from Heterorhabditis bacteriophora (Nematoda: Heterorhabditidae): Photorhabdus luminescens subsp. kayaii subsp. Nov. and Photorhabdus luminescens subsp. thraciaensis subsp. nov. Syst. Appl. Microbiol. 2004, 27, 36–42. [Google Scholar] [CrossRef]
- Loulou, A.; Mastore, M.; Caramella, S.; Bhat, A.H.; Brivio, M.F.; Machado, R.A.; Kallel, S. Entomopathogenic potential of bacteria associated with soil-borne nematodes and insect immune responses to their infection. PLoS ONE 2023, 18, e0280675. [Google Scholar] [CrossRef]
- Shapiro-Ilan, D.I.; Hazir, S.; Glaser, I. Advances in Use of Entomopathogenic Nematodes in Integrated Pest Management. In Integrated Management of Insect Pests: Current and Future Developments; Burleigh Dodds Science Publishing: Cambridge, UK, 2020; pp. 91–105. [Google Scholar]
- Abd-Elgawad, M.M.M. Xenorhabdus spp.: An Overview of the Useful Facets of Mutualistic Bacteria of Entomopathogenic Nematodes. Life 2022, 12, 1360. [Google Scholar] [CrossRef] [PubMed]
- Yimthin, T.; Fukruksa, C.; Muangpat, P.; Dumidae, A.; Wattanachaiyingcharoen, W.; Vitta, A.; Thanwisai, A. A study on Xenorhabdus and Photorhabdus isolates from Northeastern Thailand: Identification, antibacterial activity, and association with entomopathogenic nematode hosts. PLoS ONE 2021, 16, e0255943. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, X.; Cheng, Y.; Zhang, X. Manipulation of pH Shift to Enhance the Growth and Antibiotic Activity of Xenorhabdus nematophila. J. Biomed. Biotechnol. 2011, 2011, 672369. [Google Scholar] [CrossRef]
- Lulamba, T.E.; Green, E.; Serepa-Dlamini, M.H. Photorhabdus sp. ETL antimicrobial properties and characterization of its secondary metabolites by gas chromatography–mass spectrometry. Life 2021, 11, 787. [Google Scholar] [CrossRef] [PubMed]
- Heermann, R.; Fuchs, T.M. Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: Uncovering candidate genes involved in insect pathogenicity. BMC Genom. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Webster, J.M.; Chen, G.; Hu, K.; Li, J. Bacterial metabolites. In Entomopathogenic Nematology; CABI Publishing, CAB International: Wallingford, UK, 2002; pp. 99–114. [Google Scholar]
- Sahin, N.; Kula, I.; Erdogan, Y. Investigation of antimicrobial activities of nonanoic acid derivatives. Fresenius Environ. Bull. 2006, 15, 141–143. [Google Scholar]
- Pantel, L.; Florin, T.; Dobosz-Bartoszek, M.; Racine, E.; Sarciaux, M.; Serri, M.; Houard, J.; Campagne, J.M.; de Figueiredo, R.M.; Midrier, C.; et al. Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. Mol. Cell 2018, 70, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Reimer, D.; Luxenburger, E.; Brachmann, A.O.; Bode, H.B. A new type of pyrrolidine biosynthesis is involved in the late steps of xenocoumacin production in Xenorhabdus nematophila. ChemBioChem 2009, 10, 1997–2001. [Google Scholar] [CrossRef] [PubMed]
- Gad, H.A.; Mukhammadiev, E.A.; Zengen, G.; Musayeib, N.M.A.; Hussain, H.; Bin Ware, I.; Ashour, M.L.; Mamadalieva, N.Z. Chemometric Analysis Based on GC-MS Chemical Profiles of Three Stachys Species from Uzbekistan and Their Biological Activity. Plants 2022, 11, 1215. [Google Scholar] [CrossRef] [PubMed]
- Taher, M.A.; Laboni, A.A.; Shompa, S.A.; Rahman, M.M.; Hasan, M.M.; Hasnat, H.; Mala, K.H.A.N. Bioactive compounds extracted from leaves of G. cyanocarpa using various solvents in chromatographic separation showed anti-cancer and anti-microbial potentiality in in silico approach. Chin. J. Anal. Chem. 2023, 51, 100336. [Google Scholar] [CrossRef]
- Baiome, B.A.; Ye, X.; Yuan, Z.; Gaafar, Y.Z.; Melak, S.; Cao, H. Identification of volatile organic compounds produced by Xenorhabdus indica strain AB and investigation of their antifungal activities. Appl. Environ. Microbiol. 2022, 88, e00155-22. [Google Scholar] [CrossRef] [PubMed]
- Karthik, Y.; Ishwara Kalyani, M.; Krishnappa, S.; Devappa, R.; Anjali Goud, C.; Ramakrishna, K.; Wani, M.A.; Alkafafy, M.; Hussen Abduljabbar, M.; Alswat, A.S.; et al. Antiproliferative activity of antimicrobial peptides and bioactive compounds from the mangrove Glutamicibacter mysorens. Front. Microbiol. 2023, 14, 1096826. [Google Scholar] [CrossRef]
- Hawar, S.N.; Taha, Z.K.; Hamied, A.S.; Al-Shmgani, H.S.; Sulaiman, G.M.; Elsilk, S.E. Antifungal Activity of Bioactive Compounds Produced by the Endophytic Fungus Paecilomyces sp. (JN227071.1) against Rhizoctonia solani. Int. J. Biomater. 2023, 2023, 2411555. [Google Scholar] [CrossRef]
- Awori, R.M. Nematophilic bacteria associated with entomopathogenic nematodes and drug development of their biomolecules. Front. Microbiol. 2022, 13, 993688. [Google Scholar] [CrossRef] [PubMed]
- Joyce, S.A.; Lango, L.; Clarke, D.J. The regulation of secondary metabolism and mutualism in the insect pathogenic bacterium Photorhabdus luminescens. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 2011; Volume 76, pp. 1–25. [Google Scholar]
- Breijyeh, Z.; Karaman, R. Design and Synthesis of Novel Antimicrobial Agents. Antibiotics 2023, 12, 628. [Google Scholar] [CrossRef] [PubMed]
- Sanda, N.B.; Hou, Y. The Symbiotic Bacteria—Xenorhabdus nematophila All and Photorhabdus luminescens H06 Strongly Affected the Phenoloxidase Activation of Nipa Palm Hispid, Octodonta nipae (Coleoptera: Chrysomelidae) Larvae. Pathogens 2023, 12, 506. [Google Scholar] [CrossRef] [PubMed]
- Yellamanda, B.; Vijayalakshmi, M.; Kavitha, A.; Reddy, D.K.; Venkateswarlu, Y. Extraction and bioactive profile of the compounds produced by Rhodococcus sp. VLD-10. 3 Biotech 2016, 6, 261. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Yoshiga, T.; Hasegawa, K. Involvement of Vitamin B6 Biosynthesis Pathways in the Insecticidal Activity of Photorhabdus luminescens. Appl. Environ. Microbiol. 2016, 82, 3546–3553. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wareth, M.T.A.; Ali, E.A.M.; El-Shazly, M.A. Biological Activity and GC-MS/MS Analysis of Extracts of Endophytic Fungi Isolated from Eichhornia crassipes (Mart.) Solms. J. Appl. Biotechnol. Rep. 2023, 10, 895–909. [Google Scholar]
- Koilybayeva, M.; Shynykul, Z.; Ustenova, G.; Abzaliyeva, S.; Alimzhanova, M.; Amirkhanova, A.; Turgumbayeva, A.; Mustafina, K.; Yeleken, G.; Raganina, K.; et al. Molecular Characterization of Some Bacillus Species from Vegetables and Evaluation of Their Antimicrobial and Antibiotic Potency. Molecules 2023, 28, 3210. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Gad, M.Z. Omega-9 fatty acids: Potential roles in inflammation and cancer management. J. Genet. Eng. Biotechnol. 2022, 20, 48. [Google Scholar] [CrossRef]
- Sangeetha, B.G.; Jayaprakas, C.A.; Siji, J.V.; Rajitha, M.; Shyni, B.; Mohandas, C. Molecular characterization and amplified ribosomal DNA restriction analysis of entomopathogenic bacteria associated with Rhabditis (Oscheius) spp. 3 Biotech 2016, 6, 32. [Google Scholar] [CrossRef]
- Forst, S.; Nealson, K. Molecular Biology of the Symbiotic-Pathogenic Bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 1996, 60, 21–43. [Google Scholar] [CrossRef]
- Clarke, D.J. Photorhabdus: A model for the analysis of pathogenicity and mutualism. Cell. Microbiol. 2008, 10, 2159–2167. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshini, S.; Gopinath, V.; Meera Priyadharsshini, N.; Mubarak Ali, D.; Velusamy, P. Synthesis of anisotropic silver nanoparticles using novel strain, Bacillus flexus and its biomedical application. Colloids Surf. B Biointerfaces 2013, 102, 232–237. [Google Scholar] [CrossRef] [PubMed]
- San-Blas, E.; Cubillán, N.; Guerra, M.; Portillo, E.; Esteves, I. Characterization of Xenorhabdus and Photorhabdus bacteria by Fourier transform mid-infrared spectroscopy with attenuated total reflection (FT-IR/ATR). Spectrochim. Acta A Mol. Biomol. Spectrosc. 2012, 93, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, K.; Gowri, S.; Arumugam, A. Phytosynthesis of silver nanoparticles using Pterocarpus santalinus leaf extract and their antibacterial properties. J. Nanostruct. Chem. 2013, 3, 68. [Google Scholar] [CrossRef]
- Cimen, H.; Touray, M.; Gulsen, S.H.; Erincik, O.; Wenski, S.L.; Bode, H.B.; Shapiro-Ilan, D.; Hazir, S. Antifungal activity of different Xenorhabdus and Photorhabdus species against various fungal phytopathogens and identification of the antifungal compounds from X. szentirmaii. Appl. Microbiol. Biotechnol. 2021, 105, 5517–5528. [Google Scholar] [CrossRef] [PubMed]
- Paul, V.J.; Frautschy, S.; Fenical, W.; Nealson, K.H. Antibiotic in microbial ecology, isolation and structure assignment of several new antibacterial compounds from the insect-symbiotic bacteria Xenorhabdus spp. J. Chem. Ecol. 1981, 7, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, J.; Malan, A.P.; Dicks, L.M.T. Bacteria of the genus Xenorhabdus, a novel source of bioactive compounds. Front. Microbiol. 2018, 9, 3177. [Google Scholar] [CrossRef] [PubMed]
- Elbrense, H.; Elmasry, A.M.A.; Seleiman, M.F.; Al-Harbi, M.S.; Abd El-Raheem, A.M. Can Symbiotic Bacteria (Xenorhabdus and Photorhabdus) Be More Efficient than Their Entomopathogenic Nematodes against Pieris rapae and Pentodon algerinus Larvae? Biology 2021, 10, 999. [Google Scholar] [CrossRef]
- Brachmann, A.O.; Bode, H.B. Identification and bioanalysis of natural products from insect symbionts and pathogens. Adv. Biochem. Eng. Biotechnol. 2013, 135, 123–155. [Google Scholar]
- Parihar, R.D.; Dhiman, U.; Bhushan, A.; Gupta, P.K.; Gupta, P. Heterorhabditis and Photorhabdus symbiosis: A natural mine of bioactive compounds. Front. Microbiol. 2022, 13, 790339. [Google Scholar] [CrossRef]
- Gulsen, S.H.; Tileklioglu, E.; Bode, E.; Cimen, H.; Ertabaklar, H.; Ulug, D.; Ertug, S.; Wenski, S.L.; Touray, M.; Hazir, C.; et al. Antiprotozoal activity of different Xenorhabdus and Photorhabdus bacterial secondary metabolites and identification of bioactive compounds using the easyPACId approach. Sci. Rep. 2022, 12, 10779. [Google Scholar] [CrossRef] [PubMed]
- Mollah, M.M.I.; Kim, Y. Virulent secondary metabolites of entomopathogenic bacteria genera, Xenorhabdus and Photorhabdus, inhibit phospholipase A2 to suppress host insect immunity. BMC Microbiol. 2020, 20, 359. [Google Scholar] [CrossRef] [PubMed]
- Isaacson, P.J.; Webster, J.M. Antimicrobial activity of Xenorhabdus sp. RIO (Enterobacteriaceae), symbiont of the entomopathogenic nematode, Steinernema riobrave (Rhabditida: Steinernematidae). J. Invertebr. Pathol. 2002, 79, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Li, J.X.; Chen, G.H.; Webster, J.M.; Czyceskwa, E. Antimicrobial metabolites from a bacterial symbiont. J. Nat. Prod. 1995, 58, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Thaler, J.O.; Baghdiguian, S.; Boemare, N.E. Purification and characterization of nematophilicin, phage tail like bacteriocin, from the lysogenic strain FI of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 1995, 61, 2049–2052. [Google Scholar] [CrossRef] [PubMed]
- Muangpat, P.; Yooyangket, T.; Fukruksa, C.; Suwannaroj, M.; Yimthin, T.; Sitthisak, S.; Chantratita, N.; Vitta, A.; Tobias, N.J.; Bode, H.B.; et al. Screening of the antimicrobial activity against drug resistant bacteria of Photorhabdus and Xenorhabdus associated with entomopathogenic nematodes from Mae Wong National Park, Thailand. Front. Microbiol. 2017, 8, 1142. [Google Scholar] [CrossRef] [PubMed]
- León-Buitimea, A.; Garza-Cárdenas, C.R.; Garza-Cervantes, J.A.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. The demand for new antibiotics: Antimicrobial peptides, nanoparticles, and combinatorial therapies as future strategies in antibacterial agent design. Front. Microbiol. 2020, 11, 551136. [Google Scholar] [CrossRef] [PubMed]
- Le, C.F.; Fang, C.M.; Sekaran, S.D. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 2017, 61, e02340-16. [Google Scholar] [CrossRef] [PubMed]
- Eshboev, F.; Mamadalieva, N.; Nazarov, P.A.; Hussain, H.; Katanaev, V.; Egamberdieva, D.; Azimova, S. Antimicrobial action mechanisms of natural compounds isolated from endophytic microorganisms. Antibiotics 2024, 13, 271. [Google Scholar] [CrossRef]
- Hu, K.J.; Li, J.X.; Li, B.; Webster, J.M.; Chen, G. A novel antimicrobial epoxide isolated from larval Galleria mellonella infected by the nematode symbiont, Photorhabdus luminescens (Enterobacteriaceae). Bioorg. Med. Chem. 2006, 14, 4677–4681. [Google Scholar] [CrossRef]
- Booysen, E.; Dicks, L.M.T. Does the future of antibiotics lie in secondary metabolites produced by Xenorhabdus spp.? A Review. Probiotics Antimicrob. Proteins 2020, 12, 1310–1320. [Google Scholar] [CrossRef] [PubMed]
- Böszörményi, E.; Érsek, T.; Fodor, A.; Fodor, A.M.; Földes, L.S.; Hevesi, M.; Hogan, J.S.; Katona, Z.; Klein, M.G.; Kormány, A.; et al. Isolation and activity of Xenorhabdus antimicrobial compounds against the plant pathogens Erwinia amylovora and Phytophthora nicotianae. J. Appl. Microbiol. 2009, 107, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Furgani, G.; Böszörményi, E.; Fodor, A.; Máthé-Fodor, A.; Forst, S.; Hogan, J.S.; Katona, Z.; Klein, M.G.; Stackebrandt, E.; Szentirmai, A.; et al. Xenorhabdus antibiotics: A comparative analysis and potential utility for controlling mastitis caused by bacteria. J. Appl. Microbiol. 2008, 104, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Kim, Y. Development of “Bt-Plus” biopesticide using entomopathogenic bacteria (Xenorhabdus nematophila, Photorhabdus temperata ssp. temperata) metabolites. Korean J. Appl. Entomol. 2011, 50, 171–178. [Google Scholar] [CrossRef]
- Gobinath, C.; Seetharaman, P.; Gnanasekar, S.; Kadarkarai, M.; Sivaperumal, S. Xenorhabdus stockiae KT835471-mediated feasible biosynthesis of metal nanoparticles for their antibacterial and cytotoxic activities. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1675–1684. [Google Scholar]
- Muangpat, P.; Suwannaroj, M.; Yimthin, T.; Fukruksa, C.; Sitthisak, S.; Chantratita, N.; Vitta, A.; Thanwisai, A. Antibacterial activity of Xenorhabdus and Photorhabdus isolated from entomopathogenic nematodes against antibiotic-resistant bacteria. PLoS ONE 2020, 15, e0234129. [Google Scholar] [CrossRef] [PubMed]
- Orozco, R.A.; Molnár, I.; Bode, H.; Stock, S.P. Bioprospecting for secondary metabolites in the entomopathogenic bacterium Photorhabdus luminescens subsp. sonorensis. J. Invertebr. Pathol. 2016, 141, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Watson, R.J.; Millichap, P.; Joyce, S.; Reynolds, S.; Clarke, D. The role of iron uptake in pathogenicity and symbiosis in Photorhabdus luminescens TT01. BMC Microbiol. 2010, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Yüksel, E.; Yıldırım, A.; İmren, M.; Canhilal, R.; Dababat, A.A. Xenorhabdus and Photorhabdus bacteria as potential candidates for the control of Culex pipiens L. (Diptera: Culicidae), the principal vector of west Nile virus and Lymphatic Filariasis. Pathogens 2023, 12, 1095. [Google Scholar] [CrossRef]
- Akhurst, R.J. Morphological and functional dimorphism in Xenorhabditis spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. J. Gen. Microbiol. 1980, 121, 303–309. [Google Scholar]
- Tailliez, P.; Pages, S.; Ginibre, N.; Boemare, N.E. New insight into diversity in the genus Xenorhabdus, including the description of ten novel species. Int. J. Syst. Evol. Microbiol. 2006, 56, 2805–2818. [Google Scholar] [CrossRef] [PubMed]
- Chandrakasan, G.; Ayala, M.T.; Trejo, J.F.; Marcus, G.; Maruthupandy, M.; Kanisha, C.C.; Murugan, M.; Al-Mekhlafi, F.A.; Wadaan, M.A. Bio controlled efficacy of Bacillus thuringiensis cry protein protection against tomato fruit borer Helicoverpa armigera in a laboratory environment. Physiol. Mol. Plant Pathol. 2022, 119, 101827. [Google Scholar] [CrossRef]
- Forst, S.; Clarke, D. Bacteria-nematode symbiosis. In Entomopathogenic Nematology; CABI Publishing, CAB International: Wallingford, UK, 2002; pp. 57–77. [Google Scholar]
- Shapiro-Ilan, D.I.; Cottrell, T.E.; Gardner, W.A.; Behle, R.W.; Ree, B.; Harris, M.K. Efficacy of entomopathogenic fungi in suppressing pecan weevil, Curculio caryae (Coleoptera: Curculionidae), in commercial pecan orchards. Southwest. Entomol. 2009, 34, 111–120. [Google Scholar] [CrossRef]
- Cappuccino, J.G.; Sherman, N. Microbiology: A Laboratory Manual; Benjamin: Harlow, UK, 2002; pp. 263–264. [Google Scholar]
- Olano, C.; Wilkinson, B.; Sánchez, C.; Moss, S.J.; Sheridan, R.; Math, V.; Weston, A.J.; Braña, A.F.; Martin, C.J.; Oliynyk, M.; et al. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tü4055: Cluster analysis and assignment of functions. Chem. Biol. 2004, 11, 87–97. [Google Scholar]
- Bonev, B.; Hooper, J. Principles of assessing bacterial susceptibility to antibiotics using the agar diffusion method. J. Antimicrob. Chemother. 2008, 61, 1295–1301. [Google Scholar] [CrossRef]









| Sampling Site | Agriculture Crops/ Land | Recovered | Soil Type | Soil Temperature (°C) | Organic Content (%) | pH | Electrical Conductivity (mS/cm) | Symbiotic Bacteria Characteristics | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Steinernema spp. | Heterorhabditis spp. | Xenorhabdus spp. (Amz 05) | Photorhabdus spp. (Amz 10) | |||||||
| Zone I (agricultural land) | Corn | ++ | - | sandy | 29 | 4.9 | 7.25 | 0.93 | Blue-color colony | Greenish yellow-color colony |
| Rice | - | ++ | Loam clay | 31 | 4.8 | 7.13 | 1.29 | Bioluminescence (-) tive | Bioluminescence (+) tive | |
| wheat | ++ | - | loam | 30 | 3.9 | 7.40 | 1.32 | Catalase (-) tive | Catalase (+) tive | |
| Zone II (uncultured land) | Forest | + | - | Silt | 28 | 4.9 | 7.70 | 1.74 | Absorption of bromothymol blue | Absorption of bromothymol blue |
| Grasses | + | - | Silt | 29 | 3.8 | 7.41 | 1.59 | Growth on 28 °C | Growth on 28 °C | |
| Wasteland | ++ | Silt | 30 | 4.1 | 7.23 | 1.03 | Insect pathogenicity | Insect pathogenicity | ||
| S. No. | FTIR Peaks (cm−1) | Possible Assigned Functional Groups |
|---|---|---|
| 1 | 3443 | Amine, N-H Stretch |
| 2 | 1637 | NH amide bend C-Br Bend |
| 3 | 1100 | ANAH and carbonyl (CAOA) |
| 4 | 621 | Carbonyl stretching (OC) |
| S. No. | FTIR Peaks (cm−1) | Possible Assigned Functional Groups |
|---|---|---|
| 1 | 3436 | Amine, N-H Stretch |
| 2 | 2078 | C≡N, Nitriles |
| 3 | 1078 | Carbonyl stretch |
| 4 | 692 | Alkyl Halides |
| Fraction | Ret. Time (min) | Width (min) | Area (mAU*s) | Height (mAU) | Area% |
|---|---|---|---|---|---|
| 1 | 9.393 | 0.3524 | 70.87936 | 3.20061 | 0.4908 |
| 2 | 10.122 | 0.3722 | 49.71435 | 1.83400 | 0.3443 |
| 3 | 14.197 | 0.0953 | 135.46420 | 19.44917 | 0.9380 |
| 4 | 17.607 | 0.1898 | 389.49295 | 26.14004 | 2.6971 |
| 5 | 18.755 | 1.2428 | 1.02867 | 114.86192 | 71.2308 |
| 6 | 23.703 | 0.4298 | 144.59134 | 4.28346 | 1.0012 |
| 7 | 28.894 | 0.5881 | 1209.27771 | 25.73598 | 8.3737 |
| 8 | 31.89 | 1.6023 | 2149.97168 | 15.93304 | 14.8876 |
| Fraction | Ret. Time (min) | Width (min) | Area (mAU*s) | Height (mAU) | Area% |
|---|---|---|---|---|---|
| 1 | 10.637 | 0.2511 | 312.39853 | 16.05020 | 1.2641 |
| 2 | 11.795 | 0.5186 | 177.14165 | 4.21222 | 0.7168 |
| 3 | 17.842 | 0.0837 | 719.59143 | 120.40215 | 2.9119 |
| 4 | 18.842 | 1.2975 | 1.67736 | 182.29234 | 67.8757 |
| 5 | 23.503 | 0.2292 | 75.46230 | 4.55771 | 0.3054 |
| 6 | 23.715 | 0.2964 | 139.74609 | 6.30963 | 0.5655 |
| 7 | 28.887 | 0.8533 | 1907.49695 | 27.23028 | 7.7188 |
| 8 | 31.938 | 1.7550 | 4606.81006 | 31.15800 | 18.6418 |
| S. No. | R. Time | % of Area | Compound Name | Biological Activities | Reference |
|---|---|---|---|---|---|
| 1 | 5.631 | 3.89 | Nonanoic acid derivatives | Antimicrobial properties | [16] |
| 2 | 5.823 | 4.27 | Paromycin | Antibacterial agents | [17] |
| 3 | 7.428 | 42.01 | Pyrrolidinone | Bioactive compounds | [18] |
| 4 | 8.848 | 3.98 | Octodecanal derivatives | Bioactive compounds | [19] |
| 5 | 13.198 | 4.53 | Trioxa-5-aza-1-silabicyclo | Antimicrobial compounds | [20] |
| 6 | 15.671 | 26.78 | 4-Octadecenal | Antifungal activity | [21] |
| 7 | 22.981 | 2.84 | Cyclopentanetridecanoic acid, Methyl ester | Antimicrobial peptide | [22] |
| 8 | 23.978 | 4.629 | Oleic Acid | Antifungal compound | [23] |
| 9 | 31.98 | 1.876 | 1,2-benzenedicarboxylicacid | Bioactive molecules | [24] |
| S. No | R. Time | % of Area | Compound Name | Biological Activities | Reference |
|---|---|---|---|---|---|
| 1 | 6.328 | 2.95 | Indole-3-acetic acid, methyl ester | Bioactive compound | [25] |
| 2 | 7.810 | 59.43 | Piperidinol derivatives | Antimicrobial agents | [26] |
| 3 | 8.302 | 5.78 | Phthalic acid | Bioactive compound | [27] |
| 4 | 15.114 | 49.43 | Nemorosonol | Bioactive metabolites | [28] |
| 5 | 16.831 | 5.187 | Octahydro-7-methyl-3-methylene | Insecticidal activity | [29] |
| 6 | 20.302 | 3.651 | 1-eicosanol | Bioactive compound | [30] |
| 7 | 25.220 | 2.284 | Octadecanoic acid, Methyl ester | Antimicrobial activity | [31] |
| 8 | 26.897 | 1.211 | unsaturated fatty acids | Anti-inflammation | [32] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chandrakasan, G.; García-Trejo, J.F.; Feregrino-Pérez, A.A.; Aguirre-Becerra, H.; García, E.R.; Nieto-Ramírez, M.I. Preliminary Screening on Antibacterial Crude Secondary Metabolites Extracted from Bacterial Symbionts and Identification of Functional Bioactive Compounds by FTIR, HPLC and Gas Chromatography–Mass Spectrometry. Molecules 2024, 29, 2914. https://doi.org/10.3390/molecules29122914
Chandrakasan G, García-Trejo JF, Feregrino-Pérez AA, Aguirre-Becerra H, García ER, Nieto-Ramírez MI. Preliminary Screening on Antibacterial Crude Secondary Metabolites Extracted from Bacterial Symbionts and Identification of Functional Bioactive Compounds by FTIR, HPLC and Gas Chromatography–Mass Spectrometry. Molecules. 2024; 29(12):2914. https://doi.org/10.3390/molecules29122914
Chicago/Turabian StyleChandrakasan, Gobinath, Juan Fernando García-Trejo, Ana Angelica Feregrino-Pérez, Humberto Aguirre-Becerra, Enrique Rico García, and María Isabel Nieto-Ramírez. 2024. "Preliminary Screening on Antibacterial Crude Secondary Metabolites Extracted from Bacterial Symbionts and Identification of Functional Bioactive Compounds by FTIR, HPLC and Gas Chromatography–Mass Spectrometry" Molecules 29, no. 12: 2914. https://doi.org/10.3390/molecules29122914





