Exploring the Potential of Oleanolic Acid Dimers–Cytostatic and Antioxidant Activities, Molecular Docking, and ADMETox Profile
Abstract
1. Introduction
2. Results
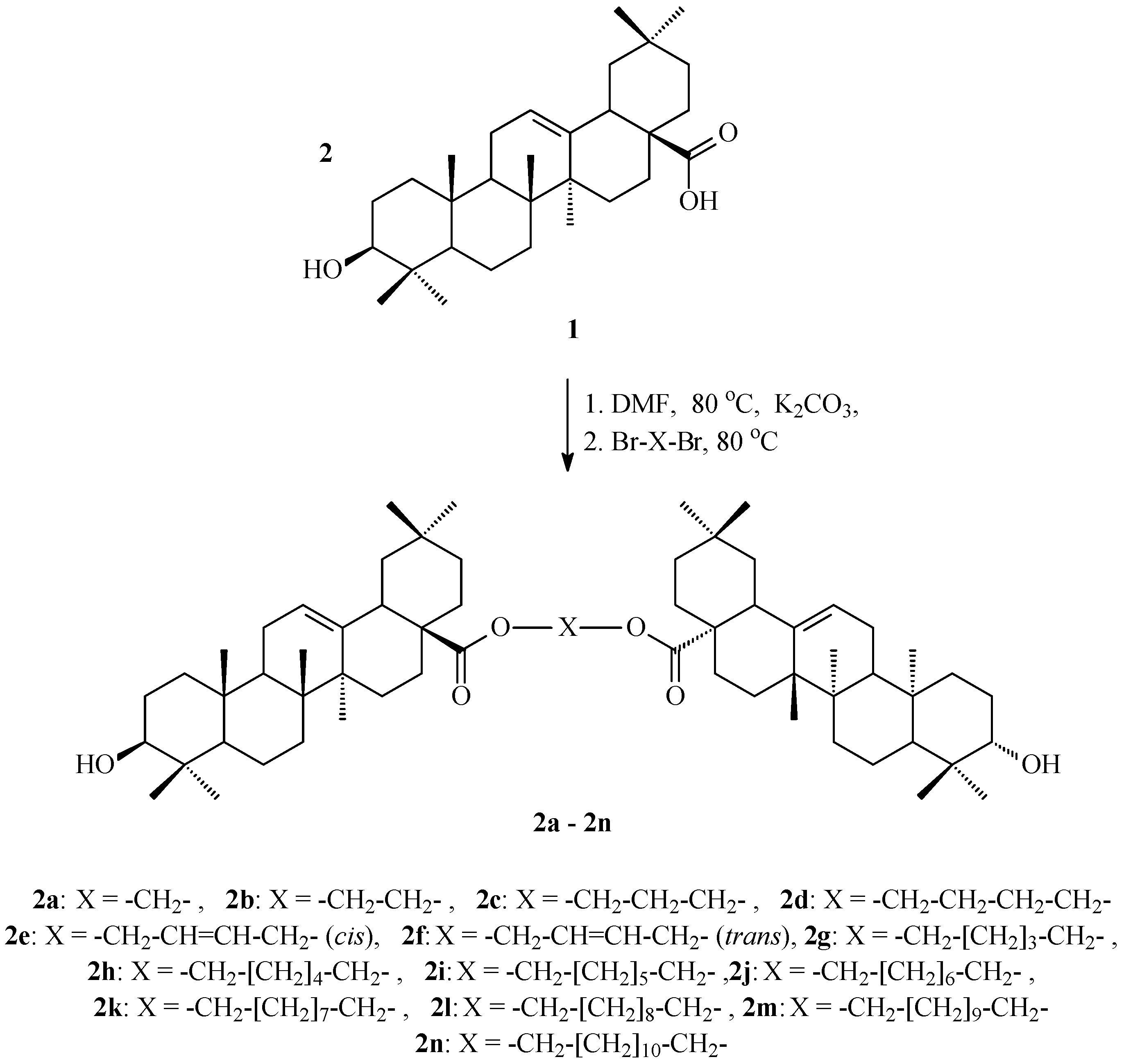
2.1. Synthesis of OADs
2.2. Potential Cytostatic Properties of OADs
2.3. Molecular Docking
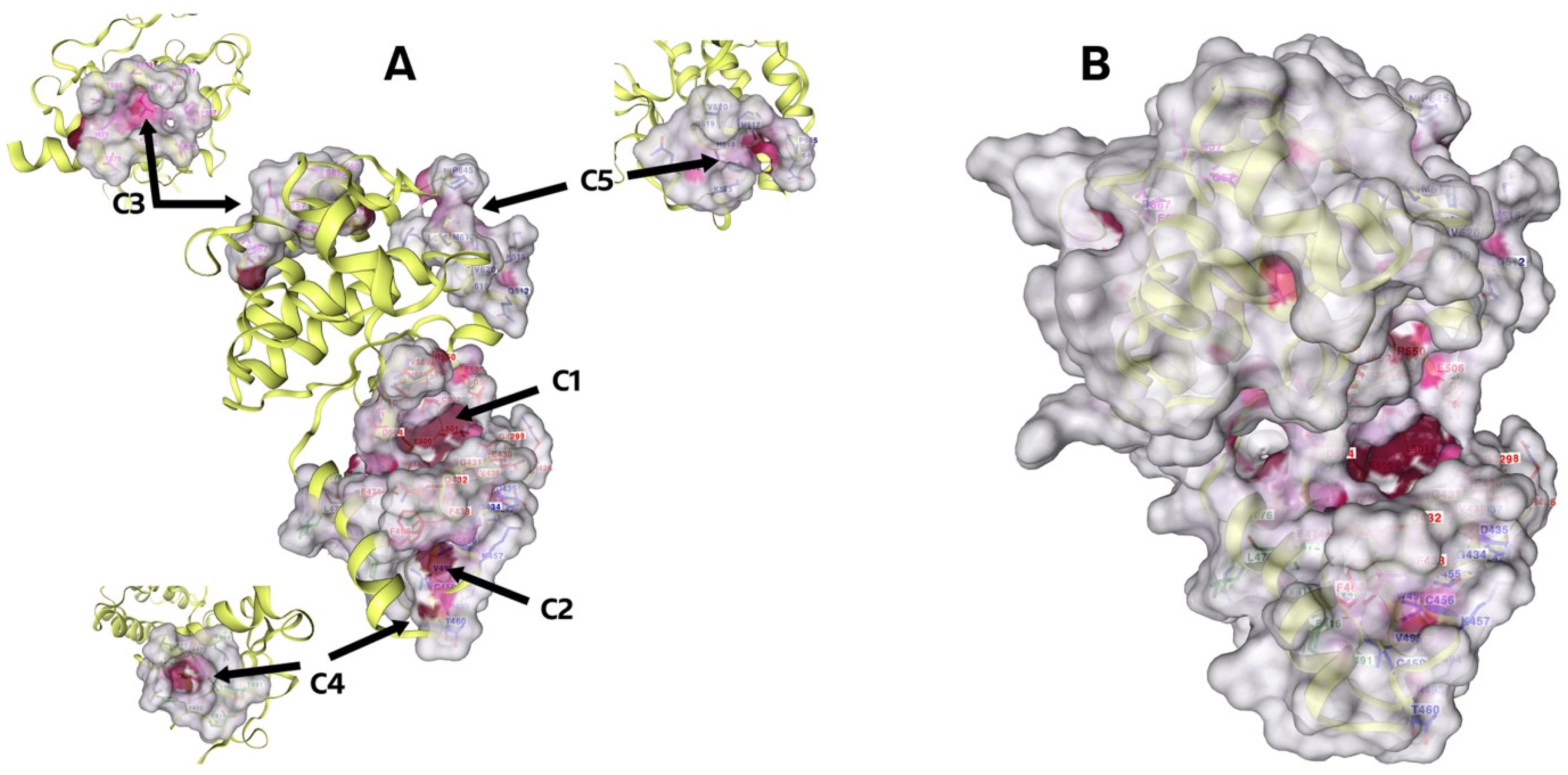
2.3.1. Detecting Cavities
2.3.2. Molecular Docking
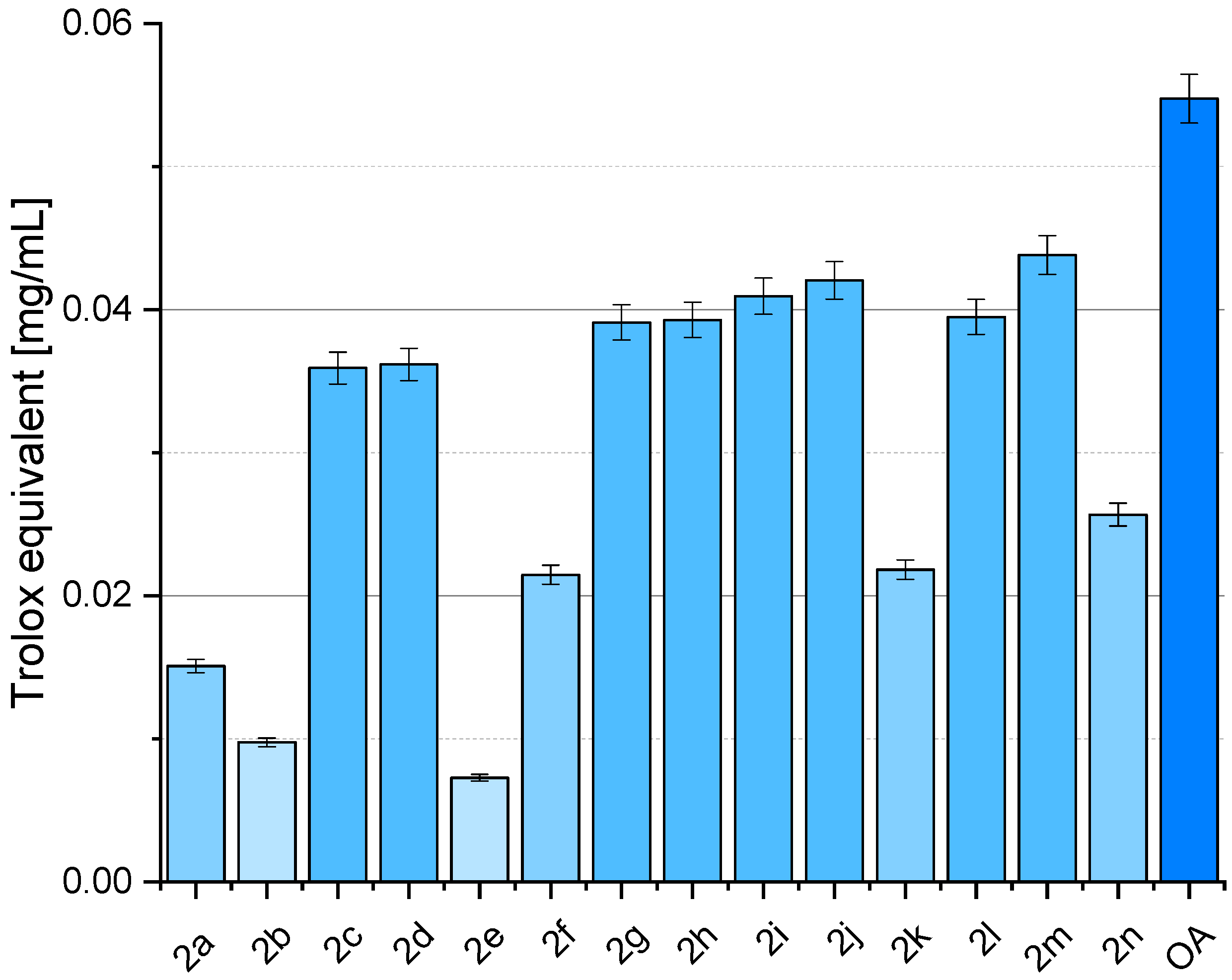
2.4. Antioxidant Activity of OADs
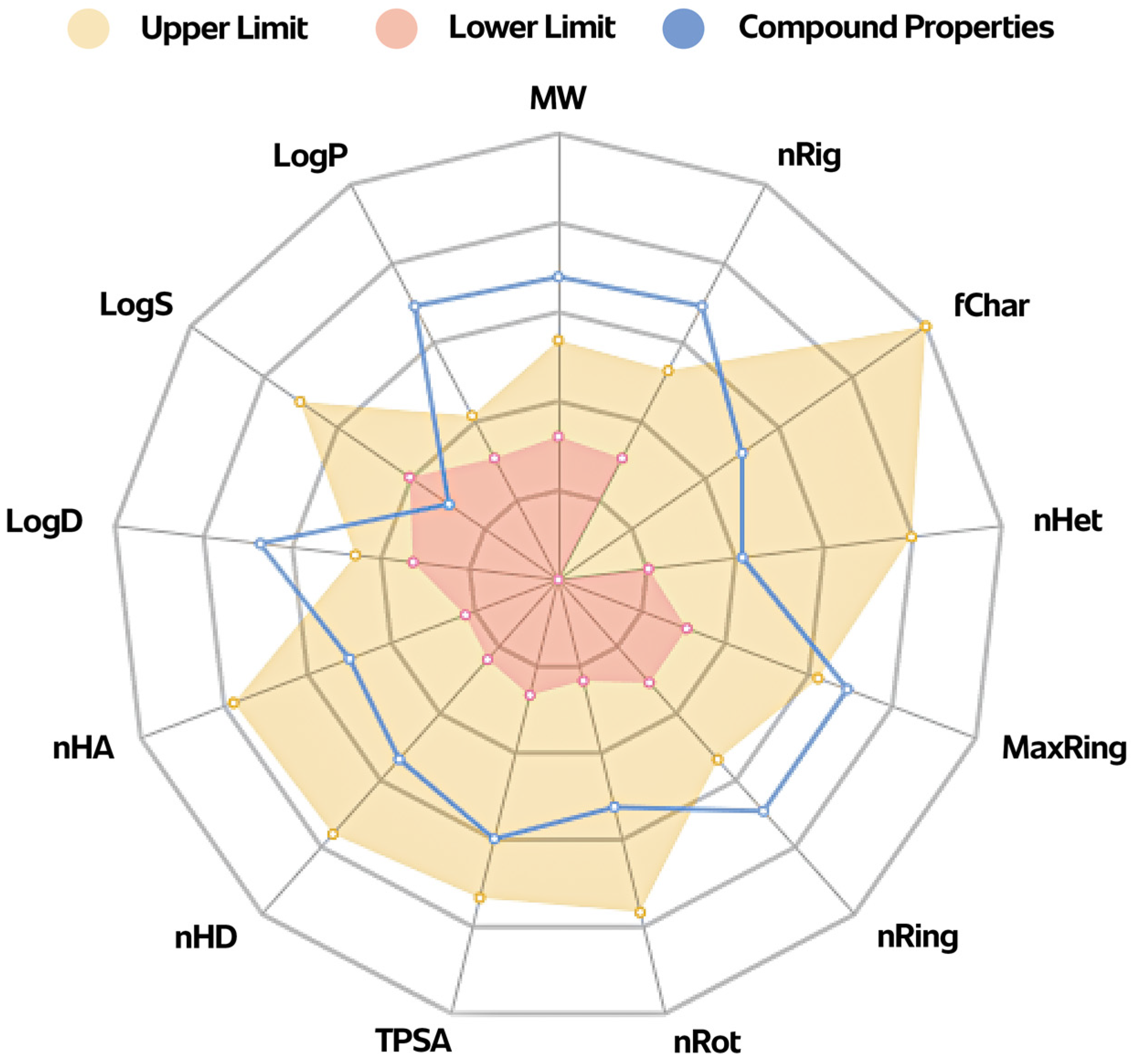
2.5. ADMETox Analysis
3. Discussion
3.1. Synthesis of OADs
3.2. Potential Cytostatic Properties of OADs
- All OADs 2a–2n will probably show a much higher level of cytostatic activity in in vivo tests than the parent oleanolic acid (1), for which only a few lines had an IC50 value ≤ 10 µM (lines: MDA-MB-468, XF-498, K-562, P388-ADR, P388, HOP-92, RXF-393, and SN12K1);
- Only in the case of four cancer cell lines did the combination of two oleanolic acid residues into a dimer derivative cause a decrease in the IC50 value, i.e., a decrease in cytotoxic activity: SNB-78, M14, M19-MEL, MDA-N, SKA-MEL-5, A-549-ATCC, and 786-0; for all remaining 69 tumour cell lines, the pdCSM-cancer program predicted the increase in cytostatic activity level;
- In the case of more than ten cancer cell lines (e.g., T-47D, SF-295, COLO-205, HT29, RPMI-8226, MALME-3M, NCI-ADR-RES, OVCAR-3, and others), it is beneficial to combine two oleanolic acid residues with a linker longer than the one-carbon one;
- It is not possible to clearly state what effect the presence of a double bond, cis or trans, in the four-carbon bridge has on the IC50 value—the number of cancer cell lines for which the IC50 value significantly increases or decreases is similar;
- The tested dimer derivatives of oleanolic acid (OADs 2a–2n) may probably be highly effective cytostatic agents against BT-549, HS-578T, MCF-7, HCC-2998, SW-620, K-562, RMPI-8226, SK-MEL28, HOP-18, HOP-92, NCI-ADR-RES, DU-145, RXF-393, RXF-631, DMS-114 (IC50 in the range of 1.01–5.00 µM), MDA-MB -468 (IC50 in the range of 0.10–0.99 µM), and P388-ADR, P388, SN12K1, and SN12K1 (IC50 ≤ 0.09 µM);
- It is hard to find a clear relationship between the structure of OADs and the level of their cytostatic activity. Probably the reason for the lack of this dependence may be the geometry of the OAD molecule—the length of the linker and the presence (or lack) of a double bond, and, even more importantly, the type of this unsaturated bond (cis/trans) may cause a given dimer to better or worse adapt to the enzymes of cancerous cells. The structure of the linker most likely involves a specific mutual arrangement of two triterpene residues, and these arrangements may be different—e.g., two triterpene residues may be arranged in a straight line, or one below the other, or in still other ways. Therefore, some dimers fit better into the enzyme pocket, while others fit worse.
3.3. Molecular Docking
3.4. Antioxidant Activity of OADs
3.5. ADMETox Analysis
4. Materials and Methods
4.1. OADs Preparation
4.2. Potential Cytostatic Properties of OADs
4.3. Molecular Docking
4.3.1. Ligands Preparation
4.3.2. Protein Preparation
4.3.3. Detecting Cavities and Uploading Ligands
4.4. Antioxidant Activity of OADs
4.5. ADMETox
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nirmala, M.J.; Samundeeswari, A.; Sankar, P.D. Natural plant resources in anticancer therapy—A review. Res. Plant Biol. 2011, 1, 1–14. [Google Scholar]
- Mali, S.B. Cancer treatment: Role of natural products. Time to have a serious rethink. Oral Oncol. Rep. 2023, 6, 100040–100043. [Google Scholar] [CrossRef]
- Dutta, S.; Mahalanobish, S.; Saha, S.; Ghosh, S.; Sil, P.C. Natural products: An upcoming therapeutic approach to cancer. Food Chem. Toxicol. 2019, 128, 240–255. [Google Scholar] [CrossRef]
- Prakash, O.; Kumar, A.; Kumar, P. Anticancer potential of plants and natural products: A review. Am. J. Pharm. Sci. 2013, 1, 104–115. [Google Scholar] [CrossRef]
- Mann, J. Natural products in cancer chemotherapy: Past, present and future. Nat. Rev. Cancer 2002, 2, 143–148. [Google Scholar] [CrossRef]
- Roaa, M.H.; Shoker, A. The Importance of the Major groups of Plants Secondary Metabolism Phenols, Alkaloids, and Terpenes. Int. J. Res. Appl. Sci. Biotechnol. 2020, 7, 354–358. [Google Scholar] [CrossRef]
- Withers, S.T.; Keasling, J.D. Biosynthesis and engineering of isoprenoid small molecules. Appl. Microbiol. Biotechnol. 2007, 73, 980–990. [Google Scholar] [CrossRef]
- Yeung, M.F. A review on the presence of oleanolic acid in natural products. Nat. Proda Medica 2009, 2, 77–290. [Google Scholar]
- Wang, X.; Ye, X.; Liu, R.; Chen, H.; Bai, H.; Liang, X.; Zhang, X.; Wang, Z.; Li, W.; Hai, C. Antioxidant activities of oleanolic acid in vitro: Possible role of Nrf2 and MAP kinases. Chem. Biol. Interact. 2010, 184, 328–337. [Google Scholar] [CrossRef]
- Mahmoud, A.H.; Mahmoud, B.N.; Samy, M.N.; Fouad, M.A.; Kamel, M.S.; Matsunami, K. Cytotoxic and antileishmanial triterpenes of Tabebuia aurea (Silva Manso) leaves. Nat. Prod. Res. 2022, 36, 6181–6185. [Google Scholar] [CrossRef]
- Szakiel, A.; Ruszkowski, D.; Grudniak, A.; Kurek, A.; Wolska, K.I.; Doligalska, M.; Janiszowska, W. Antibacterial and Antiparasitic Activity of Oleanolic Acid and its Glycosides isolated from Marigold (Calendula officinalis). Planta Med. 2008, 74, 1709–1715. [Google Scholar] [CrossRef] [PubMed]
- Oboh, M.; Govender, L.; Siwela, M.; Mkhwanazi, B.N. Antidiabetic Potential of Plant-Based Pentacyclic Triterpene Derivatives: Progress Made to Improve Efficacy and Bioavailability. Molecules 2021, 26, 7243. [Google Scholar] [CrossRef]
- Shan, T.; Ye, J.; Jia, J.; Wang, Z.; Jiang, Y.; Wang, Y.; Wang, Y.; Zheng, K.; Ren, Z. Viral UL8 Is Involved in the Antiviral Activity of Oleanolic Acid Against HSV-1 Infection. Front. Microbiol. 2021, 12, 689607–689618. [Google Scholar] [CrossRef]
- Somova, L.; Shode, F.; Ramnanan, P.; Nadar, A. Antihypertensive, antiatherosclerotic and antioxidant activity of triterpenoids isolated from Olea europaea, subspecies africana leaves. J. Ethnopharm. 2003, 84, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Gudoityte, E.; Arandarcikaite, O.; Mazeikiene, I.; Bendokas, V.; Liobikas, J. Ursolic and Oleanolic Acids: Plant Metabolites with Neuroprotective Potential. Int. J. Mol. Sci. 2021, 22, 4599. [Google Scholar] [CrossRef] [PubMed]
- Xin, C.; Liu, S.; Qu, H.; Wang, Z. The novel nanocomplexes containing deoxycholic acid-grafted chitosan and oleanolic acid displays the hepatoprotective effect against CCl4-induced liver injury in vivo. Int. J. Biol. Macromol. 2021, 185, 338–349. [Google Scholar] [CrossRef]
- Petronelli, A.; Pannitteri, G.; Testa, U. Triterpenoids as new promising anticancer drugs. Anticancer Drugs 2009, 20, 880–892. [Google Scholar] [CrossRef]
- Picheswararao, P.; Udupa, N.; Saleemulla, K. Herbal medicinal plants as an anticancer agents. Ann. Phytomed. 2015, 4, 37–45. [Google Scholar]
- Rasheed, H.M.; Farooq, U.; Bashir, K.; Wahid, F.; Khan, T.; Khusro, A.; Gajdács, M.; Alghamdi, S.; Alsaiari, A.A.; Almehmadi, M.; et al. Isolation of oleanolic acid from Lavandula stoechas and its potent anticancer properties against MCF-7 cancer cells via induced apoptosis. J. King Saud Univ. Sci. 2023, 35, 102454–102464. [Google Scholar] [CrossRef]
- Woo, J.S.; Yoo, E.S.; Kim, S.H.; Lee, J.H.; Han, S.H.; Jung, S.H.; Jung, G.H.; Jung, J.Y. Anticancer effects of oleanolic acid on human melanoma cells. Chem. Biol. Interact. 2021, 347, 109619–109627. [Google Scholar] [CrossRef]
- Zeng, Z.; Yu, J.; Jiang, Z.; Zhao, N. Oleanolic Acid (OA) Targeting UNC5B Inhibits Proliferation and EMT of Ovarian Cancer Cell and Increases Chemotherapy Sensitivity of Niraparib. Hindawi J. Oncol. 2022, 2022, 5887671. [Google Scholar] [CrossRef]
- Lucio, K.A.; da Graça Rocha, G.; Monção-Ribeiro, L.C.; Fernandes, J.; Takiya, C.M.; Gattas, C.R. Oleanolic acid initiates apoptosis in non-small cell lung cancer cell lines and reduces metastasis of a B16F10 melanoma model in vivo. PLoS ONE 2011, 6, e28596. [Google Scholar] [CrossRef] [PubMed]
- Shyu, M.H.; Kao, T.C.; Yen, G.C. Oleanolic acid and ursolic acid induce apoptosis in HuH7 human hepatocellular carcinoma cells through a mitochondrial-dependent pathway and downregulation of XIAP. J. Agric. Food Chem. 2010, 58, 6110–6118. [Google Scholar] [CrossRef] [PubMed]
- Bednarczyk-Cwynar, B.; Zaprutko, L.; Ruszkowski, P.; Hładoń, B. Anticancer effect of A-ring or/and C-ring modified oleanolic acid derivatives on KB, MCF-7 and HeLa cell lines. Org. Biomol. Chem. 2012, 10, 2201–2205. [Google Scholar] [CrossRef]
- Bednarczyk-Cwynar, B.; Ruszkowski, P.; Bobkiewicz-Hładoń, T.; Zaprutko, L. Oleanolic acid A-lactams inhibit the growth of HeLa, KB, MCF-7 and Hep-G2 cancer cell lines at micromolar concentrations. Anticancer Agents Med. Chem. 2016, 16, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Bednarczyk-Cwynar, B.; Ruszkowski, P.; Atamanyuk, D.; Lesyk, R.; Zaprutko, Z. Hybrids of Oleanolic Acid with Norbornene-2,3-dicarboximide-N-carboxylic Acids as Potential Anticancer Agents. Acta Pol. Pharm. Drug Res. 2017, 74, 827–835. [Google Scholar]
- Bednarczyk-Cwynar, B.; Ruszkowski, P.; Jarosz, T.; Krukiewicz, K. Enhancing anticancer activity through the combination of bioreducing agents and triterpenes. Future Med. Chem. 2018, 10, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Bednarczyk-Cwynar, B.; Ruszkowski, P. Acylation of Oleanolic Acid Oximes Effectively Improves Cytotoxic Activity in In Vitro Studies. Pharmaceutics 2024, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Günther, A.; Zalewski, P.; Sip, S.; Ruszkowski, P.; Bednarczyk-Cwynar, B. Oleanolic Acid Dimers with Potential Application in Medicine–Design, Synthesis, Physico-Chemical Characteristics, Cytotoxic and Antioxidant Activity. Int. J. Mol. Sci. 2024, 25, 6989. [Google Scholar] [CrossRef]
- Gülçin, I.; Elias, R.; Gepdiremen, A.; Boyer, L. Antioxidant activity of lignans from fringe 579 tree (Chionanthus virginicus L.). Eur. Food Res. Technol. 2006, 223, 759–767. [Google Scholar] [CrossRef]
- Marnett, L.J. Oxyradicals and DNA damage. Carcinogenesis 2000, 21, 361–370. [Google Scholar] [CrossRef]
- Ylä-Herttuala, S. Oxidized LDL and atherogenesis. Ann. N. Y. Acad. Sci. 1999, 874, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R.; Levine, R.L. Protein oxidation. Ann. N. Y. Acad. Sci. 2000, 899, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Günther, A.; Makuch, E.; Nowak, A.; Duchnik, W.; Kucharski, Ł.; Pełech, R.; Klimowicz, A. Enhancement of the Antioxidant and Skin Permeation Properties of Betulin and Its Derivatives. Molecules 2021, 26, 3435. [Google Scholar] [CrossRef] [PubMed]
- Oloyede, H.O.B.; Ajiboye, H.O.; Salawu, M.O.; Ajiboye, T.O. Influence of oxidative stress on the antibacterial activity of betulin, betulinic acid and ursolic acid. Microb. Pathogen. 2017, 111, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Kalra, N.; Singh, M.; Shukla, Y. Protective effects of lupeol and mango extract against androgen induced oxidative stress in Swiss albino mice. Asian J. Androl. 2008, 10, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Prasad, N.R. Effect of ursolic acid, a triterpenoid antioxidant, on ultraviolet-B radiation-induced cytotoxicity, lipid peroxidation and DNA damage in human lymphocytes. Chem. Biol. Interact. 2008, 176, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://biosig.lab.uq.edu.au/pdcsm_cancer (accessed on 15 June 2024).
- Available online: https://www.cheminfo.org/Chemistry/Cheminformatics/FormatConverter/index.html (accessed on 17 June 2024).
- Sukandar, E.R.; Kaennakam, S.; Raab, P.; Nöst, X.; Rassamee, K.; Bauer, R.; Siripong, P.; Ersam, T.; Tip-pyang, S.; Chavasiri, W. Cytotoxic and Anti-Inflammatory Activities of Dihydroisocoumarin and Xanthone Derivatives from Garcinia picrorhiza. Molecules 2021, 26, 6626. [Google Scholar] [CrossRef] [PubMed]
- Lagunin, A.A.; Rudik, A.V.; Pogodin, P.V.; Savosina, P.I.; Tarasova, O.A.; Dmitriev, A.V.; Ivanov, S.M.; Biziukova, N.Y.; Druzhilovskiy, D.S.; Filimonov, D.A.; et al. CLC-Pred 2.0: A Freely Available Web Application for In Silico Prediction of Human Cell Line Cytotoxicity and Molecular Mechanisms of Action for Druglike Compounds. Int. J. Mol. Sci. 2023, 14, 1689. [Google Scholar] [CrossRef]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for Molecular Docking: A Review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef]
- Evangelista Falcon, W.; Ellingson, S.R.; Smith, J.C.; Baudry, J. Ensemble Docking in Drug Discovery: How Many Protein Configurations from Molecular Dynamics Simulations Are Needed To Reproduce Known Ligand Binding? J. Phys. Chem. B 2019, 123, 5189–5195. [Google Scholar] [CrossRef] [PubMed]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.; Nguyen, T.H.; Pham, T.N.H.; Huy, N.T.; Bay, M.V.; Pham, M.Q.; Nam, P.C.; Vu, V.V.; Ngo, S.T. Autodock Vina Adopts More Accurate Binding Poses but Autodock4 Forms Better Binding Affinity. J. Chem. Inf. Model. 2020, 60, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Lees, D.M.; Reynolds, L.E.; Pedrosa, A.R.; Roy-Luzarraga, M.; Hodivala-Dilke, K.M. Phosphorylation of Pericyte FAK-Y861 Affects Tumour Cell Apoptosis and Tumour Blood Vessel Regression. Angiogenesis 2021, 24, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Serwe, G.; Kachaner, D.; Gagnon, J.; Plutoni, C.; Lajoie, D.; Duramé, E.; Sahmi, M.; Garrido, D.; Lefrançois, M.; Arseneault, G.; et al. CNK2 Promotes Cancer Cell Motility by Mediating ARF6 Activation Downstream of AXL Signalling. Nat. Commun. 2023, 14, 3560. [Google Scholar] [CrossRef] [PubMed]
- Brader, S.; Eccles, S.A. Phosphoinositide 3-Kinase Signalling Pathways in Tumor Progression, Invasion and Angiogenesis. Tumori 2004, 90, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Naser, R.; Aldehaiman, A.; Díaz-Galicia, E.; Arold, S.T. Endogenous Control Mechanisms of FAK and PYK2 and Their Relevance to Cancer Development. Cancers 2018, 10, 196. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qu, C.; Liu, T.; Wang, C. PFKFB3 Inhibitors as Potential Anticancer Agents: Mechanisms of Action, Current Developments, and Structure-Activity Relationships. Eur. J. Med. Chem. 2020, 203, 112612. [Google Scholar] [CrossRef] [PubMed]
- Nowakowski, J.; Cronin, C.N.; McRee, D.E.; Knuth, M.W.; Nelson, C.G.; Pavletich, N.P.; Rogers, J.; Sang, B.-C.; Scheibe, D.N.; Swanson, R.V.; et al. Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography. Structure 2002, 10, 1659–1667. [Google Scholar] [CrossRef]
- Yalçın, G. Antioxidant Capacity of a Turkish Traditional Alcoholic Drink, Raki. Pol. J. Food Nutr. Sci. 2016, 66, 167–171. [Google Scholar] [CrossRef]
- Koleva, I.I.; van Beek, T.A.; Linssen, J.P.H.; de Groot, A.; Evstatieva, L.N. Screening of Plant Extracts for Antioxidant Activity: A Comparative Study on Three Testing Methods. Phytochem. Anal. 2002, 13, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Piccinelli, A.L.; Mencherini, T.; Celano, R.; Mouhoubi, Z.; Tamendjari, A.; Aquino, R.P.; Rastrelli, L. Chemical Composition and Antioxidant Activity of Algerian Propolis. J. Agric. Food Chem. 2013, 61, 5080–5088. [Google Scholar] [CrossRef] [PubMed]
- Garbiec, E.; Rosiak, N.; Tykarska, E.; Zalewski, P.; Cielecka-Piontek, J. Sinapic Acid Co-Amorphous Systems with Amino Acids for Improved Solubility and Antioxidant Activity. Int. J. Mol. Sci. 2023, 24, 5533. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://admetmesh.scbdd.com (accessed on 20 June 2024).
- Taylor, E.; Kim, Y.; Zhang, K.; Chau, L.; Nguyen, B.C.; Rayalam, S.; Wang, X. Antiaging Mechanism of Natural Compounds: Effects on Autophagy and Oxidative Stress. Molecules 2022, 2, 4396. [Google Scholar] [CrossRef]
- Li, L.; Wei, L.; Shen, A.; Chu, J.; Lin, J.; Peng, J. Oleanolic acid modulates multiple intracellular targets to inhibit colorectal cancer growth. Int. J. Oncol. 2015, 47, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Sun, H.D.; Zhao, S.X. Triterpenoids of Isodon loxothyrsus. Phytochemistry 1996, 42, 1665–1666. [Google Scholar] [CrossRef]
- Hart, N.; Lamberton, J.; Sioumis, A.; Suares, H. New Triterpenes of Lantana camara. A Comparative Study of the Constituents of Several Taxa. Aust. J. Chem. 1976, 29, 655–671. [Google Scholar] [CrossRef]
- Cheng, K.G.; Su, C.H.; Yang, L.D.; Liu, J.; Chen, Z.F. Synthesis of oleanolic acid dimers linked at C-28 and evaluation of anti-tumor activity. Eur. J. Med. Chem. 2015, 89, 480–489. [Google Scholar] [CrossRef]



| ≥30.01 | 20.01–30.00 | 15.01–20.00 | 10.01–15.00 | 5.01–10.00 | 1.01–5.00 | 0.10–0.99 | 0.01–0.09 | |||||||||||||||
| Cancer Cell Line | IC50 (µM) | |||||||||||||||||||||
| Compound Number | ||||||||||||||||||||||
| 1 (OA) | 2a | 2b | 2c | 2d | 2e | 2f | 2g | 2h | 2i | 2j | 2k | 2l | 2m | 2n | ||||||||
| BT-549 | 26.42 | 9.93 | 3.89 | 3.89 | 3.62 | 6.78 | 6.78 | 3.54 | 3.47 | 3.43 | 3.42 | 3.46 | 3.47 | 3.40 | 3.43 | |||||||
| HS-578T | 16.75 | 7.43 | 3.42 | 3.35 | 3.29 | 3.05 | 3.05 | 3.46 | 3.57 | 3.48 | 3.52 | 3.55 | 3.55 | 3.49 | 3.51 | |||||||
| MCF-7 | 19.72 | 5.14 | 1.73 | 2.02 | 1.94 | 2.87 | 2.87 | 2.03 | 1.72 | 1.68 | 1.66 | 1.52 | 1.49 | 1.49 | 1.40 | |||||||
| MDA-MB-231-ATC | 19.45 | 7.45 | 5.44 | 4.42 | 5.87 | 5.44 | 5.44 | 5.89 | 5.912 | 5.79 | 5.78 | 5.78 | 5.82 | 5.82 | 5.85 | |||||||
| MDA-MB-468 | 3.40 | 0.35 | 0.24 | 0.63 | 0.62 | 0.23 | 0.23 | 0.62 | 0.61 | 0.61 | 0.61 | 0.61 | 0.61 | 0.61 | 0.61 | |||||||
| T-47D | 14.70 | 14.96 | 7.80 | 6.41 | 8.00 | 7.03 | 7.03 | 7.98 | 8.15 | 7.94 | 7.94 | 8.07 | 8.07 | 8.45 | 8.45 | |||||||
| SF-268 | 21.68 | 11.67 | 11.91 | 10.12 | 10.18 | 11.94 | 11.94 | 10.28 | 10.28 | 10.18 | 10.16 | 9.95 | 9.95 | 10.05 | 10.07 | |||||||
| SF-295 | 22.86 | 11.14 | 9.51 | 9.40 | 9.46 | 10.00 | 10.00 | 9.73 | 5.53 | 9.55 | 9.77 | 10.05 | 10.05 | 10.21 | 9.44 | |||||||
| SF-539 | 24.60 | 12.19 | 20.04 | 19.01 | 18.92 | 14.19 | 14.45 | 18.66 | 19.41 | 19.41 | 20.18 | 20.46 | 20.10 | 20.61 | 20.51 | |||||||
| SNB-19 | 17.82 | 14.12 | 10.79 | 12.79 | 13.83 | 17.06 | 17.06 | 14.49 | 14.35 | 14.12 | 12.65 | 11.83 | 11.45 | 11.50 | 11.43 | |||||||
| SNB-75 | 27.54 | 7.38 | 13.90 | 13.40 | 12.59 | 11.40 | 11.40 | 14.86 | 15.67 | 15.00 | 15.45 | 15.10 | 15.17 | 15.35 | 15.31 | |||||||
| SNB-78 | 15.56 | 15.78 | 20.41 | 26.49 | 26.48 | 23.93 | 23.93 | 26.48 | 26.67 | 26.67 | 26.67 | 26.67 | 26.67 | 26.67 | 26.67 | |||||||
| U251 | 14.55 | 9.27 | 9.53 | 9.64 | 10.18 | 9.22 | 9.22 | 10.35 | 9.57 | 9.77 | 9.46 | 9.37 | 9.22 | 9.08 | 8.65 | |||||||
| XF-498 | 5.99 | 3.49 | 6.08 | 5.92 | 5.89 | 4.82 | 4.82 | 5.69 | 5.57 | 5.82 | 6.29 | 6.34 | 6.14 | 6.12 | 6.18 | |||||||
| COLO-205 | 19.01 | 10.69 | 9.77 | 9.79 | 9.51 | 8.75 | 8.75 | 9.59 | 9.70 | 10.00 | 10.40 | 10.49 | 10.49 | 10.62 | 10.57 | |||||||
| DLD-1 | 14.27 | 9.62 | 9.31 | 9.98 | 11.91 | 5.14 | 5.14 | 12.39 | 13.21 | 13.21 | 13.46 | 13.46 | 13.33 | 13.03 | 13.00 | |||||||
| HCC-2998 | 15.74 | 5.32 | 4.63 | 4.32 | 4.47 | 4.25 | 4.25 | 4.25 | 4.19 | 4.21 | 4.27 | 4.33 | 4.32 | 4.23 | 4.17 | |||||||
| HCT-116 | 18.11 | 1.07 | 11.56 | 9.40 | 10.88 | 22.23 | 22.23 | 11.36 | 10.57 | 10.57 | 10.57 | 10.62 | 10.45 | 10.50 | 10.45 | |||||||
| HCT-15 | 16.90 | 1.41 | 19.63 | 16.98 | 19.91 | 17.38 | 17.38 | 20.00 | 20.51 | 20.51 | 20.47 | 19.91 | 20.00 | 20.04 | 20.00 | |||||||
| HT29 | 17.26 | 1.98 | 13.33 | 9.59 | 8.93 | 9.91 | 9.91 | 8.98 | 9.06 | 9.12 | 9.12 | 9.22 | 8.95 | 9.08 | 9.01 | |||||||
| KM112 | 13.09 | 1.67 | 11.91 | 11.30 | 10.49 | 25.82 | 25.82 | 10.89 | 10.02 | 9.12 | 8.71 | 9.08 | 8.55 | 8.73 | 8.63 | |||||||
| KM20L2 | 11.75 | 8.14 | 13.15 | 13.06 | 13.06 | 11.91 | 11.91 | 13.77 | 14.27 | 14.90 | 15.03 | 14.79 | 15.17 | 16.94 | 16.94 | |||||||
| SW-620 | 25.06 | 6.80 | 4.93 | 4.76 | 4.93 | 5.05 | 5.05 | 4.93 | 4.93 | 4.93 | 4.93 | 4.93 | 4.90 | 4.90 | 4.90 | |||||||
| CCR-CEM | 16.00 | 9.08 | 6.59 | 6.58 | 6.75 | 6.25 | 6.25 | 6.59 | 7.40 | 7.89 | 8.38 | 8.47 | 8.93 | 9.84 | 10.07 | |||||||
| HL-60TB | 17.18 | 10.37 | 10.94 | 9.20 | 10.30 | 9.20 | 9.20 | 10.57 | 10.47 | 10.45 | 10.84 | 10.84 | 10.49 | 10.42 | 10.47 | |||||||
| K-562 | 8.31 | 3.55 | 3.92 | 4.09 | 4.18 | 3.93 | 3.93 | 4.15 | 4.31 | 4.52 | 4.73 | 4.64 | 4.56 | 4.71 | 4.73 | |||||||
| MOLT-4 | 23.01 | 21.13 | 12.97 | 13.71 | 13.49 | 10.00 | 10.00 | 14.26 | 14.96 | 15.24 | 15.85 | 15.99 | 16.37 | 16.48 | 16.44 | |||||||
| P388-ADR | 3.76 | 0.03 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | |||||||
| P388 | 2.39 | 0.01 | 0.03 | 0.02 | 0.02 | 0.03 | 0.03 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | |||||||
| RPMI-8226 | 15.92 | 7.36 | 3.53 | 2.59 | 2.57 | 2.55 | 2.55 | 2.52 | 2.41 | 2.31 | 2.25 | 2.15 | 2.06 | 1.95 | 1.89 | |||||||
| SR | 16.25 | 10.14 | 13.21 | 14.29 | 16.25 | 12.02 | 12.02 | 16.59 | 18.41 | 19.23 | 17.10 | 15.27 | 14.93 | 14.52 | 12.68 | |||||||
| LOX-IMVI | 18.66 | 22.23 | 13.74 | 13.77 | 13.74 | 14.06 | 14.06 | 14.35 | 14.16 | 14.35 | 15.03 | 14.96 | 15.24 | 15.49 | 15.27 | |||||||
| M14 | 12.68 | 16.33 | 16.03 | 15.56 | 15.56 | 12.56 | 12.56 | 15.45 | 15.42 | 15.20 | 15.52 | 16.25 | 15.35 | 15.85 | 15.20 | |||||||
| M19-MEL | 10.74 | 15.10 | 16.05 | 15.03 | 16.60 | 12.88 | 12.88 | 17.02 | 15.52 | 15.20 | 15.38 | 15.30 | 15.20 | 15.27 | 15.13 | |||||||
| MALME-3M | 21.98 | 14.55 | 7.03 | 6.92 | 7.03 | 6.02 | 6.02 | 7.18 | 7.21 | 7.31 | 7.31 | 7.21 | 7.24 | 7.24 | 7.14 | |||||||
| MDA-MB-435 | 31.40 | 9.29 | 9.68 | 8.07 | 9.68 | 8.89 | 8.89 | 9.75 | 9.09 | 9.08 | 9.08 | 9.08 | 9.12 | 9.12 | 9.20 | |||||||
| MDA-N | 17.86 | 18.15 | 18.45 | 18.45 | 18.49 | 13.24 | 13.24 | 18.49 | 18.53 | 18.53 | 18.53 | 18.53 | 18.53 | 18.53 | 18.28 | |||||||
| SK-MEL-28 | 21.33 | 7.46 | 5.22 | 5.10 | 5.14 | 4.28 | 4.28 | 4.81 | 4.58 | 4.77 | 4.82 | 4.79 | 4.88 | 4.83 | 4.83 | |||||||
| SK-MEL-2 | 24.21 | 12.85 | 13.90 | 12.05 | 14.52 | 17.02 | 17.02 | 14.59 | 14.62 | 14.82 | 14.82 | 14.89 | 14.89 | 14.69 | 14.79 | |||||||
| SK-MEL-5 | 11.40 | 18.32 | 16.98 | 15.67 | 15.67 | 12.08 | 12.08 | 15.81 | 15.88 | 16.48 | 16.40 | 16.86 | 17.99 | 18.20 | 18.32 | |||||||
| UACC-257 | 47.97 | 9.77 | 6.74 | 6.89 | 7.29 | 8.30 | 8.30 | 7.52 | 7.31 | 7.26 | 7.29 | 7.36 | 7.40 | 7.05 | 7.05 | |||||||
| UACC-62 | 22.75 | 12.91 | 7.57 | 7.41 | 7.46 | 11.04 | 11.04 | 11.04 | 7.45 | 7.52 | 6.55 | 6.50 | 6.52 | 6.59 | 6.65 | |||||||
| A549-ATCC | 19.14 | 26.85 | 35.24 | 31.77 | 32.58 | 33.88 | 33.88 | 33.34 | 33.65 | 34.12 | 37.84 | 37.83 | 37.50 | 37.76 | 38.55 | |||||||
| EKVX | 30.76 | 37.84 | 18.28 | 23.77 | 23.44 | 16.57 | 16.57 | 23.01 | 23.01 | 32.01 | 22.96 | 23.12 | 23.23 | 23.23 | 23.23 | |||||||
| HOP-18 | 22.23 | 3.91 | 4.02 | 4.09 | 4.15 | 4.50 | 4.50 | 5.15 | 4.11 | 4.33 | 4.33 | 4.40 | 4.42 | 4.37 | 4.37 | |||||||
| HOP-62 | 15.74 | 17.94 | 12.36 | 14.00 | 13.71 | 12.33 | 12.33 | 13.88 | 13.12 | 13.24 | 13.64 | 13.61 | 13.58 | 13.99 | 14.19 | |||||||
| HOP_92 | 6.78 | 6.37 | 3.74 | 4.41 | 4.95 | 3.19 | 3.19 | 4.77 | 5.24 | 5.23 | 4.99 | 5.18 | 5.55 | 6.16 | 6.21 | |||||||
| LXFL-529 | 17.06 | 6.19 | 10.26 | 10.07 | 9.61 | 10.18 | 10.18 | 9.27 | 8.45 | 8.91 | 7.85 | 7.36 | 6.73 | 6.51 | 6.68 | |||||||
| NCI-H226 | 33.50 | 25.94 | 17.22 | 17.70 | 17.95 | 15.07 | 15.07 | 17.78 | 17.82 | 17.90 | 17.99 | 18.11 | 18.15 | 18.07 | 17.54 | |||||||
| NCI-H23 | 11.91 | 13.21 | 15.24 | 11.53 | 11.04 | 15.81 | 15.81 | 10.08 | 8.99 | 8.71 | 7.99 | 7.73 | 7.41 | 7.34 | 7.21 | |||||||
| NCI-H322M | 20.80 | 15.24 | 20.00 | 18.28 | 20.41 | 21.98 | 21.98 | 20.51 | 20.65 | 20.80 | 20.70 | 20.28 | 20.28 | 20.46 | 19.86 | |||||||
| NCI-H460 | 16.60 | 4.00 | 10.91 | 9.79 | 10.47 | 5.59 | 5.59 | 10.47 | 10.40 | 10.54 | 10.54 | 10.45 | 10.54 | 10.37 | 10.45 | |||||||
| NCI-H522 | 30.38 | 20.04 | 26.06 | 23.24 | 26.55 | 26.12 | 26.12 | 26.85 | 27.80 | 26.79 | 27.92 | 28.64 | 28.05 | 28.05 | 27.48 | |||||||
| IGROV1 | 19.54 | 22.13 | 17.18 | 13.21 | 13.58 | 16.56 | 16.56 | 13.87 | 13.93 | 13.93 | 13.96 | 14.16 | 14.09 | 14.16 | 14.39 | |||||||
| NCI-ADR-RES | 24.49 | 15.92 | 3.10 | 3.89 | 4.33 | 7.73 | 7.73 | 4.24 | 4.25 | 4.20 | 4.10 | 4.04 | 4.04 | 4.00 | 3.99 | |||||||
| OVCAR-3 | 15.24 | 13.37 | 10.35 | 10.66 | 10.00 | 9.70 | 9.70 | 9.95 | 9.97 | 10.00 | 9.73 | 9.68 | 9.51 | 9.42 | 9.31 | |||||||
| OVCAR-4 | 17.50 | 19.05 | 15.45 | 14.96 | 14.29 | 11.45 | 11.45 | 14.96 | 14.65 | 14.55 | 14.26 | 14.35 | 15.03 | 15.31 | 15.45 | |||||||
| OVCAR-5 | 23.39 | 36.90 | 21.63 | 17.86 | 21.43 | 19.36 | 19.36 | 21.13 | 20.80 | 21.18 | 21.48 | 21.43 | 22.23 | 22.54 | 22.85 | |||||||
| OVCAR-8 | 17.26 | 15.85 | 6.55 | 7.62 | 7.59 | 16.29 | 16.29 | 8.36 | 9.16 | 9.35 | 8.85 | 8.69 | 8.34 | 8.00 | 8.28 | |||||||
| SK-OV-3 | 22.96 | 30.34 | 20.28 | 22.33 | 24.15 | 28.71 | 28.71 | 23.39 | 24.77 | 26.12 | 26.30 | 26.73 | 26.67 | 26.00 | 25.82 | |||||||
| DU-145 | 11.45 | 11.12 | 3.03 | 2.62 | 2.10 | 6.84 | 6.84 | 2.18 | 2.15 | 2.12 | 2.04 | 2.02 | 2.02 | 2.03 | 1.99 | |||||||
| PC-3 | 15.49 | 14.39 | 16.14 | 12.79 | 17.91 | 16.52 | 16.51 | 18.24 | 17.91 | 17.02 | 16.18 | 16.07 | 15.85 | 16.11 | 16.22 | |||||||
| 786-0 | 18.20 | 19.23 | 24.72 | 25.55 | 25.18 | 23.33 | 23.33 | 24.43 | 24.27 | 24.15 | 23.33 | 23.66 | 22.59 | 21.68 | 21.13 | |||||||
| A498 | 29.85 | 10.23 | 10.18 | 7.45 | 6.81 | 10.37 | 10.37 | 6.93 | 6.90 | 7.19 | 7.11 | 7.06 | 7.08 | 6.92 | 6.87 | |||||||
| ACHN | 20.00 | 23.55 | 15.42 | 15.74 | 15.92 | 12.25 | 12.25 | 16.11 | 16.11 | 16.11 | 16.11 | 15.96 | 15.88 | 15.99 | 15.74 | |||||||
| CAKI-1 | 27.29 | 9.10 | 7.66 | 6.50 | 6.52 | 9.75 | 9.75 | 6.02 | 5.71 | 5.29 | 5.13 | 5.02 | 4.95 | 4.92 | 4.81 | |||||||
| RXF-393 | 7.69 | 2.55 | 1.60 | 1.70 | 1.65 | 2.46 | 2.46 | 1.81 | 1.99 | 2.05 | 2.03 | 2.09 | 2.09 | 2.17 | 2.20 | |||||||
| RXF-631 | 28.51 | 2.73 | 5.28 | 5.01 | 4.58 | 4.57 | 4.57 | 4.65 | 4.51 | 4.55 | 4.55 | 4.55 | 4.52 | 4.54 | 4.59 | |||||||
| SN12C | 21.53 | 11.59 | 4.77 | 4.83 | 4.84 | 7.57 | 7.57 | 4.81 | 5.08 | 5.37 | 5.44 | 5.47 | 5.52 | 5.37 | 5.37 | |||||||
| SN12K1 | 3.89 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | |||||||
| TK-10 | 27.73 | 18.84 | 16.56 | 17.74 | 20.51 | 16.90 | 16.90 | 22.18 | 24.32 | 25.94 | 27.23 | 27.48 | 2.73 | 28.64 | 28.71 | |||||||
| UO-31 | 13.37 | 20.28 | 13.96 | 13.49 | 14.93 | 21.68 | 21.68 | 14.59 | 14.49 | 14.26 | 13.06 | 13.58 | 12.88 | 12.79 | 13.06 | |||||||
| DMS-114 | 12.53 | 19.86 | 11.99 | 1.88 | 2.25 | 11.17 | 11.17 | 1.93 | 1.73 | 1.75 | 1.72 | 1.68 | 1.67 | 1.67 | 1.67 | |||||||
| DMS-273 | 28.64 | 13.35 | 4.36 | 4.32 | 4.07 | 7.53 | 7.53 | 4.61 | 5.00 | 5.22 | 5.89 | 6.68 | 8.36 | 9.35 | 8.93 | |||||||
| CurPocket ID | Cavity Volume (Å3) | Center (x, y, z) | Cavity Size (x, y, z) |
|---|---|---|---|
| C1 | 818 | 38, −3, 25 | 15, 13, 13 |
| C2 | 161 | 35, −14, 34 | 8, 11, 4 |
| C3 | 96 | 38, 13, −3 | 8, 8, 5 |
| C4 | 95 | 41, −19, 20 | 7, 4, 6 |
| C5 | 84 | 32, 18, 19 | 5, 8, 10 |
| CurPocket ID | Dimer | Vina Score (kcal⋅mol−1) | Cavity Volume (Å3) | Center (x, y, z) | Cavity Size (x, y, z) | Docking Size (x, y, z) |
|---|---|---|---|---|---|---|
| C1 | 2f | −11.6 | 818 | 38, −3, 25 | 15, 13, 13 | 30, 30, 30 |
| C2 | 2e | −8.6 | 161 | 35, −14, 34 | 8, 11, 4 | 30, 30, 30 |
| ≥30.01 | 20.01–30.00 | 15.01–20.00 | 10.01–15.00 | 5.01–10.00 | 1.01–5.00 | 0.10–0.99 | 0.01–0.09 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell Line | Compound Number IC50 [µM] | Lit. | |||||||||
| 2b [29]/11a [61] | 2d [29]/11b [61] | 2h [29]/11c [61] | 2j [29]/11d [61] | ||||||||
| Hep-G2 | 0.73 (0.06) | 5.70 (0.31) | 6.15 (0.57) | 3.99 (0.02) | [61] | ||||||
| A549 | <0.1 | 6.31 (0.55) | 0.51 (0.05) | 0.71 (0.07) | [61] | ||||||
| BGC-823 | 6.69 (0.59) | 1.49 (0.09) | 3.89 ± 0.03 | 48.34 (2.98) | [61] | ||||||
| MCF-7 | 4.74 (0.23) | <0.1 | 30.80 (4.29) | <0.1 | [61] | ||||||
| PC-3 | 1.76 (0.15) | 7.69 (0.81) | 33.24 (2.44) | 6.36 (0.56) | [61] | ||||||
| SKBR-3 | 6.67 (0.11) | 1.12 (0.03) | 6.02 (0.05) | 9.99 (0.04) | [29] | ||||||
| SKOV-3 | 6.49 (0.01) | 1.56 (0.01) | 5.39 (0.02) | 10.27 (0.05) | [29] | ||||||
| PC-3 | 6.43 (0.03) | 1.64 (0.01) | 5.34 (0.07) | 9.81 (0.02) | [29] | ||||||
| U-87 | 6.59 (0.01) | 1.20 (0.11) | 5.87 (0.09) | 10.68 (0.04) | [29] | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Günther, A.; Zalewski, P.; Sip, S.; Bednarczyk-Cwynar, B. Exploring the Potential of Oleanolic Acid Dimers–Cytostatic and Antioxidant Activities, Molecular Docking, and ADMETox Profile. Molecules 2024, 29, 3623. https://doi.org/10.3390/molecules29153623
Günther A, Zalewski P, Sip S, Bednarczyk-Cwynar B. Exploring the Potential of Oleanolic Acid Dimers–Cytostatic and Antioxidant Activities, Molecular Docking, and ADMETox Profile. Molecules. 2024; 29(15):3623. https://doi.org/10.3390/molecules29153623
Chicago/Turabian StyleGünther, Andrzej, Przemysław Zalewski, Szymon Sip, and Barbara Bednarczyk-Cwynar. 2024. "Exploring the Potential of Oleanolic Acid Dimers–Cytostatic and Antioxidant Activities, Molecular Docking, and ADMETox Profile" Molecules 29, no. 15: 3623. https://doi.org/10.3390/molecules29153623
APA StyleGünther, A., Zalewski, P., Sip, S., & Bednarczyk-Cwynar, B. (2024). Exploring the Potential of Oleanolic Acid Dimers–Cytostatic and Antioxidant Activities, Molecular Docking, and ADMETox Profile. Molecules, 29(15), 3623. https://doi.org/10.3390/molecules29153623










