Signal-On Detection of Caspase-3 with Methylene Blue-Loaded Metal-Organic Frameworks as Signal Reporters
Abstract
:1. Introduction
2. Results and Discussion
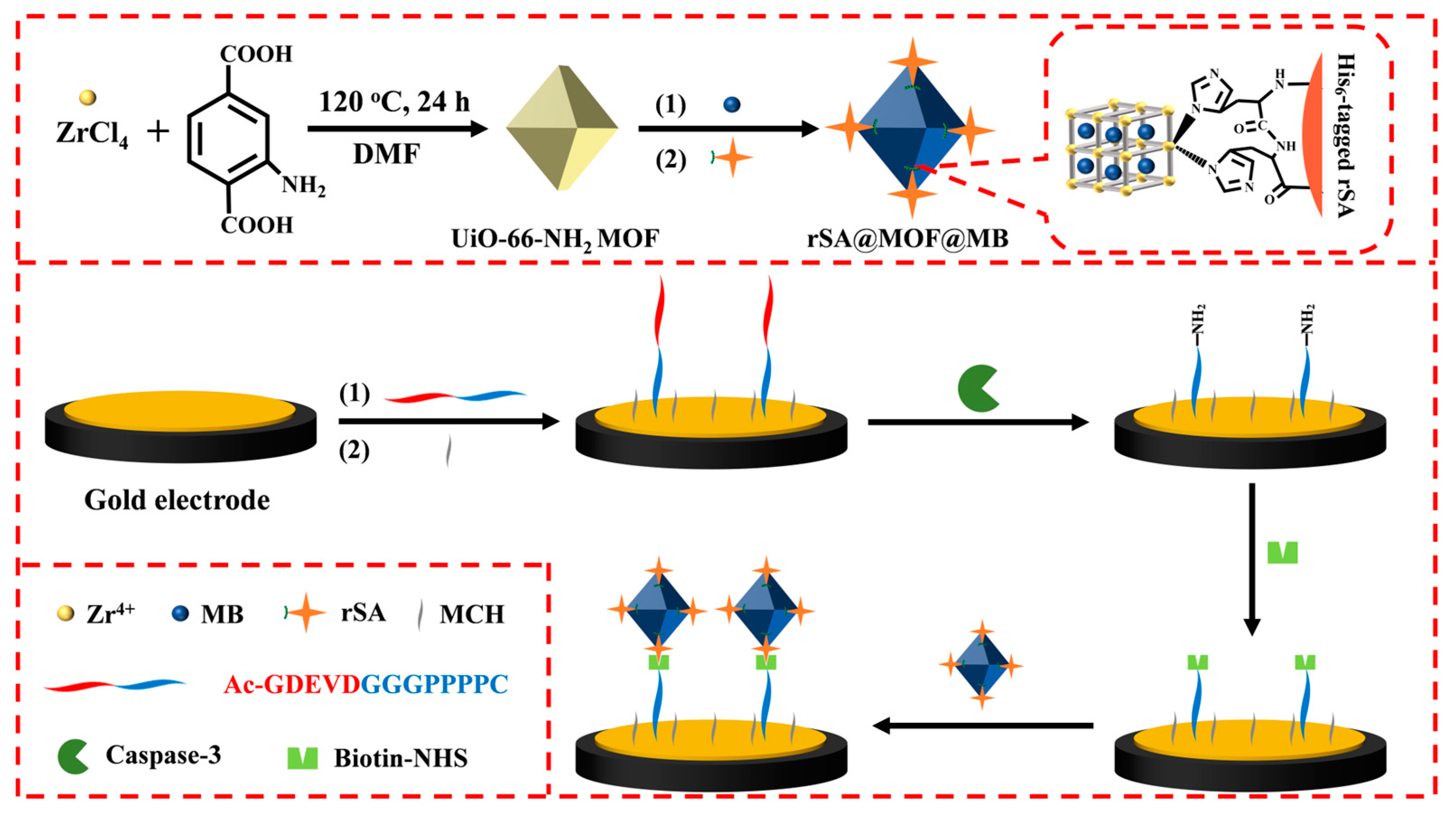
2.1. Principle of the Method
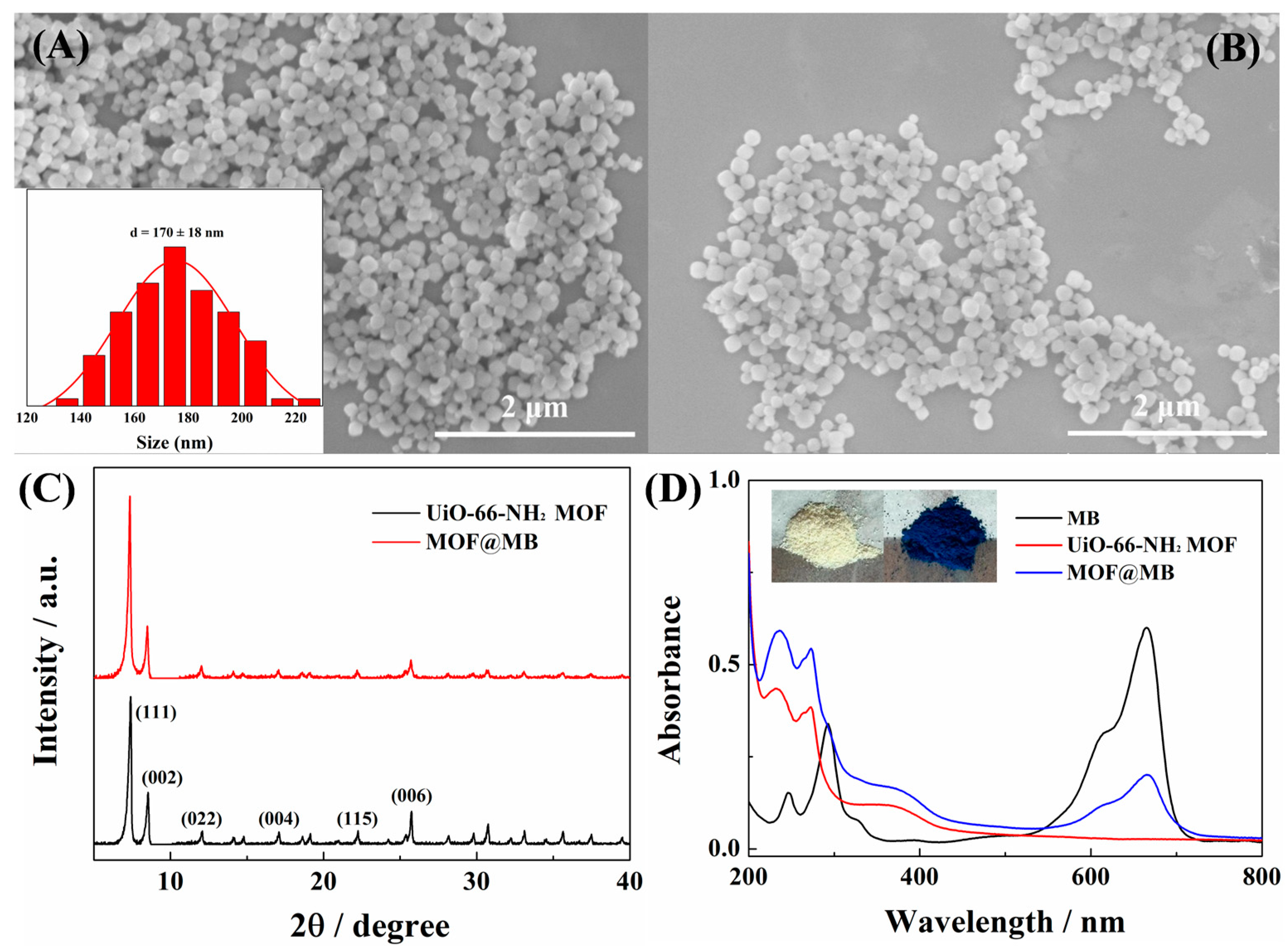
2.2. Characterization of MOFs
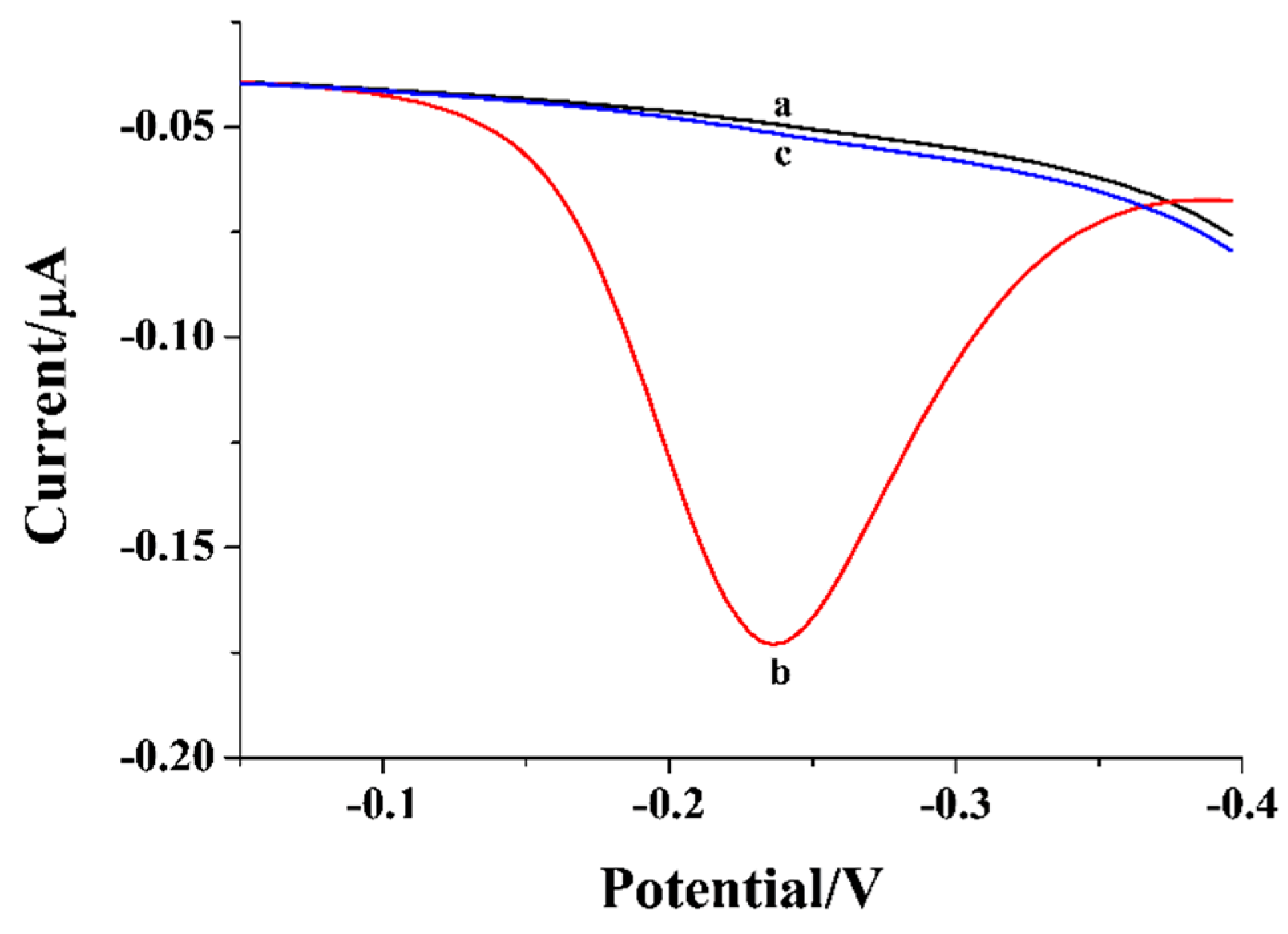
2.3. Feasibility
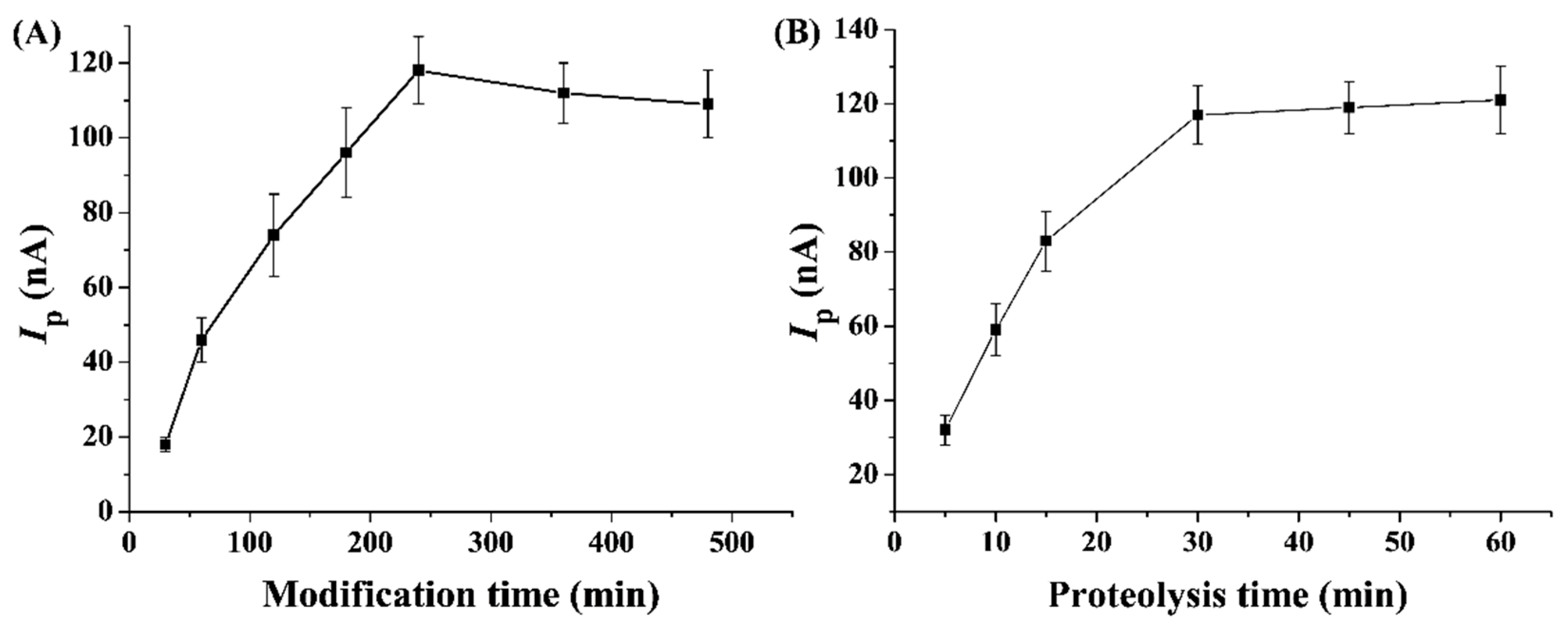
2.4. Optimization of Detection Conditions
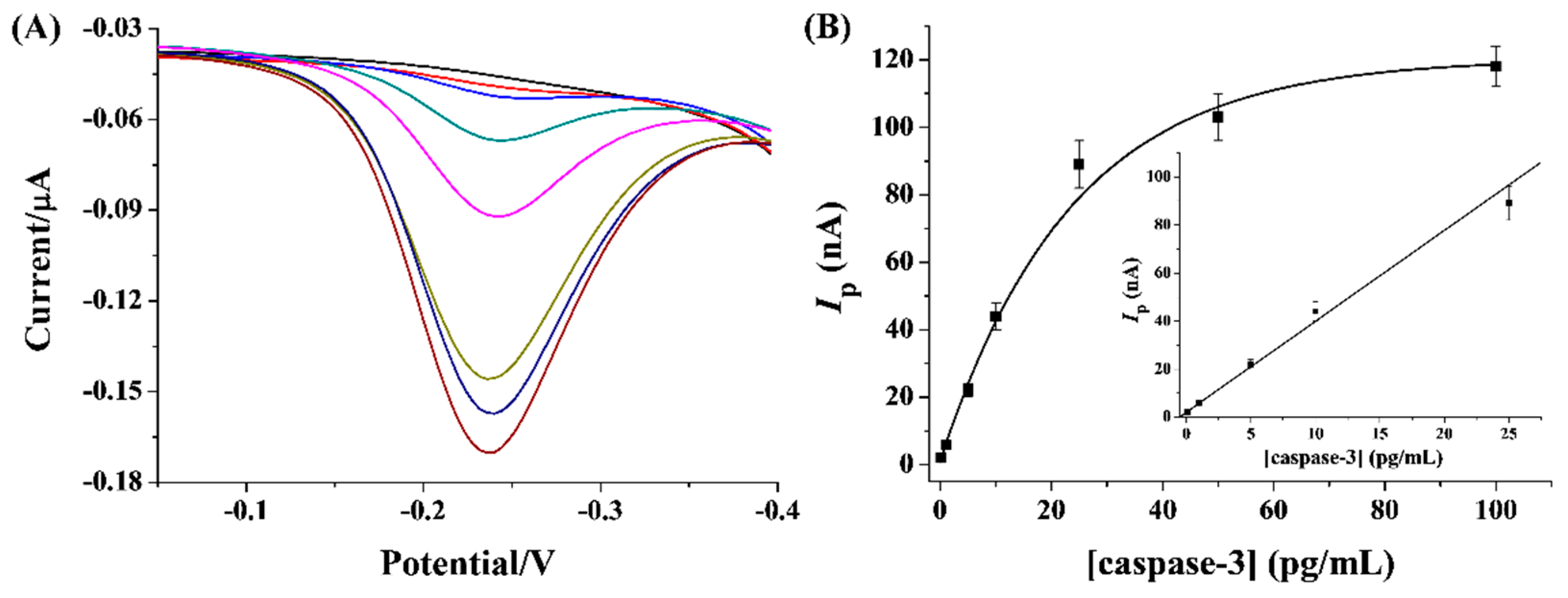
2.5. Analytical Performance of Method
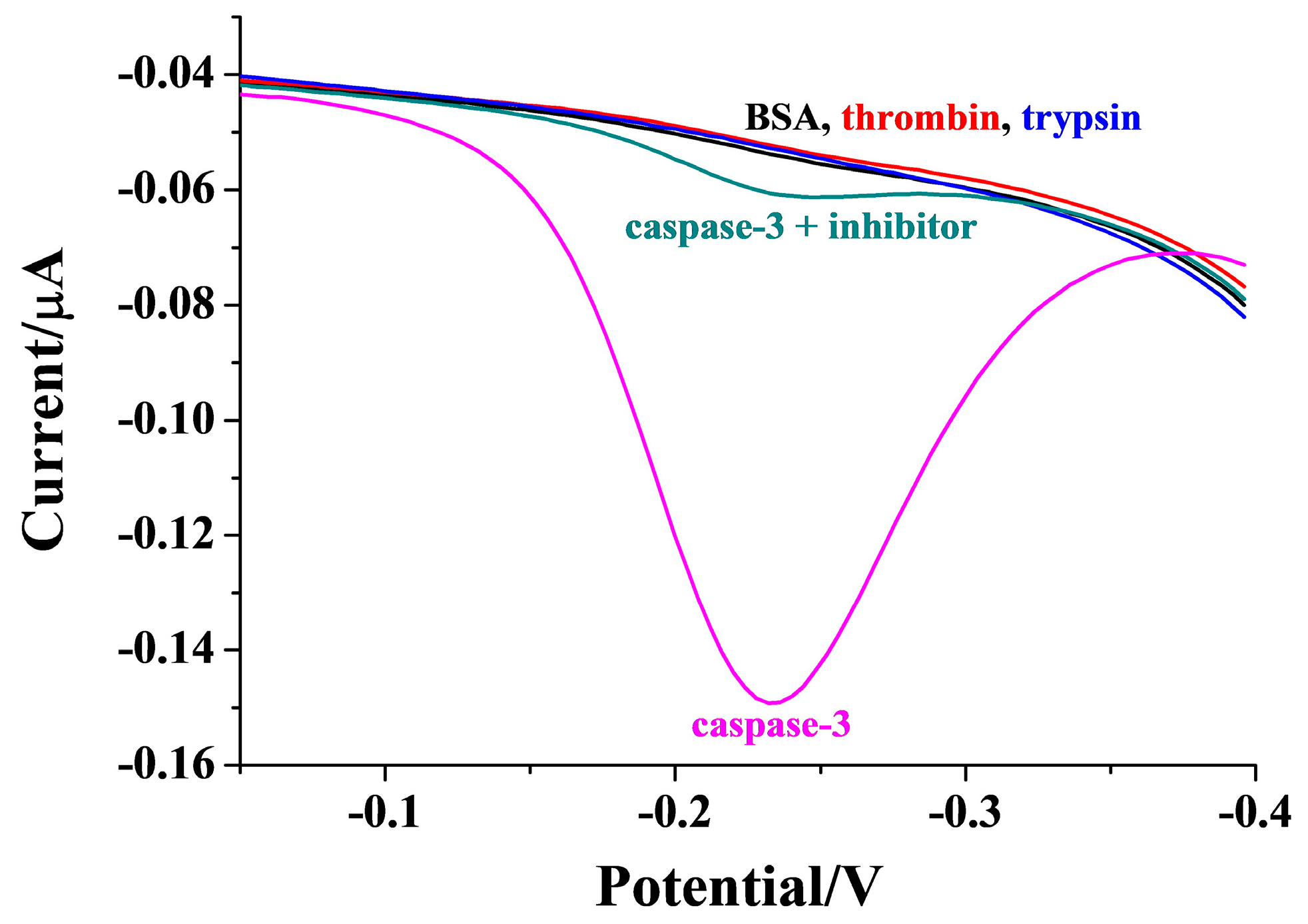
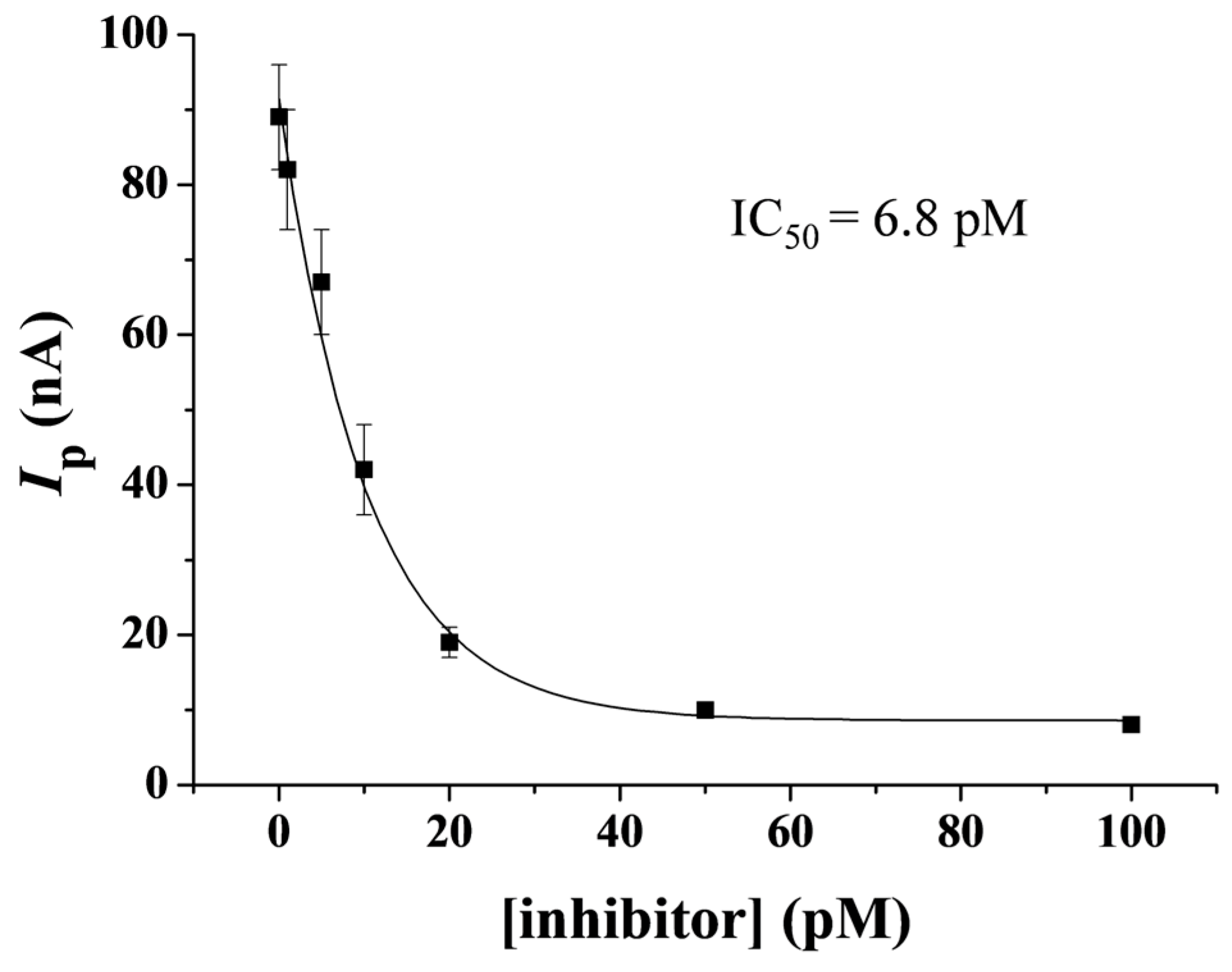
2.6. Selectivity of Method and Evaluation of Inhibition Viability
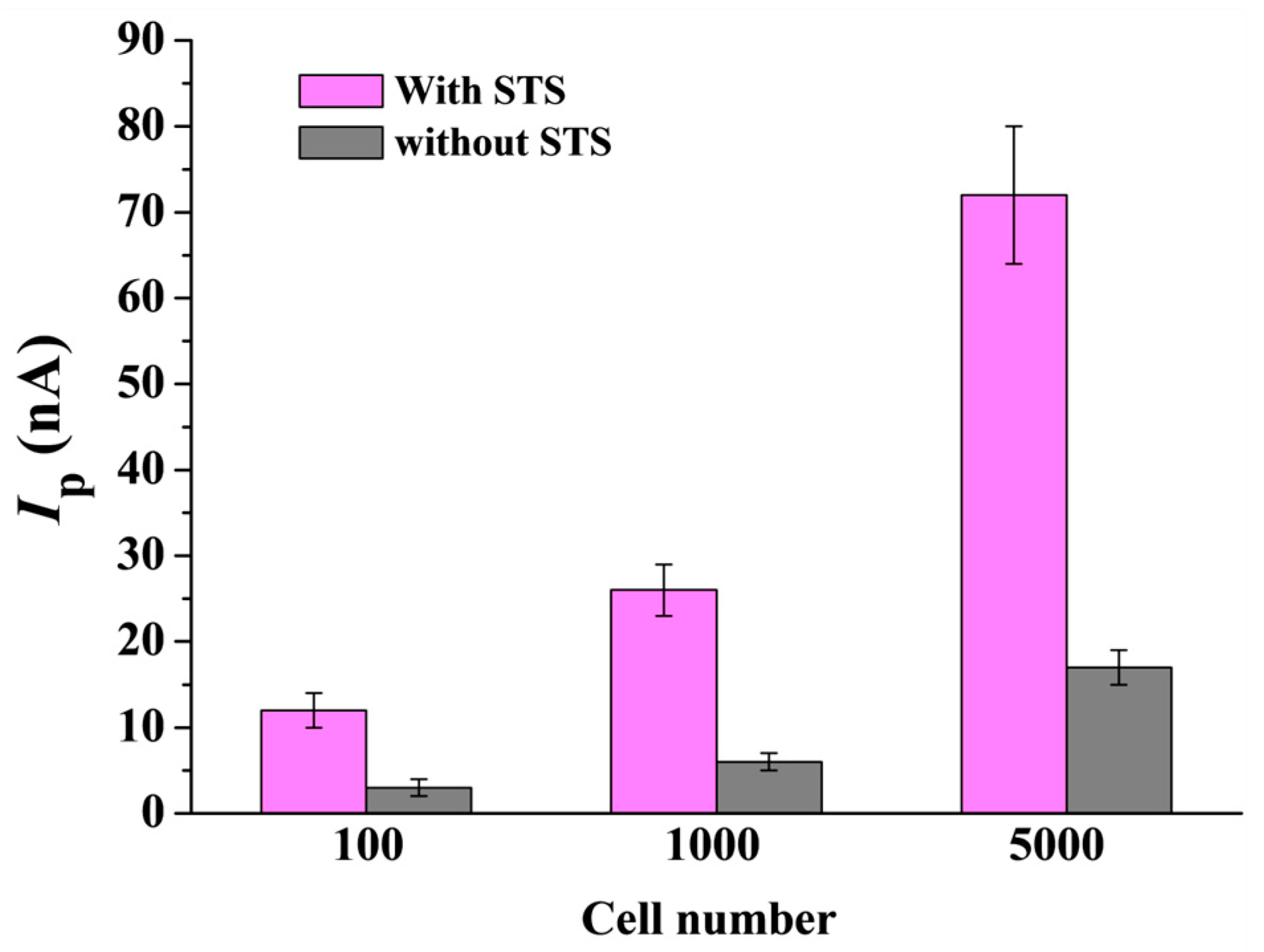
2.7. Assays of Caspase-3 in HeLa Cells
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Apparatus
3.3. Preparation of rSA@MOF@MB
3.4. Electrode Modification
3.5. Electrochemical Detection of Caspase-3
3.6. Cell Culture and Apoptosis Assays
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef]
- Favaloro, B.; Allocati, N.; Graziano, V.; Di Ilio, C.; De Laurenzi, V. Role of apoptosis in disease. Aging 2012, 4, 330–349. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death: From specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013, 332, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Shalini, S.; Dorstyn, L.; Dawar, S.; Kumar, S. Old, new and emerging functions of caspases. Cell Death Differ. 2015, 22, 526–539. [Google Scholar] [CrossRef] [PubMed]
- Julien, O.; Wells, J.A. Caspases and their substrates. Cell Death Differ. 2017, 24, 1380–1389. [Google Scholar] [CrossRef]
- Buckley, C.D.; Pilling, D.; Henriquez, N.V.; Parsonage, G.; Threlfall, K.; Scheel-Toellner, D.; Simmons, D.L.; Akbar, A.N.; Lord, J.M.; Salmon, M. RGD peptides induce apoptosis by direct caspase-3 activation. Nature 1999, 397, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Hao, Y.; Yang, S.; Han, Q.; Liu, L.; Xiang, Y.; Tu, F.; Xia, N. A signal-on electrochemical biosensor for evaluation of caspase-3 activity and cell apoptosis by the generation of molecular electrocatalysts on graphene electrode surface for water oxidation. Sens. Actuat. B Chem. 2019, 286, 415–420. [Google Scholar] [CrossRef]
- Ji, C.; Amarnath, V.; Pietenpol, J.A.; Marnett, L.J. 4-hydroxynonenal induces apoptosis via caspase-3 activation and cytochrome c release. Chem. Res. Toxicol. 2001, 14, 1090–1096. [Google Scholar] [CrossRef]
- Liu, L.; Li, X.; Zhang, H.; Chen, H.; Abualrejal, M.M.A.; Song, D.; Wang, Z. Six-in-one peptide functionalized upconversion@polydopamine nanoparticle-based ratiometric fluorescence sensing platform for real-time evaluating anticancer efficacy through monitoring caspase-3 activity. Sens. Actuat. B Chem. 2021, 333, 129554–129561. [Google Scholar] [CrossRef]
- Shen, Y.; Xin, Z.; Zhu, Y.; Wang, J. Mesoporous carbon nanospheres featured multifunctional fluorescent nanoprobe: Simultaneous activation and tracing of caspase-3 involved cell apoptosis. Sens. Actuat. B Chem. 2022, 358, 131485–131493. [Google Scholar] [CrossRef]
- Ma, X.; Lv, Y.; Liu, P.; Hao, Y.; Xia, N. Switch-on fluorescence analysis of protease activity with the assistance of a nickel ion-nitrilotriacetic acid-conjugated magnetic nanoparticle. Molecules 2023, 28, 3426. [Google Scholar] [CrossRef]
- Gurtu, V.; Kain, S.R.; Zhang, G. Fluorometric and colorimetric detection of caspase activity associated with apoptosis. Anal. Biochem. 1997, 251, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Guo, M.; Nie, Z.; Huang, Y.; Peng, Y.; Liu, A.; Qing, M.; Yao, S. Colorimetric detection of apoptosis based on caspase-3 activity assay using unmodified gold nanoparticles. Chem. Commun. 2012, 48, 997–999. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Peng, L.; Wang, X.; Xiang, Y.; Tong, A. A new colorimetric strategy for monitoring caspase 3 activity by HRP-mimicking DNAzyme-peptide conjugates. Analyst 2014, 139, 1178–1183. [Google Scholar] [CrossRef]
- Yuan, X.; Niu, Z.; Liu, L.; Zeng, Y.; Ma, L.; Nie, Z.; Tian, Z.; Kai, D.; Zhang, F.; Liu, G.; et al. Intensity interrogation-based high-sensitivity surface plasmon resonance imaging biosensor for apoptosis detection in cancer. Biosensors 2023, 13, 946. [Google Scholar] [CrossRef]
- Zhu, H.; Mao, Z.; Chen, J.; Hu, J.; Hu, X.; Koh, K.; Chen, H. Cucurbit[7]urils induced bimetallic nanoparticles network for ultra-sensitive detection of Caspase-3 based on surface plasmon resonance. Microchem. J. 2021, 171, 106792–106799. [Google Scholar] [CrossRef]
- Choi, J.H.; El-Said, W.A.; Choi, J.-W. Highly sensitive surface-enhanced Raman spectroscopy (SERS) platform using core/double shell (Ag/polymer/Ag) nanohorn for proteolytic biosensor. Appl. Surf. Sci. 2020, 506, 144669–144674. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, C.-Y.; Hu, J.-M.; Shen, A.-G. Promoted “Click” SERS detection for precise intracellular imaging of caspase-3. Anal. Chem. 2021, 93, 4876–4883. [Google Scholar] [CrossRef]
- Zhuang, Y.; Dong, H.; Liu, T.; Zhao, Y.; Xu, Y.; Zhao, X.; Sun, D. Highly sensitive and selective SERS detection of caspase-3 during cell apoptosis based on the target-induced hotspot effect. Analyst 2024, 149, 490–496. [Google Scholar] [CrossRef]
- Yu, J.; Yang, A.; Wang, N.; Ling, H.; Song, J.; Chen, X.; Lian, Y.; Zhang, Z.; Yan, F.; Gu, M. Highly sensitive detection of caspase-3 activity based on peptide-modified organic electrochemical transistor biosensors. Nanoscale 2021, 13, 2868–2874. [Google Scholar] [CrossRef]
- Shin, J.H.; Gul, A.R.; Hyun, M.S.; Choi, C.H.; Park, T.J.; Park, J.P. Electrochemical detection of caspase-3 based on a chemically modified M13 phage virus. Bioelectrochemistry 2022, 145, 108090–108097. [Google Scholar] [CrossRef]
- Dong, Y.P.; Chen, G.; Zhou, Y.; Zhu, J.J. Electrochemiluminescent Sensing for Caspase-3 Activity Based on Ru(bpy)32+-Doped Silica Nanoprobe. Anal. Chem. 2016, 88, 1922–1929. [Google Scholar] [CrossRef]
- Cheng, M.; Zhou, J.; Zhou, X.; Xing, D. Peptide cleavage induced assembly enables highly sensitive electrochemiluminescence detection of protease activity. Sens. Actuat. B Chem. 2018, 262, 516–521. [Google Scholar] [CrossRef]
- Liang, G.-X.; Zhao, K.-R.; He, Y.-S.; Liu, Z.-J.; Ye, S.-Y.; Wang, L. Carbon dots and gold nanoparticles doped metal-organic frameworks as high-efficiency ECL emitters for monitoring of cell apoptosis. Microchem. J. 2021, 171, 106787–106793. [Google Scholar] [CrossRef]
- Luo, W.; Chu, H.; Wu, X.; Ma, P.; Wu, Q.; Song, D. Disposable biosensor based on novel ternary Ru-PEI@PCN-333(Al) self-enhanced electrochemiluminescence system for on-site determination of caspase-3 activity. Talanta 2021, 239, 123083–123090. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Li, Y.; Zou, K.; Meng, L.; Zhang, X.; Chen, J. A label-free and blocker-free photoelectrochemical strategy for highly sensitive caspase-3 assay. Chem. Commun. 2018, 54, 4830–4833. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, J.; Liu, X.; Shi, X.; Dai, Z. Photoelectrochemical approach to apoptosis evaluation via multi-functional peptide- and electrostatic attraction-guided excitonic response. Anal. Chem. 2019, 91, 830–835. [Google Scholar] [CrossRef]
- Bachor, R. Peptidyl-resin substrates as a tool in the analysis of caspase activity. Molecules 2022, 27, 4107. [Google Scholar] [CrossRef]
- Hung, V.W.S.; Veloso, A.J.; Chow, A.M.; Ganesh, H.V.S.; Seo, K.; Kendüzler, E.; Brown, I.R.; Kerman, K. Electrochemical impedance spectroscopy for monitoring caspase-3 activity. Electrochim. Acta 2015, 162, 79–85. [Google Scholar] [CrossRef]
- Liu, G.; Li, Y.; Liu, M.; Cheng, J.; Yang, S.; Gao, F.; Liu, L. Overview on peptide-based electrochemical biosensors. Int. J. Electrochem. Sci. 2023, 18, 100395–100413. [Google Scholar] [CrossRef]
- Li, H.Y.; Qi, H.J.; Chang, J.F.; Gai, P.P.; Li, F. Recent progress in homogeneous electrochemical sensors and their designs and applications. TrAC-Trend. Anal. Chem. 2022, 156, 116712–116726. [Google Scholar] [CrossRef]
- Eivazzadeh-Keihan, R.; Saadatidizaji, Z.; Maleki, A.; de la Guardia, M.; Mahdavi, M.; Barzegar, S.; Ahadian, S. Recent progresses in development of biosensors for thrombin detection. Biosensors 2022, 12, 767. [Google Scholar] [CrossRef]
- Karimzadeh, A.; Hasanzadeh, M.; Shadjou, N.; Guardia, M.D.L. Peptide based biosensors. TrAC-Trend. Anal. Chem. 2018, 107, 1–20. [Google Scholar] [CrossRef]
- Yuan, L.; Liu, L. Peptide-based electrochemical biosensing. Sens. Actuat. B Chem. 2021, 344, 130232–130257. [Google Scholar] [CrossRef]
- Xiao, H.; Liu, L.; Meng, F.; Huang, J.; Li, G. Electrochemical approach to detect apoptosis. Anal. Chem. 2008, 80, 5272–5275. [Google Scholar] [CrossRef]
- Takano, S.; Shiomoto, S.; Inoue, K.Y.; Ino, K.; Shiku, H.; Matsue, T. Electrochemical approach for the development of a simple method for detecting cell apoptosis based on caspase-3 activity. Anal. Chem. 2014, 86, 4723–4728. [Google Scholar] [CrossRef]
- Sixiang, S.; Inoue, K.Y.; Shiomoto, S.; Takano, S.; Ino, K.; Shiku, H.; Matsue, T. Amperometric detection of apoptosis by using p-methoxyaniline-conjugated substrate for caspase-3. ChemElectroChem 2017, 4, 941–946. [Google Scholar] [CrossRef]
- Khalilzadeh, B.; Charoudeh, H.N.; Shadjou, N.; Mohammad-Rezaei, R.; Omidi, Y.; Velaei, K.; Aliyari, Z.; Rashidi, M.-R. Ultrasensitive caspase-3 activity detection using an electrochemical biosensor engineered by gold nanoparticle functionalized MCM-41: Its application during stem cell differentiation. Sens. Actuat. B Chem. 2016, 231, 561–575. [Google Scholar] [CrossRef]
- Gao, F.; Liu, G.; Qiao, Y.; Dong, X.; Liu, L. Streptavidin-conjugated DNA for the boronate affinity-based detection of poly(ADP-ribose) polymerase-1 with improved sensitivity. Biosensors 2023, 13, 723. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, Y.; Zhao, M.; Jiang, L.P.; Zhu, J.J. CdSeTe@CdS@ZnS Quantum-Dot-Sensitized Macroporous TiO2 Film: A Multisignal-Amplified Photoelectrochemical Platform. Chemphyschem 2015, 16, 2826–2835. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Zheng, T.T.; Cheng, F.F.; Zhu, J.J. Electrochemical sensing for caspase 3 activity and inhibition using quantum dot functionalized carbon nanotube labels. Chem. Commun. 2011, 47, 1178–1180. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Huang, Y.; Cui, Z.; Liu, S.; Deng, D.; Liu, L.; Wang, J. Impedimetric biosensor for assay of caspase-3 activity and evaluation of cell apoptosis using self-assembled biotin-phenylalanine network as signal enhancer. Sens. Actuat. B Chem. 2020, 320, 128436–128442. [Google Scholar] [CrossRef]
- Yang, R.; Zou, K.; Li, Y.; Meng, L.; Zhang, X.; Chen, J. Co3O4-Au polyhedra: A multifunctional signal amplifier for sensitive photoelectrochemical assay. Anal. Chem. 2018, 90, 9480–9486. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, J.; Gao, Y.; Liu, S.; Koh, K.; Zhu, X.; Yin, Y. Sensitive cell apoptosis assay based on caspase-3 activity detection with graphene oxide-assisted electrochemical signal amplification. Biosens. Bioelectron. 2015, 68, 777–782. [Google Scholar] [CrossRef]
- Khalilzadeh, B.; Shadjou, N.; Eskandani, M.; Charoudeh, H.N.; Omidi, Y.; Rashidi, M.-R. A reliable self-assembled peptide based electrochemical biosensor for detection of caspase 3 activity and apoptosis. RSC Adv. 2015, 5, 58316–58326. [Google Scholar] [CrossRef]
- Song, S.; Hu, X.; Li, H.; Zhao, J.; Koh, K.; Chen, H. Guests involved CB[8] capped silver nanoparticles as a means of electrochemical signal enhancement for sensitive detection of Caspase-3. Sens. Actuat. B Chem. 2018, 274, 54–59. [Google Scholar] [CrossRef]
- MacDonald, J.I.; Munch, H.K.; Moore, T.; Francis, M.B. One-step site-specific modification of native proteins with 2-pyridinecarboxyaldehydes. Nat. Chem. Biol. 2015, 11, 326–331. [Google Scholar] [CrossRef]
- Song, X.; Ren, X.; Mei, Q.; Liu, H.; Huang, H. Advancing in-depth N-terminomics detection with a cleavable 2-pyridinecarboxyaldehyde probe. J. Am. Chem. Soc. 2024, 146, 6487–6492. [Google Scholar] [CrossRef]
- Song, S.; Shang, X.; Zhao, J.; Hu, X.; Koh, K.; Wang, K.; Chen, H. Sensitive and selective determination of caspase-3 based on calixarene functionalized reduction of graphene oxide assisted signal amplification. Sens. Actuat. B Chem. 2018, 267, 357–365. [Google Scholar] [CrossRef]
- Fan, H.; Kou, J.; Han, D.; Li, P.; Zhang, D.; Wu, Q.; He, Q. Sensitive proteolysis assay based on the detection of a highly characteristic solid-state process. RSC Adv. 2015, 5, 48893–48897. [Google Scholar] [CrossRef]
- Hofmann, C.; Duerkop, A.; Baeumner, A.J. Nanocontainers for analytical applications. Angew.Chem. Int. Ed. 2019, 58, 12840–12860. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Dang, Z.; Muddassir, M.; Raza, S.; Zhong, A.; Wang, X.; Jin, J. A new type Co(II)-based photocatalyst for the nitrofurantoin antibiotic degradation. J. Mol. Struct. 2024, 1312, 138501. [Google Scholar]
- Zhao, J.; Dang, Z.; Muddassir, M.; Raza, S.; Zhong, A.; Wang, X.; Jin, J. A new Cd(II)-based coordination polymer for efficient photocatalytic removal of organic dyes. Molecules 2023, 28, 6848. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Huang, J.; Jian, Y.; Singh, A.; Kumar, A.; Liu, D.; Pan, Y.; Ouyang, Q. Metal–organic frameworks (MOFs) as apt luminescent probes for the detection of biochemical analytes. J. Mater. Chem. B 2023, 11, 6802–6822. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Kong, X.-J.; Li, J.-R. Chemically stable metal-organic frameworks: Rational construction and application expansion. Acc. Chem. Res. 2021, 54, 3083–3094. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lai, C.; Liu, X.; Li, B.; Zhang, C.; Qin, L.; Huang, D.; Yia, H.; Zhang, M.; Li, L.; et al. Metal-organic frameworks and their derivatives as signal amplification elements for electrochemical sensing. Coord. Chem. Rev. 2020, 424, 213520. [Google Scholar] [CrossRef]
- Chang, Y.; Lou, J.; Yang, L.; Liu, M.; Xia, N.; Liu, L. Design and application of electrochemical sensors with Metal-organic frameworks as the electrode materials or signal tags. Nanomaterials 2022, 12, 3248. [Google Scholar] [CrossRef] [PubMed]
- Roder, R.; Preiss, T.; Hirschle, P.; Steinborn, B.; Zimpel, A.; Hohn, M.; Radler, J.O.; Bein, T.; Wagner, E.; Wuttke, S.; et al. Multifunctional nanoparticles by coordinative self-assembly of His-tagged units with metal-organic frameworks. J. Am. Chem. Soc. 2017, 139, 2359–2368. [Google Scholar] [CrossRef]
- Jin, Z.; Ling, C.; Li, Y.; Zhou, J.; Li, K.; Yim, W.; Yeung, J.; Chang, Y.C.; He, T.; Cheng, Y.; et al. Spacer matters: All-peptide-based ligand for promoting interfacial proteolysis and plasmonic coupling. Nano Lett. 2022, 22, 8932–8940. [Google Scholar] [CrossRef]
- Nowinski, A.K.; Sun, F.; White, A.D.; Keefe, A.J.; Jiang, S. Sequence, structure, and function of peptide self-assembled monolayers. J. Am. Chem. Soc. 2012, 134, 6000–6005. [Google Scholar] [CrossRef]
- Sun, Z.; Wu, S.; Ma, J.; Shi, H.; Wang, L.; Sheng, A.; Yin, T.; Sun, L.; Li, G. Colorimetric sensor array for human semen identification designed by coupling Zirconium metal-organic frameworks with DNA-modified gold nanoparticles. ACS Appl. Mater. Interfaces 2019, 11, 36316–36323. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wang, L.; Wu, S.; Pan, Y.; Dong, Y.; Zhu, S.; Yang, J.; Yin, Y.; Li, G. An electrochemical biosensor designed by using Zr-based metal-organic frameworks for the detection of glioblastoma-derived exosomes with practical application. Anal. Chem. 2020, 92, 3819–3826. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Wang, S.; Ding, Z.; Wang, S.; Zhou, Y.; Huang, L.; Wang, X. Amine-functionalized zirconium metal-organic framework as efficient visible-light photocatalyst for aerobic organic transformations. Chem. Commun. 2012, 48, 11656–11658. [Google Scholar] [CrossRef] [PubMed]
- Helal, A.; Qamaruddin, M.; Aziz, M.A.; Shaikh, M.N.; Yamani, Z.H. MB-UiO-66-NH2 metal-organic framework as chromogenic and fluorogenic sensor for hydrazine gydrate in aqueous solution. ChemistrySelect 2017, 2, 7630–7636. [Google Scholar] [CrossRef]
- Wu, D.; He, Y.; Tong, L.; Wang, J.; Liu, L.; Yi, X.; Hu, S. Electrochemical determination of caspase-3 using signal amplification by HeLa cells modified with silver nanoparticles. Microchim. Acta 2021, 188, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Xu, H.; Cheng, L.; Yuan, Q.; Liu, C.; Gao, H.; Liang, H. Highly selective entrapment of His-tagged enzymes on superparamagnetic Zirconium-based MOFs with robust renewability to enhance pH and thermal stability. ACS Biomater. Sci. Eng. 2021, 7, 3727–3736. [Google Scholar] [CrossRef]

| Signal Labels | Linear Range | LOD | Ref. |
|---|---|---|---|
| MB-carried GO | 0.1–100 pg/mL | 0.06 pg/mL | [45] |
| AgNPs | 0.5–50 pg/mL | 0.1 pg/mL | [51] |
| HRP-modified magnetic bead | 100 pM–1 nM | 100 pM | [46] |
| CB[8]-AgNPs | 1–10 ng/mL | 24.62 pg/mL | [47] |
| MB-pSC6-GO | 10–100 pg/mL | 0.0167 pg/mL | [50] |
| ATCUN-Cu(II) complex | 0.5 pg/mL–2 ng/mL | 0.2 pg/mL | [8] |
| biotin-Phe nanoparticle | 1–125 pg/mL | 1 pg/mL | [43] |
| AgNPs-modified HeLa cells | 0.02–0.2 U/mL | 0.02 U/mL | [66] |
| rSA@MOF@MB | 0.1–25 pg/mL | 0.04 pg/mL | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Wang, J.; Xu, Y.; Zhang, J.; Xia, N. Signal-On Detection of Caspase-3 with Methylene Blue-Loaded Metal-Organic Frameworks as Signal Reporters. Molecules 2024, 29, 3700. https://doi.org/10.3390/molecules29153700
Huang Y, Wang J, Xu Y, Zhang J, Xia N. Signal-On Detection of Caspase-3 with Methylene Blue-Loaded Metal-Organic Frameworks as Signal Reporters. Molecules. 2024; 29(15):3700. https://doi.org/10.3390/molecules29153700
Chicago/Turabian StyleHuang, Yaliang, Jiaqiang Wang, Yirui Xu, Jiwen Zhang, and Ning Xia. 2024. "Signal-On Detection of Caspase-3 with Methylene Blue-Loaded Metal-Organic Frameworks as Signal Reporters" Molecules 29, no. 15: 3700. https://doi.org/10.3390/molecules29153700





