Advancements in the Heterologous Expression of Sucrose Phosphorylase and Its Molecular Modification for the Synthesis of Glycosylated Products
Abstract
:1. Introduction
2. Source Microorganisms of SPase
3. Structure and Catalytic Mechanism of SPase
3.1. Structure of SPase
3.2. Catalytic Mechanism of SPase
4. Heterologous Expression of SPase
5. Application and Molecular Modification of SPase
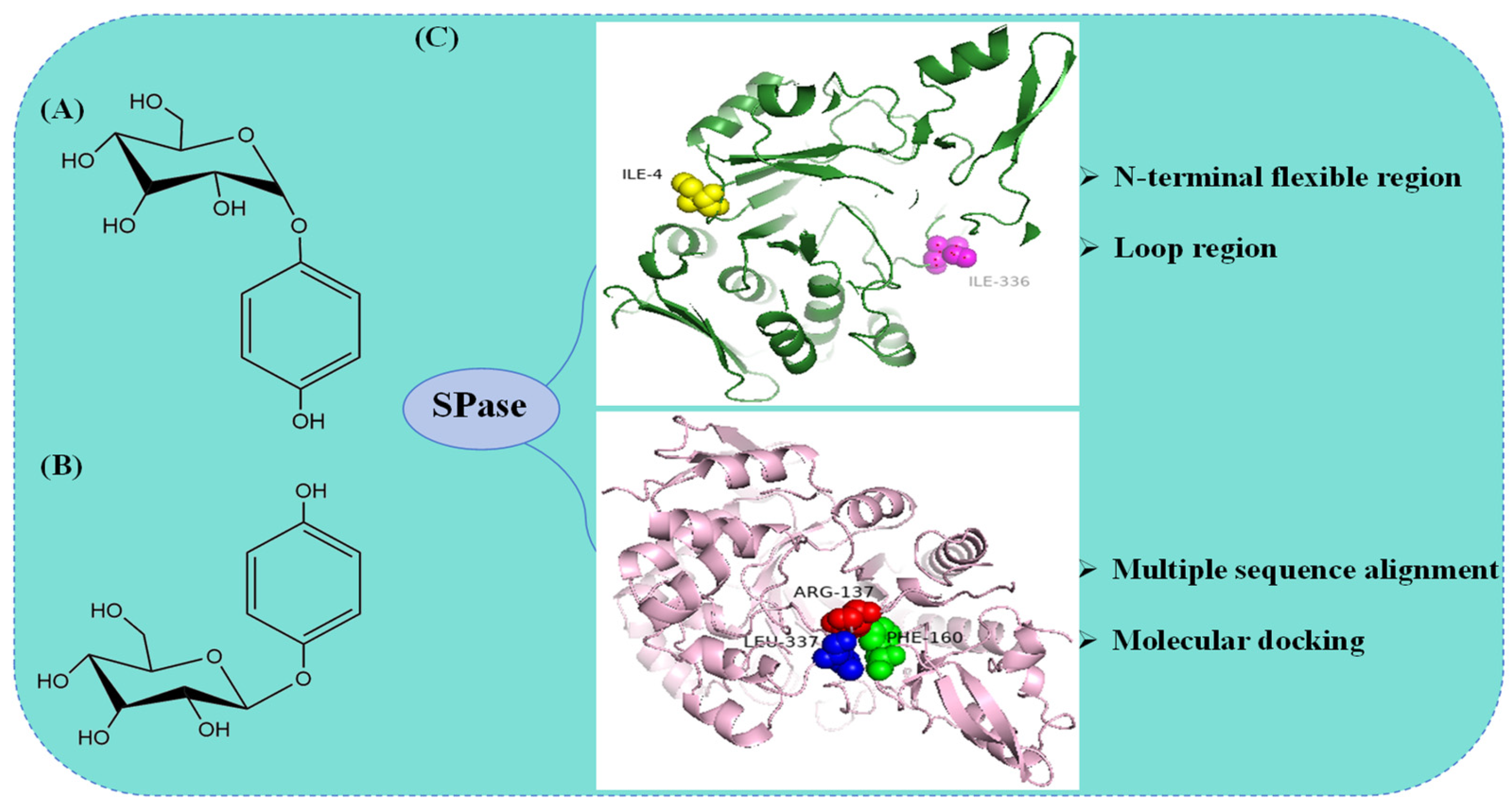
5.1. Molecular Modification of SPase in the Synthesis of α-Arbutin
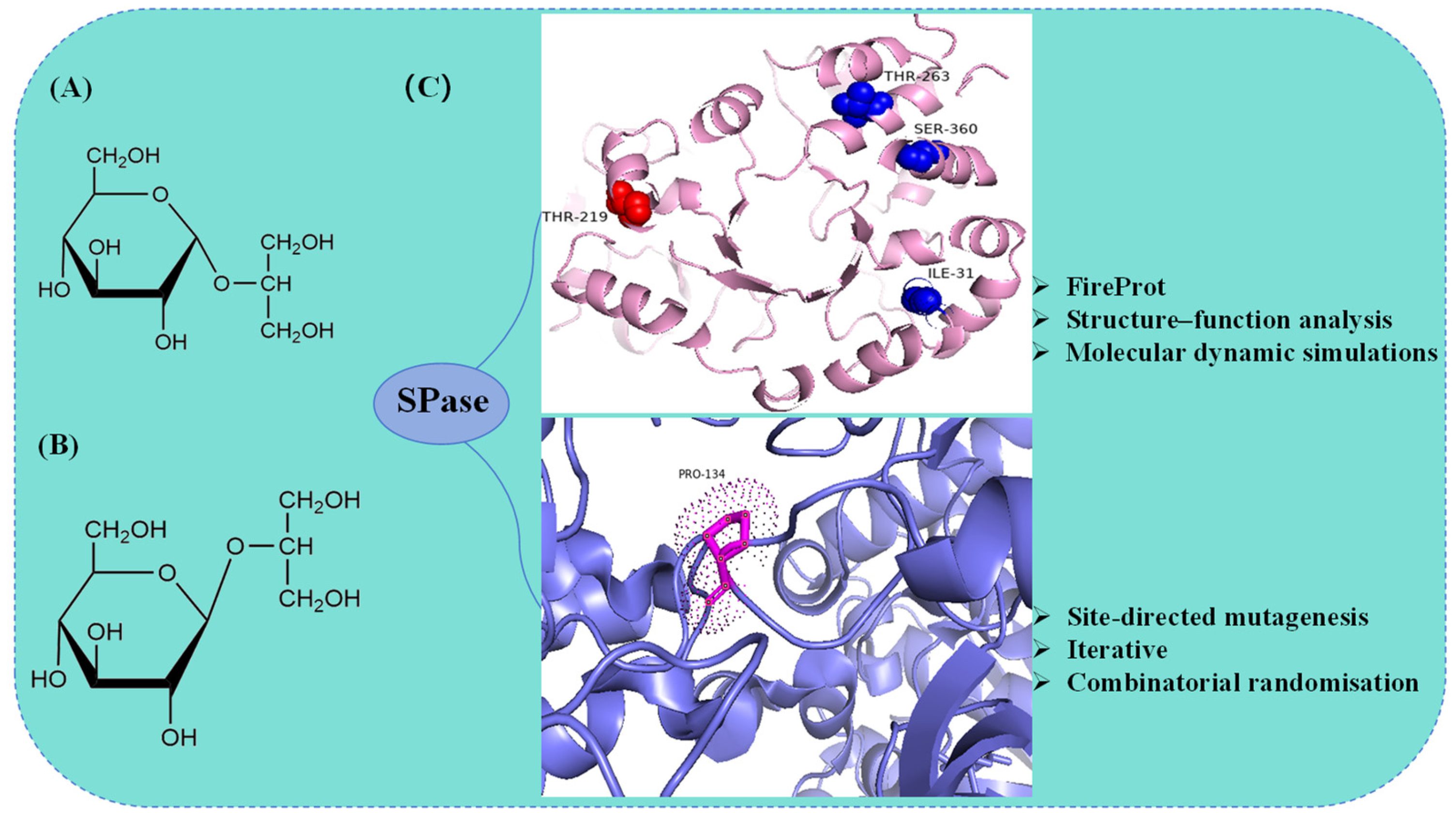
5.2. Molecular Modification of SPase in the Synthesis of α-GG
5.3. Molecular Modification of SPase in the Synthesis of AA-2G
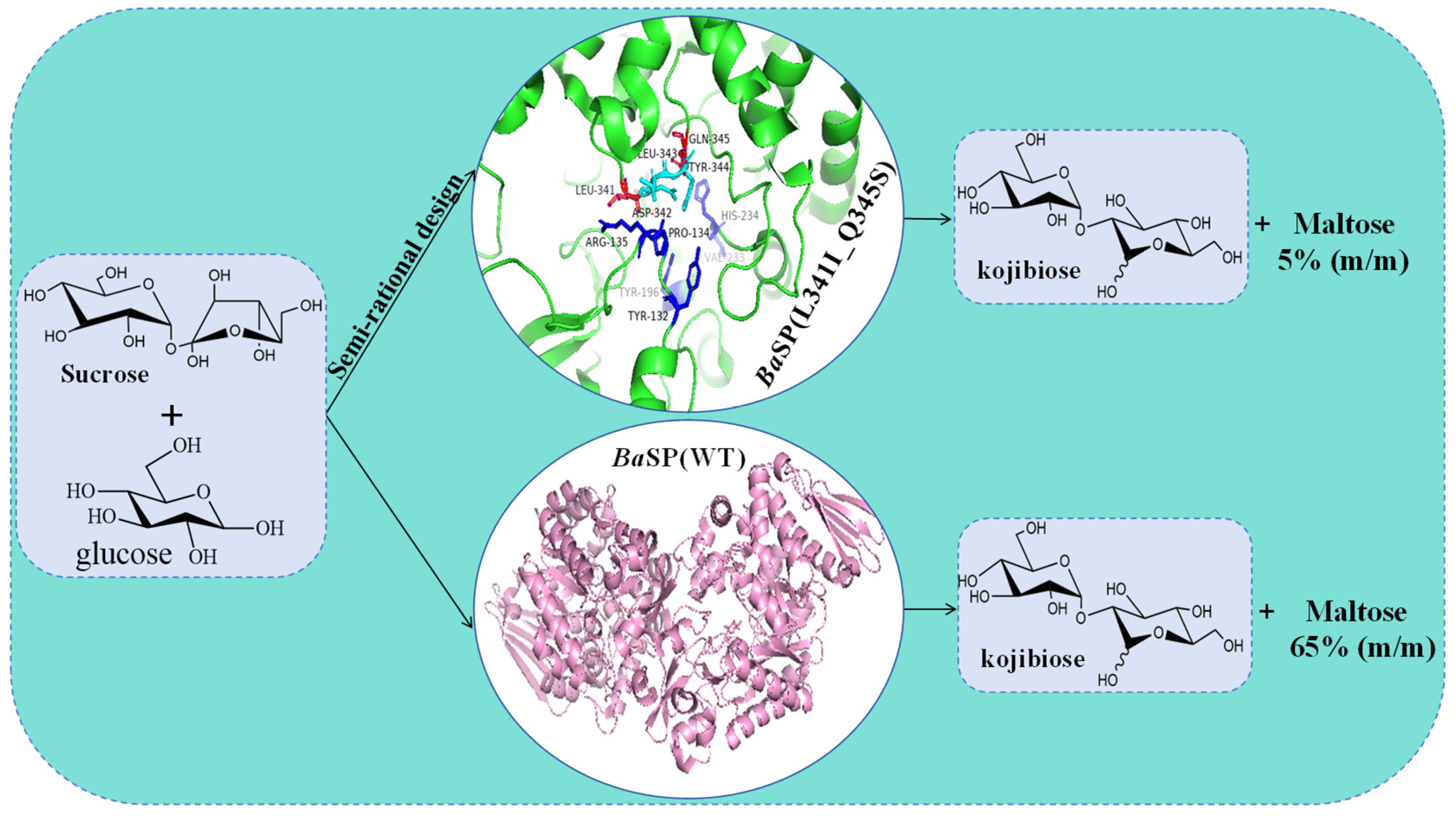
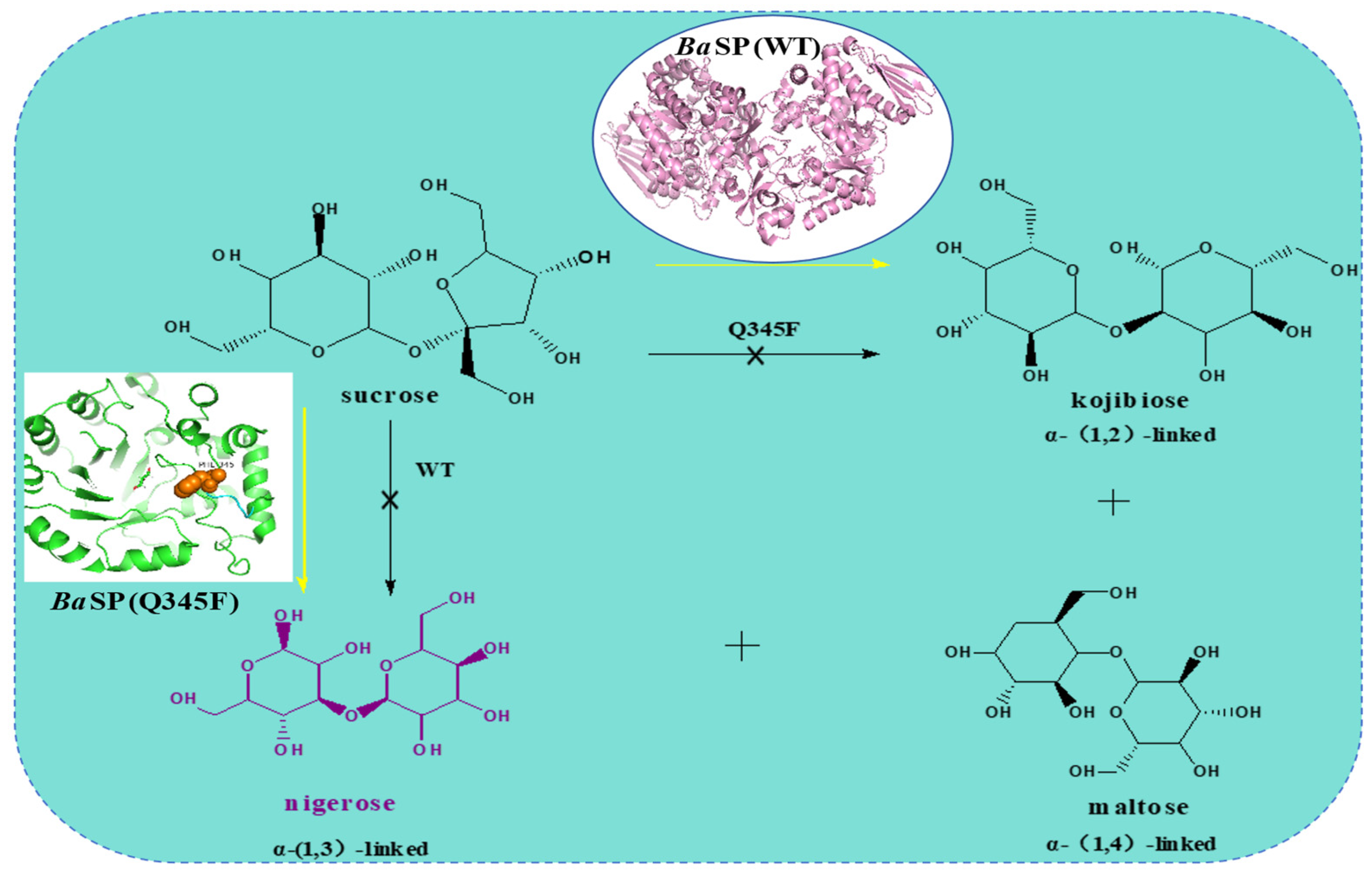
5.4. Molecular Modification of SPase in the Synthesis of Kojibiose
5.5. Molecular Modification of SPase in the Synthesis of Nigerose
5.6. Molecular Modification of SPase in the Synthesis of Polyphenol Glycosides
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bednarska, N.G.; Wren, B.W.; Willcocks, S.J. The importance of the glycosylation of antimicrobial peptides: Natural and synthetic approaches. Drug Discov. Today 2017, 22, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Christensen, H.M.; Oscarson, S.; Jensen, H.H. Common side reactions of the glycosyl donor in chemical glycosylation. Carbohydr. Res. 2015, 408, 51–95. [Google Scholar] [CrossRef] [PubMed]
- de Roode, B.M.; Franssen, M.C.R.; Padt, A.v.d.; Boom, R.M. Perspectives for the Industrial Enzymatic Production of Glycosides. Biotechnol. Prog. 2003, 19, 1391–1402. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- De Bruyn, F.; Maertens, J.; Beauprez, J.; Soetaert, W.; De Mey, M. Biotechnological advances in UDP-sugar based glycosylation of small molecules. Biotechnol. Adv. 2015, 33, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.J.; Williams, S.J. Carbohydrate-active enzymes: Sequences, shapes, contortions and cells. Biochem. Soc. Trans. 2016, 44, 79–87. [Google Scholar] [CrossRef]
- Goedl, C.; Sawangwan, T.; Wildberger, P.; Nidetzky, B. Sucrose phosphorylase: A powerful transglucosylation catalyst for synthesis of α-D-glucosides as industrial fine chemicals. Biocatal. Biotransform. 2010, 28, 10–21. [Google Scholar] [CrossRef]
- Koshland, D.E. Stereochemistry and the mechanism of enzymatic reactions. Biol. Rev. Camb. Philos. Soc. 1953, 28, 416–436. [Google Scholar] [CrossRef]
- Mirza, O.; Skov, L.K.; Sprogøe, D.; van den Broek, L.A.; Beldman, G.; Kastrup, J.S.; Gajhede, M. Structural rearrangements of sucrose phosphorylase from Bifidobacterium adolescentis during sucrose conversion. J. Biol. Chem. 2006, 281, 35576–35584. [Google Scholar] [CrossRef] [PubMed]
- Beerens, K.; De Winter, K.; Van de Wane, D.V.; Grootaer, C.; Kamiloglu, S.; Miclotte, L.; Van de Wiele, T.V.; Van Camp, J.; Dewettinck, K.; Desmet, T. Biocatalytic Synthesis of the Rare Sugar Kojibiose: Process Scale-Up and Application Testing. J. Agric. Food Chem. 2017, 65, 6030–6041. [Google Scholar] [CrossRef]
- Kraus, M.; Görl, J.; Timm, M.; Seibel, J. Synthesis of the rare disaccharide nigerose by structure-based design of a phosphorylase mutant with altered regioselectivity. Chem. Commun. 2016, 52, 4625–4627. [Google Scholar] [CrossRef]
- Wei, W.; Qi, D.; Diao, M.; Zhaoxin, L.; Lv, F.; Zhao, H. β-galactosidase-catalyzed synthesis of 3-O-β-D-galactopyranosyl-sn-glycerol: Optimization by response surface methodology. Biocatal. Biotransform. 2016, 34, 152–160. [Google Scholar] [CrossRef]
- Xu, H.; Yin, T.; Wei, B.; Su, M.; Liang, H. Turning waste into treasure: Biosynthesis of value-added 2-O-α-glucosyl glycerol and d-allulose from waste cane molasses through an in vitro synthetic biology platform. Bioresour. Technol. 2024, 391, 129982. [Google Scholar] [CrossRef]
- Kraus, M.; Grimm, C.; Seibel, J. Switching enzyme specificity from phosphate to resveratrol glucosylation. Chem. Commun. 2017, 53, 12181–12184. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.T.; Tian, Y.Q.; Zhang, W.L.; Zhang, T.; Guang, C.E.; Mu, W.M. Recent progress on biological production of α-arbutin. Appl. Microbiol. Biotechnol. 2018, 102, 8145–8152. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Alfonso, J.L.; Ubiparip, Z.; Jimenez-Ortega, E.; Poveda, A.; Alonso, C.; Coderch, L.; Jimenez-Barbero, J.; Sanz-Aparicio, J.; Ballesteros, A.O.; Desmet, T.; et al. Enzymatic Synthesis of Phloretin α-Glucosides Using a Sucrose Phosphorylase Mutant and its Effect on Solubility, Antioxidant Properties and Skin Absorption. Adv. Synth. Catal. 2021, 363, 3079–3089. [Google Scholar] [CrossRef]
- Demonceaux, M.; Goux, M.; Hendrickx, J.; Solleux, C.; Cadet, F.; Lormeau, É.; Offmann, B.; André-Miral, C. Regioselective glucosylation of (+)-catechin using a new variant of sucrose phosphorylase from Bifidobacterium adolescentis. Org. Biomol. Chem. 2023, 21, 2307–2311. [Google Scholar] [CrossRef] [PubMed]
- Bolivar, J.M.; Luley-Goedl, C.; Leitner, E.; Sawangwan, T.; Nidetzky, B.J. Production of glucosyl glycerol by immobilized sucrose phosphorylase: Options for enzyme fixation on a solid support and application in microscale flow format. J. Biotechnol. 2017, 257, 131–138. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, R.; Shi, Y.; Ma, W.; Liu, K.; Lu, Y.; Zhu, L.; Chen, X. Sucrose phosphorylase from Lactobacillus reuteri: Characterization and application of enzyme for production of 2-O-α-D-glucopyranosyl glycerol. Int. J. Biol. Macromol. 2022, 209, 376–384. [Google Scholar] [CrossRef]
- Yao, D.; Fan, J.; Han, R.; Xiao, J.; Li, Q.; Xu, G.; Dong, J.; Ni, Y. Enhancing soluble expression of sucrose phosphorylase in Escherichia coli by molecular chaperones. Protein Expr. Purif. 2020, 169, 105571. [Google Scholar] [CrossRef]
- Desmet, T.; Soetaert, W.; Bojarová, P.; Křen, V.; Dijkhuizen, L.; Eastwick-Field, V.; Schiller, A. Enzymatic Glycosylation of Small Molecules: Challenging Substrates Require Tailored Catalysts. Chem.—A Eur. J. 2012, 18, 10786–10801. [Google Scholar] [CrossRef]
- Hancock, S.M.; Vaughan, M.D.; Withers, S.G. Engineering of glycosidases and glycosyltransferases. Curr. Opin. Chem. Biol. 2006, 10, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Furubayashi, M.; Li, L.; Katabami, A.; Saito, K.; Umeno, D. Directed evolution of squalene synthase for dehydrosqualene biosynthesis. FEBS. Lett. 2014, 588, 3375–3381. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.K.; Wu, Z.; Arnold, F.H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 2019, 16, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Ye, Z.; Li, X.; Tian, Y.; Bai, Y.; Chen, L.; Qiu, C.; Xie, Z.; Jin, Z.; Svensson, B. Enzymatic preparation and potential applications of agar oligosaccharides: A review. Crit. Rev. Food Sci. Nutr. 2024, 64, 5818–5834. [Google Scholar] [CrossRef]
- Chaturvedi, S.S.; Bím, D.; Christov, C.Z.; Alexandrova, A.N. From random to rational: Improving enzyme design through electric fields, second coordination sphere interactions, and conformational dynamics. Chem. Sci. 2023, 14, 10997–11011. [Google Scholar] [CrossRef]
- Zhou, L.; Tao, C.; Shen, X.; Sun, X.; Wang, J.; Yuan, Q. Unlocking the potential of enzyme engineering via rational computational design strategies. Biotechnol. Adv. 2024, 73, 108376. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, R.; Zhang, W.; Xu, Y. Structure-based rational design of hydroxysteroid dehydrogenases for improving and diversifying steroid synthesis. Crit. Rev. Biotechnol. 2023, 43, 770–786. [Google Scholar] [CrossRef]
- Damborsky, J.; Brezovsky, J. Computational tools for designing and engineering enzymes. Curr. Opin. Chem. Biol. 2014, 19, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Kagan, B.; Latker, S.; Zfasman, E. Phosphorolysis of saccharose by cultures of Leuconostoc mesenteroides. Biokhimiya 1942, 7, 93–108. [Google Scholar]
- Zhou, Y.; Gan, T.; Jiang, R.; Chen, H.; Ma, Z.; Lu, Y.; Zhu, L.; Chen, X. Whole-cell catalytic synthesis of 2-O-α-glucopyranosyl-L-ascorbic acid by sucrose phosphorylase from Bifidobacterium breve via a batch-feeding strategy. Process Biochem. 2022, 112, 27–34. [Google Scholar] [CrossRef]
- Verhaeghe, T.; Aerts, D.; Diricks, M.; Soetaert, W.; Desmet, T. The quest for a thermostable sucrose phosphorylase reveals sucrose 6′-phosphate phosphorylase as a novel specificity. Appl. Microbiol. Biotechnol. 2014, 98, 7027–7037. [Google Scholar] [CrossRef] [PubMed]
- van den Broek, L.A.; van Boxtel, E.L.; Kievit, R.P.; Verhoef, R.; Beldman, G.; Voragen, A.G. Physico-chemical and transglucosylation properties of recombinant sucrose phosphorylase from Bifidobacterium adolescentis DSM20083. Appl. Microbiol. Biotechnol. 2004, 65, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kwon, T.; Lee, H.J.; Kim, K.H.; Chung, D.K.; Ji, G.E.; Byeon, E.S.; Lee, J.H. Cloning and expression of sucrose phosphorylase gene from Bifidobacterium longum in E. coli and characterization of the recombinant enzyme. Biotechnol. Lett. 2003, 25, 1211–1217. [Google Scholar] [CrossRef]
- Barrangou, R.; Altermann, E.; Hutkins, R.; Cano, R.; Klaenhammer, T.R. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 2003, 100, 8957–8962. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.; Mukasa, H.; Shimamura, A.; Ferretti, J. Streptococcus mutans gtfA gene specifies sucrose phosphorylase. Infect. Immun. 1988, 56, 2763–2765. [Google Scholar] [CrossRef]
- Silverstein, R.; Voet, J.; Reed, D.; Abeles, R.H. Purification and Mechanism of Action of Sucrose Phosphorylase. J. Biol. Chem. 1967, 242, 1338–1346. [Google Scholar] [CrossRef]
- Zhou, Y.; Ke, F.; Chen, L.; Lu, Y.; Zhu, L.; Chen, X. Enhancing regioselectivity of sucrose phosphorylase by loop engineering for glycosylation of L-ascorbic acid. Appl. Microbiol. Biotechnol. 2022, 106, 4575–4586. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; He, X.; Chen, L.; Cheng, Y.; Jia, H.; Yan, M.; Chen, K. Characterisation of a Thermobacillus sucrose phosphorylase and its utility in enzymatic synthesis of 2-O-α-D-glucopyranosyl-L-ascorbic acid. J. Biotechnol. 2019, 305, 27–34. [Google Scholar] [CrossRef]
- Lee, J.-H.; Yoon, S.-H.; Nam, S.-H.; Moon, Y.-H.; Moon, Y.-Y.; Kim, D. Molecular cloning of a gene encoding the sucrose phosphorylase from Leuconostoc mesenteroides B-1149 and the expression in Escherichia coli. Enzym. Microb. Technol. 2006, 39, 612–620. [Google Scholar] [CrossRef]
- Kitao, S.; Nakano, E. Cloning of the sucrose phosphorylase gene from Leuconostoc mesenteroides and its overexpression using a ‘sleeper’bacteriophage vector. J. Ferment. Bioeng. 1992, 73, 179–184. [Google Scholar] [CrossRef]
- Goux, M.; Demonceaux, M.; Hendrickx, J.; Solleux, C.; Lormeau, E.; Fredslund, F.; Tezé, D.; Offmann, B.; André-Miral, C. Sucrose phosphorylase from Alteromonas mediterranea: Structural insight into the regioselective α-glucosylation of (+)-catechin. Biochimie 2024, 221, 13–19. [Google Scholar] [CrossRef]
- Teixeira, J.S.; Abdi, R.; Su, M.S.-W.; Schwab, C.; Gänzle, M.G. Functional characterization of sucrose phosphorylase and scrR, a regulator of sucrose metabolism in Lactobacillus reuteri. Food Microbiol. 2013, 36, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Weimberg, R.; Doudoroff, M. Studies with three bacterial sucrose phosphorylases. J. Bacteriol. 1954, 68, 381–388. [Google Scholar] [CrossRef]
- Kawasaki, H.; Nakamura, N.; Ohmori, M.; Sakai, T. Cloning and expression in Escherichia coli of sucrose phosphorylase gene from Leuconostoc mesenteroides No. 165. Biosci. Biotechnol. Biochem. 1996, 60, 322–324. [Google Scholar] [CrossRef]
- Tauzin, A.S.; Bruel, L.; Laville, E.; Nicoletti, C.; Navarro, D.; Henrissat, B.; Perrier, J.; Potocki-Veronese, G.; Giardina, T.; Lafond, M. Sucrose 6F-phosphate phosphorylase: A novel insight in the human gut microbiome. Microb. Genomics 2019, 5, e000253. [Google Scholar] [CrossRef]
- Kasperowicz, A.; Stan-Glasek, K.; Guczynska, W.; Piknova, M.; Pristas, P.; Nigutova, K.; Javorsky, P.; Michalowski, T. Sucrose phosphorylase of the rumen bacterium Pseudobutyrivibrio ruminis strain A. J. Appl. Microbiol. 2009, 107, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Goedl, C.; Schwarz, A.; Minani, A.; Nidetzky, B. Recombinant sucrose phosphorylase from Leuconostoc mesenteroides: Characterization, kinetic studies of transglucosylation, and application of immobilised enzyme for production of α-D-glucose 1-phosphate. J. Biotechnol. 2007, 129, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, X.; Li, W.; Li, T.; Li, S.; Kitaoka, M. Expression and characterization of recombinant sucrose phosphorylase. Protein J. 2018, 37, 93–100. [Google Scholar]
- Choi, H.-C.; Seo, D.-H.; Jung, J.-H.; Ha, S.-J.; Kim, M.-J.; Lee, J.-H.; Chang, P.-S.; Kim, H.-Y.; Park, C.-S. Development of new assay for sucrose phosphorylase and its application to the characterization of Bifidobacterium longum SJ32 sucrose phosphorylase. Food Sci. Biotechnol. 2011, 20, 513–518. [Google Scholar] [CrossRef]
- Kullin, B.; Abratt, V.; Reid, S. A functional analysis of the Bifidobacterium longum cscA and scrP genes in sucrose utilization. Appl. Microbiol. Biotechnol. 2006, 72, 975–981. [Google Scholar] [CrossRef]
- Sprogøe, D.; van den Broek, L.A.; Mirza, O.; Kastrup, J.S.; Voragen, A.G.; Gajhede, M.; Skov, L.K. Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis. Biochemistry 2004, 43, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Koga, T.; Nakamura, K.; Shirokane, Y.; Mizusawa, K.; Kitao, S.; Kikuchi, M. Purification and some properties of sucrose phosphorylase from Leuconostoc mesenteroides. Agric. Biol. Chem. 1991, 55, 1805–1810. [Google Scholar] [CrossRef]
- Rye, C.S.; Withers, S.G. Glycosidase mechanisms. Curr. Opin. Chem. Biol. 2000, 4, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.; Nidetzky, B. The role of Asp-295 in the catalytic mechanism of Leuconostoc mesenteroides sucrose phosphorylase probed with site-directed mutagenesis. FEBS. Lett. 2007, 581, 1403–1408. [Google Scholar] [CrossRef]
- Kitao, S.; Yoshida, S.; Horiuchi, T.; Sekine, H.; Kusakabe, I. Formation of Kojibiose and Nigerose by Sucrose Phosphorylase. Biosci. Biotechnol. Biochem. 2014, 58, 790–791. [Google Scholar] [CrossRef]
- Luley-Goedl, C.; Sawangwan, T.; Brecker, L.; Wildberger, P.; Nidetzky, B. Regioselective O-glucosylation by sucrose phosphorylase: A promising route for functional diversification of a range of 1,2-propanediols. Carbohydr. Res. 2010, 345, 1736–1740. [Google Scholar] [CrossRef]
- He, X.; Li, Y.; Tao, Y.; Qi, X.; Ma, R.; Jia, H.; Yan, M.; Chen, K.; Hao, N. Discovering and efficiently promoting the extracellular secretory expression of Thermobacillus sp. ZCTH02-B1 sucrose phosphorylase in Escherichia coli. Int. J. Biol. Macromol. 2021, 173, 532–540. [Google Scholar] [CrossRef]
- Franceus, J.; Capra, N.; Desmet, T.; Thunnissen, A.-M.W. Structural comparison of a promiscuous and a highly specific sucrose 6F-phosphate phosphorylase. Int. J. Mol. Sci. 2019, 20, 3906. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.H.; Jung, M.W.; Lee, J.-H.; Kim, M.D.; Kim, K.H. Strategies for producing recombinant sucrose phosphorylase originating from Bifidobacterium longum in Escherichia coli JM109. Process Biochem. 2008, 43, 822–828. [Google Scholar] [CrossRef]
- Li, X.; Xia, Y.; Shen, W.; Yang, H.; Cao, Y.; Chen, X. Characterization of a sucrose phosphorylase from Leuconostoc mesenterides for the synthesis of α-arbutin. Chin. J. Biotechnol. 2020, 36, 1546–1555. [Google Scholar]
- He, H.H.; Lin, H.M.; Kou, L.D.; Qin, F.L.; Wei, Y.T.; Huang, R.B.; Du, L.Q. Characterization of recombinant sucrose phosphorylase from Leuconostoc mesenteroides ATCC 12291 and its molecular modification for improved transglucoside activity. Food Sci. 2019, 40, 122–129. [Google Scholar]
- Trindade, M.I.; Abratt, V.R.; Reid, S.J. Induction of sucrose utilization genes from Bifidobacterium lactis by sucrose and raffinose. Appl. Environ. Microbiol. 2003, 69, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zheng, P.; Hu, M.; Tao, Y. Inorganic phosphate self-sufficient whole-cell biocatalysts containing two co-expressed phosphorylases facilitate cellobiose production. J. Ind. Microbiol. Biotechnol. 2022, 49, 008. [Google Scholar] [CrossRef] [PubMed]
- Verhaeghe, T.; Diricks, M.; Aerts, D.; Soetaert, W.; Desmet, T. Mapping the acceptor site of sucrose phosphorylase from Bifidobacterium adolescentis by alanine scanning. J. Mol. Catal. B-Enzym. 2013, 96, 81–88. [Google Scholar] [CrossRef]
- Wang, M.; Wu, J.; Wu, D. Cloning and expression of the sucrose phosphorylase gene in Bacillus subtilis and synthesis of kojibiose using the recombinant enzyme. Microb. Cell. Fact. 2018, 17, 23. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann-Sommergruber, K.; Susani, M.; Ferreira, F.; Jertschin, P.; Ahorn, H.; Steiner, R.; Kraft, D.; Scheiner, O.; Breiteneder, H. High-level expression and purification of the major birch pollen allergen, Bet v 1. Protein Expr. Purif. 1997, 9, 33–39. [Google Scholar] [CrossRef]
- Shen, Y.; Lyu, X.-Q.; Lin, L.; Li, J.-H.; Du, G.-C.; Liu, L. Semi-rational design of sucrose phosphorylase and optimization of conditions for α-arbutin production. Food Ferment. Ind. 2020, 46, 1–9. [Google Scholar]
- Ao, J.; Pan, X.; Wang, Q.; Zhang, H.; Ren, K.; Jiang, A.; Zhang, X.; Rao, Z. Efficient whole-cell biotransformation for α-arbutin production through the engineering of sucrose phosphorylase combined with engineered cell modification. J. Agric. Food Chem. 2023, 71, 2438–2445. [Google Scholar] [CrossRef]
- Franceus, J.; Ubiparip, Z.; Beerens, K.; Desmet, T. Engineering of a Thermostable Biocatalyst for the Synthesis of 2-O-Glucosylglycerol. ChemBioChem 2021, 22, 2777–2782. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Li, X.; Yang, L.; Luo, X.; Shen, W.; Cao, Y.; Peplowski, L.; Chen, X. Development of thermostable sucrose phosphorylase by semi-rational design for efficient biosynthesis of alpha-D-glucosylglycerol. Appl. Microbiol. Biotechnol. 2021, 105, 7309–7319. [Google Scholar] [CrossRef] [PubMed]
- Kraus, M.; Grimm, C.; Seibel, J. Redesign of the Active Site of Sucrose Phosphorylase through a Clash-Induced Cascade of Loop Shifts. ChemBioChem 2016, 17, 33–36. [Google Scholar] [CrossRef]
- Verhaeghe, T.; De Winter, K.; Berland, M.; De Vreese, R.; D’Hooghe, M.; Offmann, B.; Desmet, T. Converting bulk sugars into prebiotics: Semi-rational design of a transglucosylase with controlled selectivity. Chem. Commun. 2016, 52, 3687–3689. [Google Scholar] [CrossRef] [PubMed]
- Kraus, M.; Grimm, C.; Seibel, J. Reversibility of a point mutation induced domain shift: Expanding the conformational space of a sucrose phosphorylase. Sci. Rep. 2018, 8, 10490. [Google Scholar] [CrossRef] [PubMed]
- Dirks-Hofmeister, M.E.; Verhaeghe, T.; De Winter, K.; Desmet, T. Creating space for large acceptors: Rational biocatalyst design for resveratrol glycosylation in an aqueous system. Angew. Chem. Int. Edit. 2015, 127, 9421–9424. [Google Scholar] [CrossRef]
- Seo, D.-H.; Jung, J.-H.; Lee, J.-E.; Jeon, E.-J.; Kim, W.; Park, C.-S. Biotechnological production of arbutins (α-and β-arbutins), skin-lightening agents, and their derivatives. Appl. Microbiol. Biotechnol. 2012, 95, 1417–1425. [Google Scholar] [CrossRef]
- Nikulin, A.V.; Okuneva, M.V.; Goryainov, S.V.; Potanina, O.G. Development and Validation of an HPLC-UV Method for Arbutin Determination in Bearberry Leaves. Pharm. Chem. J. 2019, 53, 736–740. [Google Scholar] [CrossRef]
- Liu, P.; Lindstedt, A.; Markkinen, N.; Sinkkonen, J.; Suomela, J.-P.; Yang, B. Characterization of Metabolite Profiles of Leaves of Bilberry (Vaccinium myrtillus L.) and Lingonberry (Vaccinium vitis-idaea L.). J. Agric. Food Chem. 2014, 62, 12015–12026. [Google Scholar] [CrossRef]
- Cho, J.-Y.; Park, K.Y.; Lee, K.H.; Lee, H.J.; Lee, S.-H.; Cho, J.A.; Kim, W.-S.; Shin, S.-C.; Park, K.-H.; Moon, J.-H. Recovery of arbutin in high purity from fruit peels of pear (Pyrus pyrifolia Nakai). Food Sci. Biotechnol. 2011, 20, 801–807. [Google Scholar] [CrossRef]
- Bang, S.H.; Han, S.J.; Kim, D.H. Hydrolysis of arbutin to hydroquinone by human skin bacteria and its effect on antioxidant activity. J. Cosmet. Dermatol. 2008, 7, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Nishimura, T.; Nomura, K.; Sugimoto, K.; Kuriki, T. Syntheses of arbutin-α-glycosides and a comparison of their inhibitory effects with those of α-arbutin and arbutin on human tyrosinase. Chem. Pharm. Bull. 2003, 51, 798–801. [Google Scholar] [CrossRef]
- Kitao, S.; Sekine, H. α-D-Glucosyl transfer to phenolic compounds by sucrose phosphorylase from Leuconostoc mesenteroides and production of α-arbutin. Biosci. Biotechnol. Biochem. 1994, 58, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Zheng, W.; Li, A.; Wu, H.; He, Y.; Tao, H.; Zhang, W.; Zheng, H.; Zhao, Z.; Li, S. Characterization of a novel sucrose phosphorylase from Paenibacillus elgii and its use in biosynthesis of α-arbutin. World J. Microbiol. Biotechnol. 2024, 40, 24. [Google Scholar] [CrossRef]
- Takenaka, F.; Uchiyama, H.; Imamura, T. Identification of α-D-glucosylglycerol in sake. Biosci. Biotechnol. Biochem. 2000, 64, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Luley-Goedl, C.; Sawangwan, T.; Mueller, M.; Schwarz, A.; Nidetzky, B. Biocatalytic process for production of α-glucosylglycerol using sucrose phosphorylase. Food Technol. Biotechnol. 2010, 48, 276–283. [Google Scholar]
- Sawangwan, T. Glucosylglycerol on performance of prebiotic potential. Funct. Foods Health Dis. 2015, 5, 427–436. [Google Scholar] [CrossRef]
- Sawangwan, T.; Goedl, C.; Nidetzky, B. Glucosylglycerol and glucosylglycerate as enzyme stabilizers. Biotechnol. J. 2010, 5, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, F.; Uchiyama, H. Synthesis of α-D-glucosylglycerol by α-glucosidase and some of its characteristics. Biosci. Biotechnol. Biochem. 2000, 64, 1821–1826. [Google Scholar] [CrossRef]
- Roder, A.; Hoffmann, E.; Hagemann, M.; Berg, G. Synthesis of the compatible solutes glucosylglycerol and trehalose by salt-stressed cells of Stenotrophomonas strains. FEMS Microbiol. Lett. 2005, 243, 219–226. [Google Scholar] [CrossRef]
- Goedl, C.; Sawangwan, T.; Mueller, M.; Schwarz, A.; Nidetzky, B. A high-yielding biocatalytic process for the production of 2-O-(α-D-glucopyranosyl)-sn-glycerol, a natural osmolyte and useful moisturizing ingredient. Angew. Chem. Int. Ed. Engl. 2008, 47, 10086–10089. [Google Scholar] [CrossRef]
- Kruschitz, A.; Peinsipp, L.; Pfeiffer, M.; Nidetzky, B. Continuous process technology for glucoside production from sucrose using a whole cell-derived solid catalyst of sucrose phosphorylase. Appl. Microbiol. Biotechnol. 2021, 105, 5383–5394. [Google Scholar] [CrossRef]
- Kwon, T.; Kim, C.T.; Lee, J.-H. Transglucosylation of ascorbic acid to ascorbic acid 2-glucoside by a recombinant sucrose phosphorylase from Bifidobacterium longum. Biotechnol. Lett. 2007, 29, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Wakamiya, H.; Suzuki, E.; Yamamoto, I.; Akiba, M.; Otsuka, M.; Arakawa, N. Vitamin C activity of 2-O-α-D-glucopyranosyl-L-ascorbic acid in guinea pigs. J. Nutr. Sci. Vitaminol. 1992, 38, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, I.; Tanaka, M.; Muto, N. Enhancement of in vitro antibody production of murine splenocytes by ascorbic acid 2-O-α-glucoside. Int. J. Immunopharmacol. 1993, 15, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Miyai, E.; Yanagida, M.; Akiyama, J.-I.; Yamamoto, I. Ascorbic acid 2-O-α-glucoside-induced redox modulation in human keratinocyte cell line, SCC: Mechanisms of photoprotective effect against ultraviolet light B. Biol. Pharm. Bull. 1997, 20, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, I.; Muto, N.; Murakami, K.; Suga, S.; Yamaguchi, H. L-Ascorbic acid α-glucoside formed by regioselective transglucosylation with rat intestinal and rice seed α-glucosidases: Its improved stability and structure determination. Chem. Pharm. Bull. 1990, 38, 3020–3023. [Google Scholar] [CrossRef]
- Kumano, Y.; Sakamoto, T.; Egawa, M.; Iwai, I.; Tanaka, M.; Yamamoto, I. In vitro and in vivo prolonged biological activities of novel vitamin C derivative, 2-O-α-D-glucopyranosyl-L-ascorbic acid (AA-2G), in cosmetic fields. J. Nutr. Sci. Vitaminol. 1998, 44, 345–359. [Google Scholar] [CrossRef]
- Gallie, D.R. Increasing vitamin C content in plant foods to improve their nutritional value—Successes and challenges. Nutrients 2013, 5, 3424–3446. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-B.; Nam, K.-C.; Lee, S.-J.; Lee, J.-H.; Inouye, K.; Park, K.-H. Antioxidative effects of glycosyl-ascorbic acids synthesized by maltogenic amylase to reduce lipid oxidation and volatiles production in cooked chicken meat. Biosci. Biotechnol. Biochem. 2004, 68, 36–43. [Google Scholar] [CrossRef]
- Muto, N.; Nakamura, T.; Yamamoto, I. Enzymatic formation of a nonreducing L-ascorbic acid α-glucoside: Purification and properties of α-glucosidases catalyzing site-specific transglucosylation from rat small intestine. J. Biochem. 1990, 107, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Gudiminchi, R.K.; Nidetzky, B. Walking a Fine Line with Sucrose Phosphorylase: Efficient Single-Step Biocatalytic Production of l-Ascorbic Acid 2-Glucoside from Sucrose. ChemBioChem 2017, 18, 1387–1390. [Google Scholar] [CrossRef]
- Mukai, K.; Tsusaki, K.; Kubota, M.; Fukuda, S.; Miyake, T. Process for producing 2-O-alpha-D-glucopyranosyl-L-ascorbic acid. E.U. Patent 1553186A1, 7 July 2003. [Google Scholar]
- Tanaka, M.; Muto, N.; Yamamoto, I. Characterization of Bacillus stearothermophilus cyclodextrin glucanotransferase in ascorbic acid 2-O-alpha-glucoside formation. Biochim. Biophysi. Acta 1991, 1078, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Yeon, M.J.; Choi, N.-S.; Chang, Y.-H.; Jung, M.Y.; Song, J.J.; Kim, J.S. Purification and characterization of a novel glucansucrase from Leuconostoc lactis EG001. Microbiol. Res. 2010, 165, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lv, X.; Chen, L.; Zhang, H.; Zhu, L.; Lu, Y.; Chen, X.J. Identification of Process-Related Impurities and Corresponding Control Strategy in Biocatalytic Production of 2-O-α-D-Glucopyranosyl-L-ascorbic Acid Using Sucrose Phosphorylase. J. Agric. Food Chem. 2022, 70, 5066–5076. [Google Scholar] [CrossRef] [PubMed]
- Desmet, T.; Soetaert, W.J.P.B. Broadening the synthetic potential of disaccharide phosphorylases through enzyme engineering. Process Biochem. 2012, 47, 11–17. [Google Scholar] [CrossRef]
- Hodoniczky, J.; Morris, C.A.; Rae, A.L. Oral and intestinal digestion of oligosaccharides as potential sweeteners: A systematic evaluation. Food Chem. 2012, 132, 1951–1958. [Google Scholar] [CrossRef]
- Laparra, J.M.; Díez-Municio, M.; Herrero, M.; Moreno, F.J. Structural differences of prebiotic oligosaccharides influence their capability to enhance iron absorption in deficient rats. Food Funct. 2014, 5, 2430–2437. [Google Scholar] [CrossRef]
- García-Cayuela, T.; Díez-Municio, M.; Herrero, M.; Martínez-Cuesta, M.C.; Peláez, C.; Requena, T.; Moreno, F.J. Selective fermentation of potential prebiotic lactose-derived oligosaccharides by probiotic bacteria. Int. Dairy J. 2014, 38, 11–15. [Google Scholar] [CrossRef]
- Miyazaki, T.; Matsumoto, Y.; Matsuda, K.; Kurakata, Y.; Matsuo, I.; Ito, Y.; Nishikawa, A.; Tonozuka, T. Heterologous expression and characterization of processing α-glucosidase I from Aspergillus brasiliensis ATCC 9642. Glycoconj. J. 2011, 28, 563–571. [Google Scholar] [CrossRef] [PubMed]
- de Melo, E.B.; da Silveira Gomes, A.; Carvalho, I. α-and β-Glucosidase inhibitors: Chemical structure and biological activity. Tetrahedron 2006, 62, 10277–10302. [Google Scholar] [CrossRef]
- Consonni, R.; Cagliani, L.R.; Cogliati, C. NMR characterization of saccharides in Italian honeys of different floral sources. J. Agric. Food Chem. 2012, 60, 4526–4534. [Google Scholar] [CrossRef] [PubMed]
- Briggs, D.E.; Stevens, R.; Young, T.W.; Hough, J.S. Malting and Brewing Science: Malt and Sweet Wort, 3rd ed.; Springer Science & Business Media: Birmingham, UK, 1981; Volume 1. [Google Scholar]
- Watanabe, T.; Aso, K. Studies on honeyII. Isolation of kojibiose, nigerose, maltose and isomaltose from honey. Tohoku J. Agric. Res. 1960, 11, 109–115. [Google Scholar]
- Aso, K.; Shibasaki, K.; Nakamura, M. Preparation of kojibiose. Nature 1958, 182, 1303–1304. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Furui, S.; Sasaki, K.; Suzuki, Y. Transglucosylation activities of multiple forms of α-glucosidase from spinach. Biosci. Biotechnol. Biochem. 2003, 67, 1160–1163. [Google Scholar] [CrossRef]
- Díez-Municio, M.; Montilla, A.; Moreno, F.J.; Herrero, M. A sustainable biotechnological process for the efficient synthesis of kojibiose. Green Chem. 2014, 16, 2219–2226. [Google Scholar] [CrossRef]
- Chaen, H.; Nishimoto, T.; Nakada, T.; Fukuda, S.; Kurimoto, M.; Tsujisaka, Y. Enzymatic synthesis of kojioligosaccharides using kojibiose phosphorylase. J. Biosci. Bioeng. 2001, 92, 177–182. [Google Scholar] [CrossRef]
- Chen, H.M.; Borjesson, U.; Engkvist, O.; Kogej, T.; Svensson, M.A.; Blomherg, N.; Weigelt, D.; Burrows, J.N.; Lange, T. ProSAR: A New Methodology for Combinatorial Library Design. J. Chem. Inf. Model. 2009, 49, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Barker, S.A.; Carrington, T.R. 721. Studies of Aspergillus niger. Part II. Transglycosidation by Aspergillus niger. Chem. Ind. 1953, 3588–3593. [Google Scholar] [CrossRef]
- Murosaki, S.; Muroyama, K.; Yamamoto, Y.; Kusaka, H.; Liu, T.; Yoshikai, Y. Immunopotentiating activity of nigerooligosaccharides for the T helper 1-like immune response in mice. Biosci. Biotech. Bioch. 1999, 63, 373–378. [Google Scholar] [CrossRef]
- Al-Otaibi, N.A.; Cassoli, J.S.; Martins-de-Souza, D.; Slater, N.K.; Rahmoune, H. Human Leukaemia cells (HL-60) proteomic and biological signatures underpinning cryo-damage are differentially modulated by novel cryo-additives. Gigascience 2019, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.; Takeuchi, K.; Shibata, K.; Yoshikawa, S.; Kitahata, S.; Okada, S.; Araya, S.; Nisizawa, T. Screening of Sugars Inhibitory against Sucrose-dependent Synthesis and Adherence of Insoluble Glucan and Acid Production by Streptococcus mutans. J. Dent. Res. 2016, 63, 1293–1297. [Google Scholar] [CrossRef] [PubMed]
- Nihira, T.; Nishimoto, M.; Nakai, H.; Ohtsubo, K.I.; Kitaoka, M. Characterization of Two α-1,3-Glucoside Phosphorylases from Clostridium phytofermentans. J. Appl. Glyosci. 2014, 61, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Franceus, J.; Dhaene, S.; Decadt, H.; Vandepitte, J.; Caroen, J.; Van der Eycken, J.; Beerens, K.; Desmet, T. Rational design of an improved transglucosylase for production of the rare sugar nigerose. Chem. Commun. 2019, 55, 4531–4533. [Google Scholar] [CrossRef]
- Kitao, S.; Ariga, T.; Matsudo, T.; Sekine, H. The syntheses of catechin-glucosides by transglycosylation with Leuconostoc mesenteroides sucrose phosphorylase. Biosci. Biotechnol. Biochem. 1993, 57, 2010–2015. [Google Scholar] [CrossRef]




| GenBank No. | Substrate | Product | Source Microorganisms | Ref. |
|---|---|---|---|---|
| AAO33821.1 | Suc; Glu | kojibiose | B. adolescentis DSM20083 | [34] |
| — | Suc | α-G-1-P | P. saccharophila ATCC9114 | [38] |
| AY795566.1 | Suc; Glu | kojibiose; nigerose | L. mesenteroides NRRLB-1149 | [41] |
| BAA14344.1 | Suc; Glu | kojibiose; nigerose | L. mesenteroides ATCC 12291 | [42] |
| AAA26937.1 | Suc | α-G-1-P | Streptococcus mutans | [37] |
| AGP78457.1 | Suc | glucosylated(+)-catechin | Alteromonas mediterranea | [43] |
| AGK37834.1 | Suc | levan | Lactobacillus reuteri LTH5448 | [44] |
| — | Suc | α-G-1-P | P. puterfaciens | [45] |
| 2207198A | Suc; Glu | kojibiose; nigerose | L. mesenteroides No. 165 | [46] |
| AIE44773.1 | Suc | α-G-1-P; α-glycosides | T. thermosaccharolyticum | [33] |
| CCA61958.1 | Suc; phosphate | α-G-1-P; Fru | Ruminococcus gnavus E1 | [47] |
| — | Suc | α-G-1-P | Pseudobutyrivibrio ruminis | [48] |
| — | Suc; phosphate | α-G-1-P | L. mesenteroides DSM 20193 | [49] |
| AAO84039.1 | Suc | Glu;Fru;by-products | B. longum SJ32 | [35] |
| BAF62433.1 | Suc | α-G-1-P | B. longum JCM1217 | [50] |
| AAO84039.1 | Suc; orthophosphate | Fru; α-G-1-P | B. longum SJ32 | [51] |
| — | Suc | Glu; Fru | B. longum NCIMB 702259T | [52] |
| GenBank No. | Source Microorganism | Expression Host | Expression Vector | Ref. |
|---|---|---|---|---|
| 2207198A | L. mesenteroides No. 165 | E. coli BL21(DE3) | pET-28 | [20] |
| A0A1Y3Q6Q6 | Thermobacillus sp. ZCTH02-B1 | E. coli BL21(DE3) | pRSFDute1; pET-5(+) | [40,59] |
| BAN03569.1 | Ilumatobacter coccineus YM16-304 | E. coli CGSC 8974 | pET21a; pCXP34h | [33,59] |
| ADL69407.1 | T. thermosaccharolyticum DSM 571 | E. coli BL21(DE3) | pET21a | [60] |
| WP_094046414.1 | T. thermosaccharolyticum | E. coli BL21(DE3) | pET20b+chaperone | [21] |
| BAF62433.1 | B. longum JCM1217 | E. coli BL21(DE3) | pET30 | [50] |
| AAO84039.1 | B. longum SJ32 | E. coli JM109; E. coli BL21 | p6×His119 | [35,51,61] |
| BAA14344.1 | L. mesenteroides ATCC 12291 | E. coli BL21(DE3); E. coli M15 | pET-28a; pQE30 | [62,63] |
| AAN01605.1 | B. lactis | E. coli JM109 | pSuc | [64] |
| AAO33821.1 | B. adolescentis | E. coli BW25113; E. coli BL21(DE3) | pYB1s; pCXP34h; | [65,66] |
| AAO33821.1 | B. adolescentis | Bacillus subtilis | pBSMuL3 | [67] |
| AGP78457.1 | Alteromonas mediterranea | E. coli BL21(DE3) | pET28b | [43] |
| AGK37834.1 | Lactobacillus reuteri LTH5448 | E. coli JM109 | pGEMTeasy | [44] |
| AY795566.1 | L. mesenteroides NRRLB-1149 | E. coli BL21(DE3) | pGEM-TEasy | [41] |
| 2207198A | L. mesenteroides No. 165 | E. coli JM109 | — | [46] |
| AAA26937.1 | Streptococcus mutans | E. coli RX4, | — | [37] |
| CCA61958.1 | Ruminococcus gnavus E1 | E. coli BL21(DE3) | pOPINE | [47] |
| — | L. mesenteroides DSM 20193 | E. coli DH10B | pQE30 | [49] |
| AAO33821.1 | B. adolescentis DSM20083 | E. coli XL1-Blue | — | [34] |
| BAF62433.1 | B. longum JCM1217 | E. coli BL21(DE3) | pET30 | [50] |
| — | B. longum NCIMB 702259 | E. coli | pSK | [52] |
| GenBank No. | Source Microorganism | Mutant Site | Substrate | Products | Ref. |
|---|---|---|---|---|---|
| AAN58596.1 | Streptococcus mutans UA159 | I4; I336 | HQ | α-arbutin | [69] |
| BAA14344.1 | L. mesenteroides ATCC 12291 | R137; F160; L337 | HQ | α-arbutin | [70] |
| AAO33821.1 | B. adolescentis DSM 20083 | P134 | Suc; glycerol | α-GG | [71] |
| BAA14344.1 | L. mesenteroides ATCC 12291 | T219; I31F/T219L/T263L/S360A | Suc; glycerol | α-GG | [72] |
| POO08785 | B. breve | P134; L341; L343 | Suc; l-ascorbic acid | AA-2G | [39] |
| AAO33821.1 | B. adolescentis DSM 20083 | Q345 | Resveratrol; Suc | Resveratrol-3-O-α-d-glucoside | [73] |
| AAO33821.1 | B. adolescentis DSM 20083 | Q345 | Suc | nigerose | [12] |
| AIE44773.1 | T. thermosaccharolyticum | R134; H344 | sucrose 6′-phosphate | α-G-1-P; Fructose 6-phosphate | [33] |
| AAO33821.1 | B. adolescentis DSM 20083 | Arg135; Leu343; Tyr344; Tyr132; Asp342; Pro134; Tyr196; His234; Gln345 | Phosphate; Fru; D-Arabitol and pyridoxine | α-G-1-P; Fru; glycoconjugates | [66] |
| AAO33821.1 | B. adolescentis DSM 20083 | L341; D342; L343; Y344; Q345; Y132; P134; R135; Y196; V233; H234 | Suc; Glu | kojibiose | [74] |
| AAO33821.1 | B. adolescentis DSM 20083 | Q345 | Suc; polyphenols | resveratrol-3-α-d-glucoside and nigerose | [75] |
| ADL69407.1 | T. thermosaccharolyticum | R134 | Suc; polyphenol | Phloretin α-Glucosides and resveratrol-3-α-d-glucoside | [17,76] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Zhu, L.; Zhou, Z.; Wang, D.; Yang, J.; Wang, S.; Lou, T. Advancements in the Heterologous Expression of Sucrose Phosphorylase and Its Molecular Modification for the Synthesis of Glycosylated Products. Molecules 2024, 29, 4086. https://doi.org/10.3390/molecules29174086
Zhang H, Zhu L, Zhou Z, Wang D, Yang J, Wang S, Lou T. Advancements in the Heterologous Expression of Sucrose Phosphorylase and Its Molecular Modification for the Synthesis of Glycosylated Products. Molecules. 2024; 29(17):4086. https://doi.org/10.3390/molecules29174086
Chicago/Turabian StyleZhang, Hongyu, Leting Zhu, Zixuan Zhou, Danyun Wang, Jinshan Yang, Suying Wang, and Tingting Lou. 2024. "Advancements in the Heterologous Expression of Sucrose Phosphorylase and Its Molecular Modification for the Synthesis of Glycosylated Products" Molecules 29, no. 17: 4086. https://doi.org/10.3390/molecules29174086






