Coumarins: Quorum Sensing and Biofilm Formation Inhibition
Abstract
1. Introduction
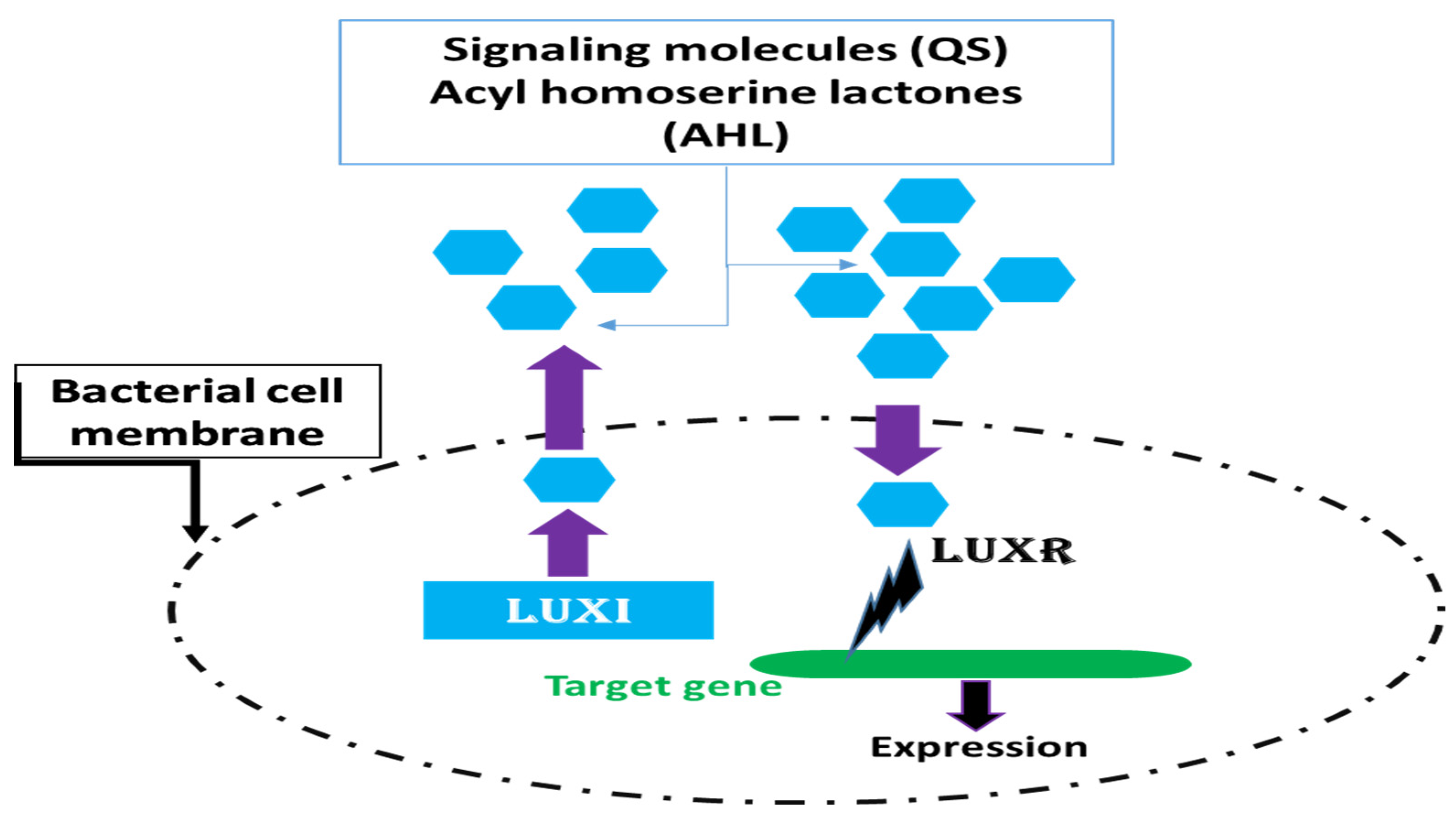
2. Quorum Sensing (QS)
2.1. Quorum Sensing in Gram-Negative Bacteria
2.2. Quorum Sensing in Gram-Positive Bacteria
2.3. Detection of Quorum Sensing Signaling Molecules
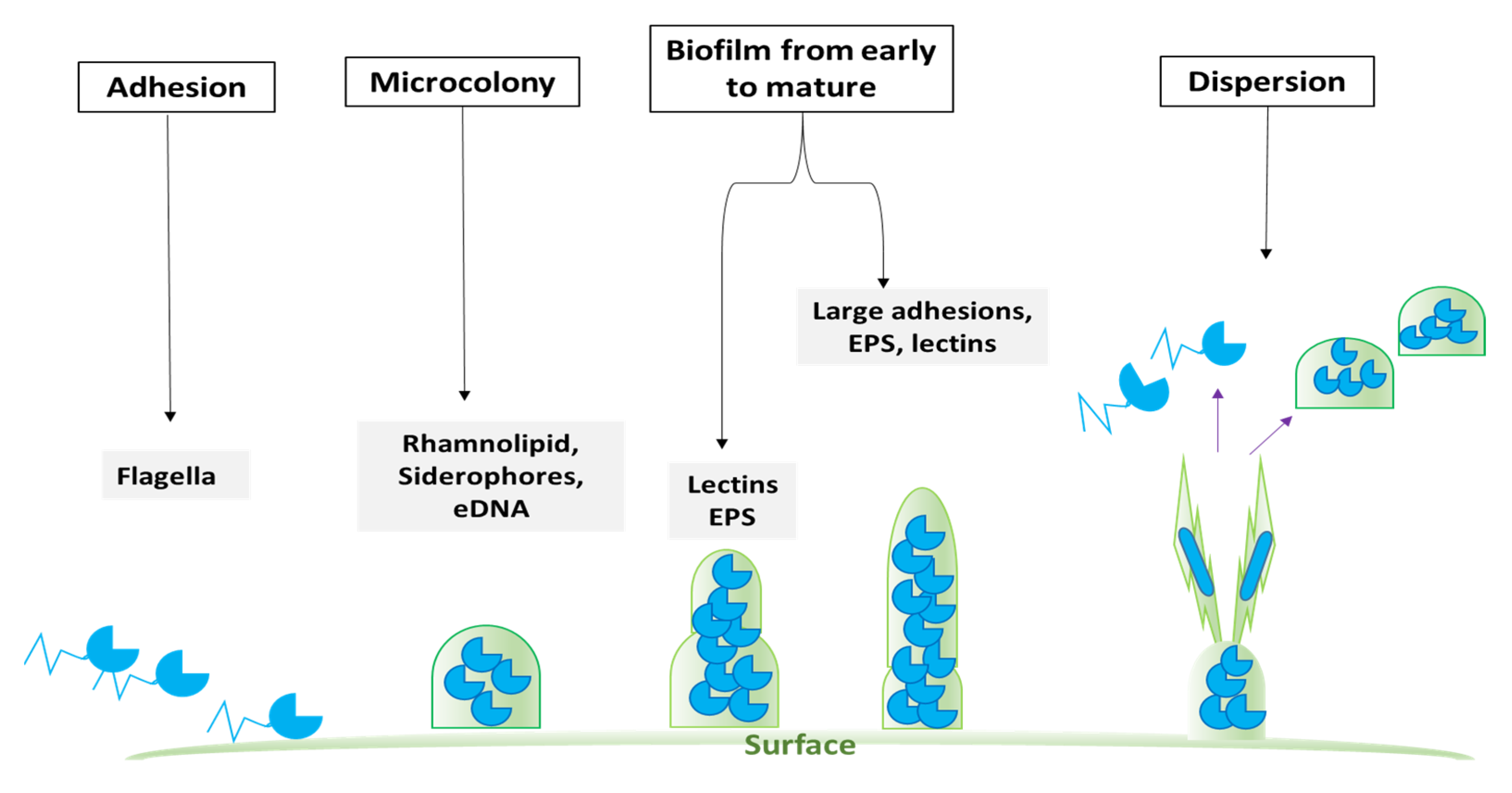
2.4. Qs as a Chemical Tool in Enhancing Biofilm Formation in Bacteria
3. QS Activity and Biofilm Formation Inhibition of Natural Coumarins
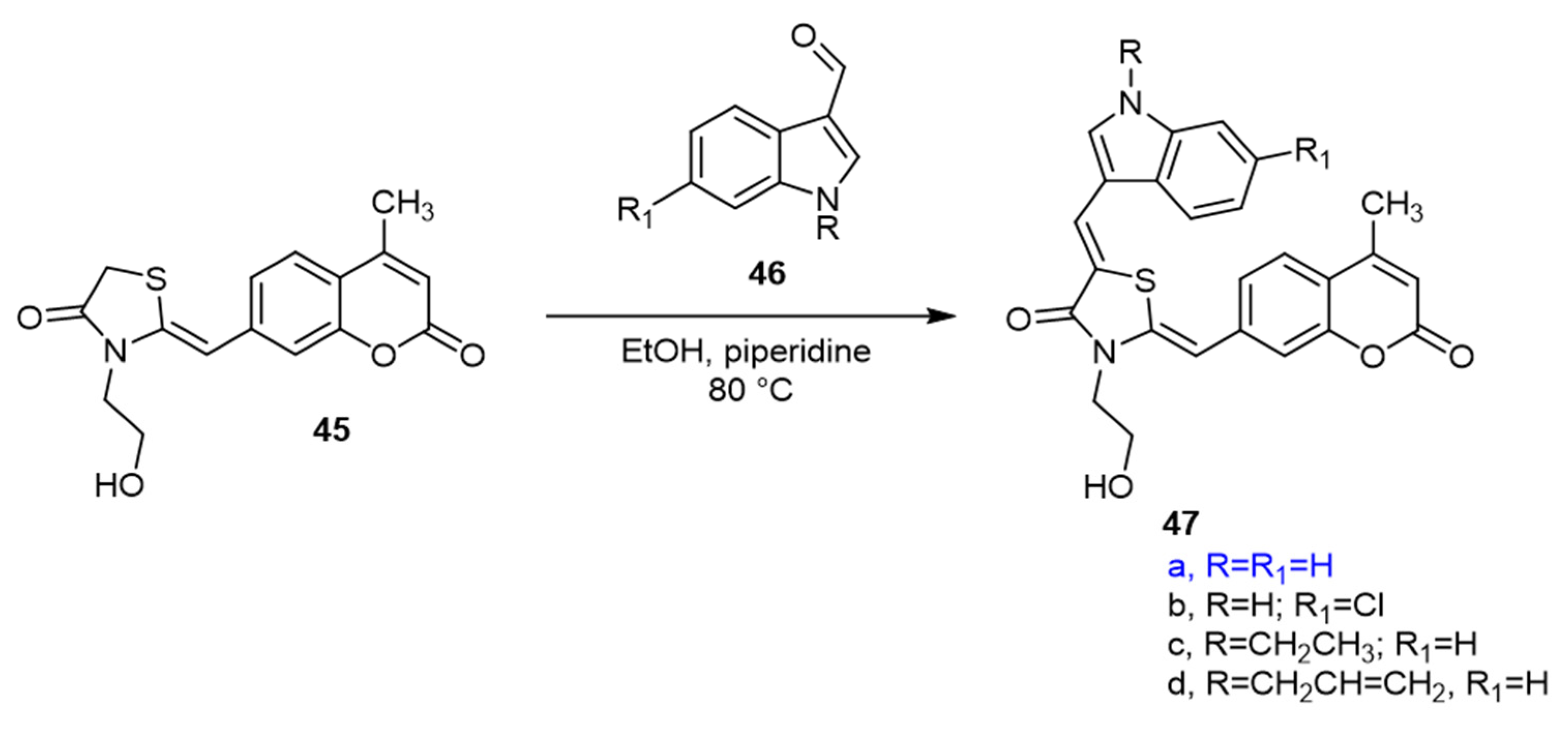
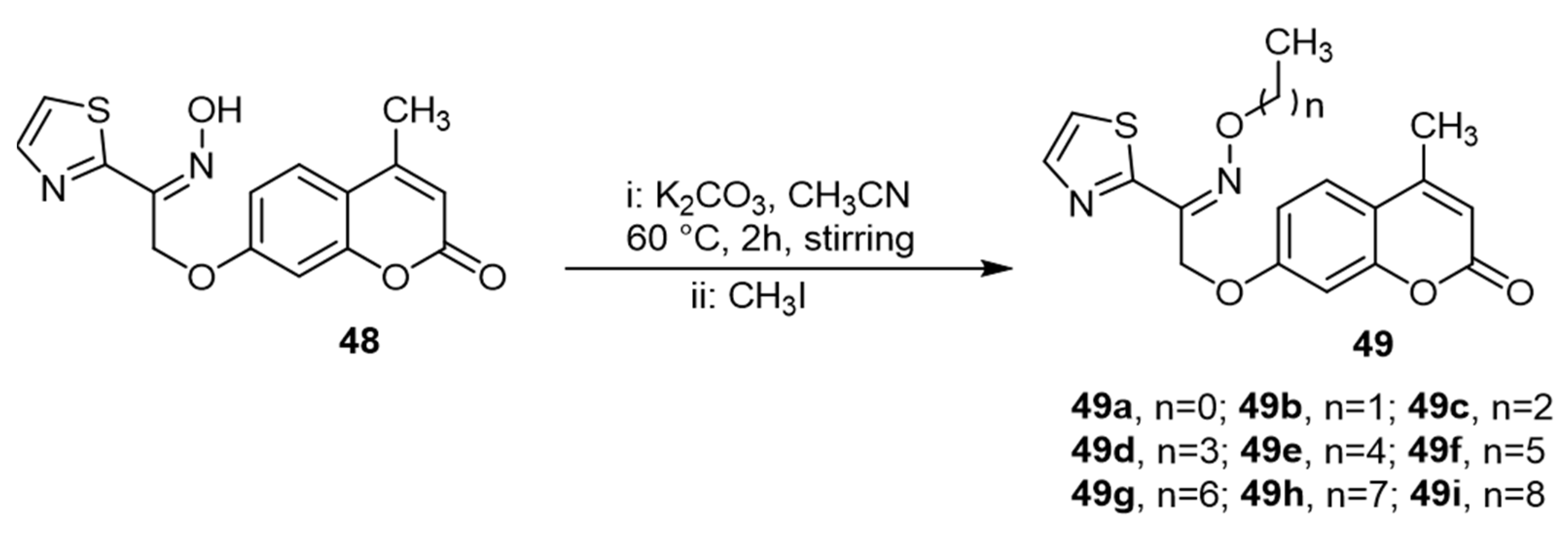
4. QS Activity and Biofilm Formation Inhibition of Synthetic Coumarin Compounds
5. Synergy between Antibiotics and Coumarin Compounds
Quorum Sensing and Antibiofilm Formation: Theoretical Studies of Coumarin Compounds
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pierson, L.S.; Maier, R.M.; Pepper, I.L. Microbial Communication: Bacteria–Bacteria and Bacteria–Host. In Environmental Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 335–346. ISBN 978-0-12-370519-8. [Google Scholar]
- Moreno-Gámez, S.; Hochberg, M.E.; Van Doorn, G.S. Quorum Sensing as a Mechanism to Harness the Wisdom of the Crowds. Nat. Commun. 2023, 14, 3415. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bian, Z.; Wang, Y. Biofilm Formation and Inhibition Mediated by Bacterial Quorum Sensing. Appl. Microbiol. Biotechnol. 2022, 106, 6365–6381. [Google Scholar] [CrossRef] [PubMed]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9, e02331-17. [Google Scholar] [CrossRef] [PubMed]
- Bouyahya, A.; Chamkhi, I.; Balahbib, A.; Rebezov, M.; Shariati, M.A.; Wilairatana, P.; Mubarak, M.S.; Benali, T.; El Omari, N. Mechanisms, Anti-Quorum-Sensing Actions, and Clinical Trials of Medicinal Plant Bioactive Compounds against Bacteria: A Comprehensive Review. Molecules 2022, 27, 1484. [Google Scholar] [CrossRef]
- Juszczuk-Kubiak, E. Molecular Aspects of the Functioning of Pathogenic Bacteria Biofilm Based on Quorum Sensing (QS) Signal-Response System and Innovative Non-Antibiotic Strategies for Their Elimination. Int. J. Mol. Sci. 2024, 25, 2655. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, Z.; Ding, T. Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms 2020, 8, 425. [Google Scholar] [CrossRef]
- Hense, B.A.; Schuster, M. Core Principles of Bacterial Autoinducer Systems. Microbiol. Mol. Biol. Rev. 2015, 79, 153–169. [Google Scholar] [CrossRef]
- De Kievit, T.R.; Iglewski, B.H. Bacterial Quorum Sensing in Pathogenic Relationships. Infect. Immun. 2000, 68, 4839–4849. [Google Scholar] [CrossRef]
- Gilbert, P.; Maira-Litran, T.; McBain, A.J.; Rickard, A.H.; Whyte, F.W. The Physiology and Collective Recalcitrance of Microbial Biofilm Communities. Adv. Microb. Physiol. 2002, 46, 202–256. [Google Scholar]
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The Role of Biofilms as Environmental Reservoirs of Antibiotic Resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef]
- Nugbemado, I.N.; Danquah, C.A.; Ofori, M. Hydroethanolic Stem Bark Extract of Treculia africana Decne (Moraceae) Shows Antimicrobial Resistance Modulatory Effects. South. Afr. J. Bot. 2022, 148, 546–551. [Google Scholar] [CrossRef]
- Mali, G.; Maji, S.; Chavan, K.A.; Shukla, M.; Kumar, M.; Bhattacharyya, S.; Erande, R.D. Effective Synthesis and Biological Evaluation of Functionalized 2,3-Dihydrofuro[3,2-c]Coumarins via an Imidazole-Catalyzed Green Multicomponent Approach. ACS Omega 2022, 7, 36028–36036. [Google Scholar] [CrossRef] [PubMed]
- El-Sawy, E.R.; Abdelwahab, A.B.; Kirsch, G. Synthetic Routes to Coumarin(Benzopyrone)-Fused Five-Membered Aromatic Heterocycles Built on the α-Pyrone Moiety. Part II: Five-Membered Aromatic Rings with Multi Heteroatoms. Molecules 2021, 26, 3409. [Google Scholar] [CrossRef] [PubMed]
- El-Sawy, E.R.; Abdelwahab, A.B.; Kirsch, G. Synthetic Routes to Coumarin(Benzopyrone)-Fused Five-Membered Aromatic Heterocycles Built on the α-Pyrone Moiety. Part 1: Five-Membered Aromatic Rings with One Heteroatom. Molecules 2021, 26, 483. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, J.; Liu, Y.; Zeng, Y.; Wu, G. A Review on Anti-Tumor Mechanisms of Coumarins. Front. Oncol. 2020, 10, 592853. [Google Scholar] [CrossRef]
- Rostom, B.; Karaky, R.; Kassab, I.; Sylla-Iyarreta Veitía, M. Coumarins Derivatives and Inflammation: Review of Their Effects on the Inflammatory Signaling Pathways. Eur. J. Pharmacol. 2022, 922, 174867. [Google Scholar] [CrossRef]
- Kasperkiewicz, K.; Ponczek, M.B.; Owczarek, J.; Guga, P.; Budzisz, E. Antagonists of Vitamin K—Popular Coumarin Drugs and New Synthetic and Natural Coumarin Derivatives. Molecules 2020, 25, 1465. [Google Scholar] [CrossRef]
- Mishra, S.; Pandey, A.; Manvati, S. Coumarin: An Emerging Antiviral Agent. Heliyon 2020, 6, e03217. [Google Scholar] [CrossRef]
- Hassan, M.Z.; Osman, H.; Ali, M.A.; Ahsan, M.J. Therapeutic Potential of Coumarins as Antiviral Agents. Eur. J. Med. Chem. 2016, 123, 236–255. [Google Scholar] [CrossRef]
- Patil, S.A.; Nesaragi, A.R.; Rodríguez-Berrios, R.R.; Hampton, S.M.; Bugarin, A.; Patil, S.A. Coumarin Triazoles as Potential Antimicrobial Agents. Antibiotics 2023, 12, 160. [Google Scholar] [CrossRef]
- Ranjan Sahoo, C.; Sahoo, J.; Mahapatra, M.; Lenka, D.; Kumar Sahu, P.; Dehury, B.; Nath Padhy, R.; Kumar Paidesetty, S. Coumarin Derivatives as Promising Antibacterial Agent(s). Arab. J. Chem. 2021, 14, 102922. [Google Scholar] [CrossRef]
- Xia, T.; Liu, Y.; Lu, Z.; Yu, H. Natural Coumarin Shows Toxicity to Spodoptera Litura by Inhibiting Detoxification Enzymes and Glycometabolism. Int. J. Mol. Sci. 2023, 24, 13177. [Google Scholar] [CrossRef]
- Qais, F.A.; Khan, M.S.; Ahmad, I.; Husain, F.M.; Khan, R.A.; Hassan, I.; Shahzad, S.A.; AlHarbi, W. Coumarin Exhibits Broad-Spectrum Antibiofilm and Antiquorum Sensing Activity against Gram-Negative Bacteria: In Vitro and In Silico Investigation. ACS Omega 2021, 6, 18823–18835. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Barranquero, J.A.; Reen, F.J.; McCarthy, R.R.; O’Gara, F. Deciphering the Role of Coumarin as a Novel Quorum Sensing Inhibitor Suppressing Virulence Phenotypes in Bacterial Pathogens. Appl. Microbiol. Biotechnol. 2015, 99, 3303–3316. [Google Scholar] [CrossRef]
- Reen, F.J.; Gutiérrez-Barranquero, J.A.; Parages, M.L.; O Gara, F. Coumarin: A Novel Player in Microbial Quorum Sensing and Biofilm Formation Inhibition. Appl. Microbiol. Biotechnol. 2018, 102, 2063–2073. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bassler, B.L. Bacterial Quorum Sensing in Complex and Dynamically Changing Environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Li, J.; Zhao, X. Effects of Quorum Sensing on the Biofilm Formation and Viable but Non-Culturable State. Food Res. Int. 2020, 137, 109742. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zou, Y.; Zheng, J.; Qiu, S.; Liu, L.; Wei, C. Quorum Sensing-Mediated Microbial Interactions: Mechanisms, Applications, Challenges and Perspectives. Microbiol. Res. 2023, 273, 127414. [Google Scholar] [CrossRef]
- Fan, Q.; Wang, H.; Mao, C.; Li, J.; Zhang, X.; Grenier, D.; Yi, L.; Wang, Y. Structure and Signal Regulation Mechanism of Interspecies and Interkingdom Quorum Sensing System Receptors. J. Agric. Food Chem. 2022, 70, 429–445. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Arciola, C.R.; Campoccia, D.; Speziale, P.; Montanaro, L.; Costerton, J.W. Biofilm Formation in Staphylococcus Implant Infections. A Review of Molecular Mechanisms and Implications for Biofilm-Resistant Materials. Biomaterials 2012, 33, 5967–5982. [Google Scholar] [CrossRef] [PubMed]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef] [PubMed]
- Brexó, R.P.; Sant’Ana, A. de S. Microbial Interactions during Sugar Cane Must Fermentation for Bioethanol Production: Does Quorum Sensing Play a Role? Crit. Rev. Biotechnol. 2018, 38, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Minervini, F.; Limitone, A. Cell-Cell Communication in Food Related Bacteria. Int. J. Food Microbiol. 2007, 120, 34–45. [Google Scholar] [CrossRef]
- Soto-Aceves, M.P.; Diggle, S.P.; Greenberg, E.P. Microbial Primer: LuxR-LuxI Quorum Sensing. Microbiology 2023, 169, 001343. [Google Scholar] [CrossRef]
- Borges, A.; Sousa, P.; Gaspar, A.; Vilar, S.; Borges, F.; Simões, M. Furvina Inhibits the 3-Oxo-C12-HSL-Based Quorum Sensing System of Pseudomonas aeruginosa and QS-Dependent Phenotypes. Biofouling 2017, 33, 156–168. [Google Scholar] [CrossRef]
- Nazzaro, F.; Fratianni, F.; Coppola, R. Quorum Sensing and Phytochemicals. Int. J. Mol. Sci. 2013, 14, 12607–12619. [Google Scholar] [CrossRef]
- Kang, B.H.; Xia, F.; Pop, R.; Dohi, T.; Socolovsky, M.; Altieri, D.C. Developmental Control of Apoptosis by the Immunophilin Aryl Hydrocarbon Receptor-Interacting Protein (AIP) Involves Mitochondrial Import of the Survivin Protein. J. Biol. Chem. 2011, 286, 16758–16767. [Google Scholar] [CrossRef]
- Liu, L.-P.; Huang, L.-H.; Ding, X.-T.; Yan, L.; Jia, S.-R.; Dai, Y.-J.; Xie, Y.-Y.; Zhong, C. Identification of Quorum-Sensing Molecules of N-Acyl-Homoserine Lactone in Gluconacetobacter Strains by Liquid Chromatography-Tandem Mass Spectrometry. Molecules 2019, 24, 2694. [Google Scholar] [CrossRef]
- Chorianopoulos, N.G.; Giaouris, E.D.; Kourkoutas, Y.; Nychas, G.-J.E. Inhibition of the Early Stage of Salmonella enterica Serovar Enteritidis Biofilm Development on Stainless Steel by Cell-Free Supernatant of a Hafnia Alvei Culture. Appl. Environ. Microbiol. 2010, 76, 2018–2022. [Google Scholar] [CrossRef]
- Shaw, P.D.; Ping, G.; Daly, S.L.; Cha, C.; Cronan, J.E.; Rinehart, K.L.; Farrand, S.K. Detecting and Characterizing N-Acyl-Homoserine Lactone Signal Molecules by Thin-Layer Chromatography. Proc. Natl. Acad. Sci. USA 1997, 94, 6036–6041. [Google Scholar] [CrossRef] [PubMed]
- Passos da Silva, D.; Schofield, M.C.; Parsek, M.R.; Tseng, B.S. An Update on the Sociomicrobiology of Quorum Sensing in Gram-Negative Biofilm Development. Pathogens 2017, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Rickard, A.H.; Palmer, R.J.; Blehert, D.S.; Campagna, S.R.; Semmelhack, M.F.; Egland, P.G.; Bassler, B.L.; Kolenbrander, P.E. Autoinducer 2: A Concentration-Dependent Signal for Mutualistic Bacterial Biofilm Growth. Mol. Microbiol. 2006, 60, 1446–1456. [Google Scholar] [CrossRef]
- Silagyi, K.; Kim, S.-H.; Lo, Y.M.; Wei, C. Production of Biofilm and Quorum Sensing by Escherichia coli O157:H7 and Its Transfer from Contact Surfaces to Meat, Poultry, Ready-to-Eat Deli, and Produce Products. Food Microbiol. 2009, 26, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Machado, I.; Silva, L.R.; Giaouris, E.D.; Melo, L.F.; Simões, M. Quorum Sensing in Food Spoilage and Natural-Based Strategies for Its Inhibition. Food Res. Int. 2020, 127, 108754. [Google Scholar] [CrossRef] [PubMed]
- Gohil, N.; Ramírez-García, R.; Panchasara, H.; Patel, S.; Bhattacharjee, G.; Singh, V. Book Review: Quorum Sensing vs. Quorum Quenching: A Battle With No End in Sight. Front. Cell. Infect. Microbiol. 2018, 8, 106. [Google Scholar] [CrossRef]
- Smith, R.S.; Iglewski, B.H.P. Aeruginosa Quorum-Sensing Systems and Virulence. Curr. Opin. Microbiol. 2003, 6, 56–60. [Google Scholar] [CrossRef]
- Tielker, D.; Hacker, S.; Loris, R.; Strathmann, M.; Wingender, J.; Wilhelm, S.; Rosenau, F.; Jaeger, K.-E. Pseudomonas aeruginosa Lectin LecB Is Located in the Outer Membrane and Is Involved in Biofilm Formation. Microbiology 2005, 151, 1313–1323. [Google Scholar] [CrossRef]
- Winzer, K.; Hardie, K.R.; Williams, P. LuxS and Autoinducer-2: Their Contribution to Quorum Sensing and Metabolism in Bacteria. Adv. Appl. Microbiol. 2003, 53, 291–396. [Google Scholar] [CrossRef]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel Is a Cationic Exopolysaccharide That Cross-Links Extracellular DNA in the Pseudomonas aeruginosa Biofilm Matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The Connections between Quorum Sensing and Biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Cirioni, O.; Mocchegiani, F.; Cacciatore, I.; Vecchiet, J.; Silvestri, C.; Baldassarre, L.; Ucciferri, C.; Orsetti, E.; Castelli, P.; Provinciali, M.; et al. Quorum Sensing Inhibitor FS3-Coated Vascular Graft Enhances Daptomycin Efficacy in a Rat Model of Staphylococcal Infection. Peptides 2013, 40, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Zhao, L.; Sun, B. LuxS/AI-2 System Is Involved in Antibiotic Susceptibility and Autolysis in Staphylococcus aureus NCTC 8325. Int. J. Antimicrob. Agents 2013, 41, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Ingremeau, F.; Zhao, A.; Bassler, B.L.; Stone, H.A. Local and Global Consequences of Flow on Bacterial Quorum Sensing. Nat. Microbiol. 2016, 1, 15005. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-G.; Cho, H.S.; Ryu, S.Y.; Cho, M.H.; Lee, J. Coumarins Reduce Biofilm Formation and the Virulence of Escherichia coli O157:H7. Phytomedicine 2014, 21, 1037–1042. [Google Scholar] [CrossRef]
- Thakur, S.; Ray, S.; Jhunjhunwala, S.; Nandi, D. Insights into Coumarin-Mediated Inhibition of Biofilm Formation in Salmonella Typhimurium: Biofouling. Biofouling 2020, 36, 479–491. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Y.; Li, S.; Ding, W. Resveratrol and Coumarin: Novel Agricultural Antibacterial Agent against Ralstonia solanacearum In Vitro and In Vivo. Molecules 2016, 21, 1501. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wang, J.L.; Chu, M.P.; Jia, C. Activity of Coumarin against Candida albicans Biofilms. J. De. Mycol. Méd. 2019, 29, 28–34. [Google Scholar] [CrossRef]
- Marquis, A.; Genovese, S.; Epifano, F.; Grenier, D. The Plant Coumarins Auraptene and Lacinartin as Potential Multifunctional Therapeutic Agents for Treating Periodontal Disease. BMC Complement. Altern. Med. 2012, 12, 80. [Google Scholar] [CrossRef]
- Monte, J.; Abreu, A.C.; Borges, A.; Simões, L.C.; Simões, M. Antimicrobial Activity of Selected Phytochemicals against Escherichia coli and Staphylococcus aureus and Their Biofilms. Pathogens 2014, 3, 473–498. [Google Scholar] [CrossRef]
- Swetha, T.K.; Pooranachithra, M.; Subramenium, G.A.; Divya, V.; Balamurugan, K.; Pandian, S.K. Umbelliferone Impedes Biofilm Formation and Virulence of Methicillin-Resistant Staphylococcus epidermidis via Impairment of Initial Attachment and Intercellular Adhesion. Front. Cell. Infect. Microbiol. 2019, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Kasthuri, T.; Barath, S.; Nandhakumar, M.; Karutha Pandian, S. Proteomic Profiling Spotlights the Molecular Targets and the Impact of the Natural Antivirulent Umbelliferone on Stress Response, Virulence Factors, and the Quorum Sensing Network of Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2022, 12, 998540. [Google Scholar] [CrossRef] [PubMed]
- Sushmitha, T.J.; Rajeev, M.; Kathirkaman, V.; Shivam, S.; Rao, T.S.; Pandian, S.K. 3-Hydroxy Coumarin Demonstrates Anti-Biofilm and Anti-Hyphal Efficacy against Candida albicans via Inhibition of Cell-Adhesion, Morphogenesis, and Virulent Genes Regulation. Sci. Rep. 2023, 13, 11687. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wei, Z.; Li, S.; Xiao, R.; Xu, Q.; Ran, Y.; Ding, W. Plant Secondary Metabolite, Daphnetin Reduces Extracellular Polysaccharides Production and Virulence Factors of Ralstonia solanacearum. Pestic. Biochem. Physiol. 2021, 179, 104948. [Google Scholar] [CrossRef]
- Yang, L.; Ding, W.; Xu, Y.; Wu, D.; Li, S.; Chen, J.; Guo, B. New Insights into the Antibacterial Activity of Hydroxycoumarins against Ralstonia solanacearum. Molecules 2016, 21, 468. [Google Scholar] [CrossRef]
- Lemos, A.S.O.; Florêncio, J.R.; Pinto, N.C.C.; Campos, L.M.; Silva, T.P.; Grazul, R.M.; Pinto, P.F.; Tavares, G.D.; Scio, E.; Apolônio, A.C.M.; et al. Antifungal Activity of the Natural Coumarin Scopoletin Against Planktonic Cells and Biofilms From a Multidrug-Resistant Candida tropicalis Strain. Front. Microbiol. 2020, 11, 1525. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; He, X.; Xiao, Q.; Han, S.; Jia, Z.; Li, S.; Ding, W. Discovery of a Novel Plant-Derived Agent against Ralstonia solanacearum by Targeting the Bacterial Division Protein FtsZ. Pestic. Biochem. Physiol. 2021, 177, 104892. [Google Scholar] [CrossRef]
- Bajire, S.K.; Jain, S.; Johnson, R.P.; Shastry, R.P. 6-Methylcoumarin Attenuates Quorum Sensing and Biofilm Formation in Pseudomonas aeruginosa PAO1 and Its Applications on Solid Surface Coatings with Polyurethane. Appl. Microbiol. Biotechnol. 2021, 105, 8647–8661. [Google Scholar] [CrossRef]
- da Cunha, M.G.; de Cássia Orlandi Sardi, J.; Freires, I.A.; Franchin, M.; Rosalen, P.L. Antimicrobial, Anti-Adherence and Antibiofilm Activity against Staphylococcus aureus of a 4-Phenyl Coumarin Derivative Isolated from Brazilian Geopropolis. Microb. Pathog. 2020, 139, 103855. [Google Scholar] [CrossRef]
- Hou, H.M.; Jiang, F.; Zhang, G.L.; Wang, J.Y.; Zhu, Y.H.; Liu, X.Y. Inhibition of Hafnia Alvei H4 Biofilm Formation by the Food Additive Dihydrocoumarin. J. Food Prot. 2017, 80, 842–847. [Google Scholar] [CrossRef]
- Clatworthy, A.E.; Pierson, E.; Hung, D.T. Targeting Virulence: A New Paradigm for Antimicrobial Therapy. Nat. Chem. Biol. 2007, 3, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Girennavar, B.; Cepeda, M.L.; Soni, K.A.; Vikram, A.; Jesudhasan, P.; Jayaprakasha, G.K.; Pillai, S.D.; Patil, B.S. Grapefruit Juice and Its Furocoumarins Inhibits Autoinducer Signaling and Biofilm Formation in Bacteria. Int. J. Food Microbiol. 2008, 125, 204–208. [Google Scholar] [CrossRef]
- Luciardi, M.C.; Blázquez, M.A.; Alberto, M.R.; Cartagena, E.; Arena, M.E. Grapefruit Essential Oils Inhibit Quorum Sensing of Pseudomonas aeruginosa. Food Sci. Technol. Int. 2020, 26, 231–241. [Google Scholar] [CrossRef] [PubMed]
- D’Almeida, R.E.; Molina, R.D.I.; Viola, C.M.; Luciardi, M.C.; Nieto Peñalver, C.; Bardón, A.; Arena, M.E. Comparison of Seven Structurally Related Coumarins on the Inhibition of Quorum Sensing of Pseudomonas aeruginosa and Chromobacterium Violaceum. Bioorg Chem. 2017, 73, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Alain, K.Y.; Tamfu, A.N.; Kucukaydin, S.; Ceylan, O.; Cokou Pascal, A.D.; Félicien, A.; Koko Dominique, S.C.; Duru, M.E.; Dinica, R.M. Phenolic Profiles, Antioxidant, Antiquorum Sensing, Antibiofilm and Enzyme Inhibitory Activities of Selected Acacia Species Collected from Benin. LWT 2022, 171, 114162. [Google Scholar] [CrossRef]
- Qu, D.; Hou, Z.; Li, J.; Luo, L.; Su, S.; Ye, Z.; Bai, Y.; Zhang, X.; Chen, G.; Li, Z.; et al. A New Coumarin Compound DCH Combats Methicillin-Resistant Staphylococcus aureus Biofilm by Targeting Arginine Repressor. Sci. Adv. 2020, 6, eaay9597. [Google Scholar] [CrossRef]
- Das, T.; Das, M.C.; Das, A.; Bhowmik, S.; Sandhu, P.; Akhter, Y.; Bhattacharjee, S.; De, U.C. Modulation of S. Aureus and P. Aeruginosa Biofilm: An in Vitro Study with New Coumarin Derivatives. World J. Microbiol. Biotechnol. 2018, 34, 170. [Google Scholar] [CrossRef]
- Sullivan, M.; Kia, A.F.-A.; Long, M.; Walsh, M.; Kavanagh, K.; McClean, S.; Creaven, B.S. Isolation and Characterisation of Silver(I) Complexes of Substituted Coumarin-4-Carboxylates Which Are Effective against Pseudomonas aeruginosa Biofilms. Polyhedron 2014, 67, 549–559. [Google Scholar] [CrossRef]
- Emmadi, N.R.; Atmakur, K.; Bingi, C.; Godumagadda, N.R.; Chityal, G.K.; Nanubolu, J.B. Regioselective Synthesis of 3-Benzyl Substituted Pyrimidino Chromen-2-Ones and Evaluation of Anti-Microbial and Anti-Biofilm Activities. Bioorg. Med. Chem. Lett. 2014, 24, 485–489. [Google Scholar] [CrossRef]
- Alnufaie, R.; Raj KC, H.; Alsup, N.; Whitt, J.; Andrew Chambers, S.; Gilmore, D.; Alam, M.A. Synthesis and Antimicrobial Studies of Coumarin-Substituted Pyrazole Derivatives as Potent Anti-Staphylococcus aureus Agents. Molecules 2020, 25, 2758. [Google Scholar] [CrossRef]
- Yang, X.-C.; Zeng, C.-M.; Avula, S.R.; Peng, X.-M.; Geng, R.-X.; Zhou, C.-H. Novel Coumarin Aminophosphonates as Potential Multitargeting Antibacterial Agents against Staphylococcus aureus. Eur. J. Med. Chem. 2023, 245, 114891. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bhasme, P.; Reddy, D.S.; Liu, D.; Yu, Z.; Zhao, T.; Zheng, Y.; Kumar, A.; Yu, H.; Ma, L.Z. Dual Functions: A Coumarin-Chalcone Conjugate Inhibits Cyclic-Di-GMP and Quorum-Sensing Signaling to Reduce Biofilm Formation and Virulence of Pathogens. mLife 2023, 2, 283–294. [Google Scholar] [CrossRef]
- Yang, X.-C.; Zhang, P.-L.; Kumar, K.V.; Li, S.; Geng, R.-X.; Zhou, C.-H. Discovery of Unique Thiazolidinone-Conjugated Coumarins as Novel Broad Spectrum Antibacterial Agents. Eur. J. Med. Chem. 2022, 232, 114192. [Google Scholar] [CrossRef]
- Yang, X.-C.; Hu, C.-F.; Zhang, P.-L.; Li, S.; Hu, C.-S.; Geng, R.-X.; Zhou, C.-H. Coumarin Thiazoles as Unique Structural Skeleton of Potential Antimicrobial Agents. Bioorg. Chem. 2022, 124, 105855. [Google Scholar] [CrossRef]
- Chamsaz, E.A.; Mankoci, S.; Barton, H.A.; Joy, A. Nontoxic Cationic Coumarin Polyester Coatings Prevent Pseudomonas aeruginosa Biofilm Formation. ACS Appl. Mater. Interfaces 2017, 9, 6704–6711. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhu, B.; Chen, W.; Hu, J.; Xue, Y.; Yin, H.; Hu, X.; Liu, W. Pseudolaric Acid A: A Promising Antifungal Agent Against Prevalent Non-Albicans Candida Species. Infect. Drug Resist. 2023, 16, 5953–5964. [Google Scholar] [CrossRef] [PubMed]
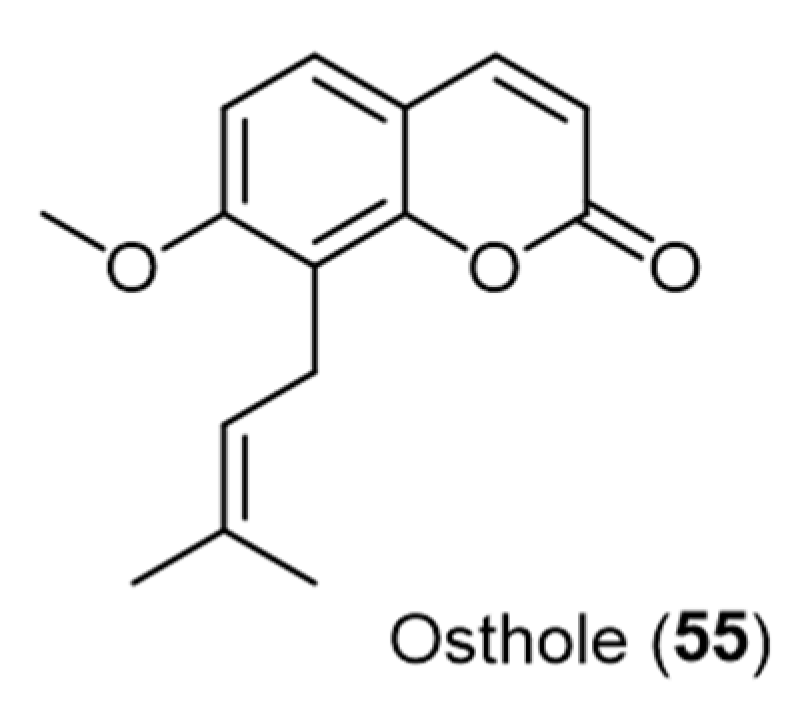
- Li, D.-D.; Chai, D.; Huang, X.-W.; Guan, S.-X.; Du, J.; Zhang, H.-Y.; Sun, Y.; Jiang, Y.-Y. Potent In Vitro Synergism of Fluconazole and Osthole against Fluconazole-Resistant Candida albicans. Antimicrob. Agents Chemother. 2017, 61, e00436-17. [Google Scholar] [CrossRef]
- Bonincontro, G.; Scuderi, S.A.; Marino, A.; Simonetti, G. Synergistic Effect of Plant Compounds in Combination with Conventional Antimicrobials against Biofilm of Staphylococcus aureus, Pseudomonas aeruginosa, and Candida spp. Pharmaceuticals 2023, 16, 1531. [Google Scholar] [CrossRef]
- Inchagova, K.S.; Duskaev, G.K.; Deryabin, D.G. Quorum Sensing Inhibition in Chromobacterium Violaceum by Amikacin Combination with Activated Charcoal or Small Plant-Derived Molecules (Pyrogallol and Coumarin). Microbiology 2019, 88, 63–71. [Google Scholar] [CrossRef]
- Elmaidomy, A.H.; Shady, N.H.; Abdeljawad, K.M.; Elzamkan, M.B.; Helmy, H.H.; Tarshan, E.A.; Adly, A.N.; Hussien, Y.H.; Sayed, N.G.; Zayed, A.; et al. Antimicrobial Potentials of Natural Products against Multidrug Resistance Pathogens: A Comprehensive Review. RSC Adv. 2022, 12, 29078–29102. [Google Scholar] [CrossRef]
- Zou, J.; Liu, Y.; Guo, R.; Tang, Y.; Shi, Z.; Zhang, M.; Wu, W.; Chen, Y.; Hou, K. An In Vitro Coumarin-Antibiotic Combination Treatment of Pseudomonas aeruginosa Biofilms. Nat. Prod. Commun. 2021, 16, 1934578X2098774. [Google Scholar] [CrossRef]
- Qais, F.A.; Ahmad, I.; Husain, F.M.; Arshad, M.; Khan, A.; Adil, M. Umbelliferone modulates the quorum sensing and biofilm of Gram–ve bacteria: In vitro and in silico investigations. J. Biomol. Struct. Dyn. 2023, 42, 5827–5840. [Google Scholar] [CrossRef] [PubMed]
- Fatima, M.; Amin, A.; Alharbi, M.; Ishtiaq, S.; Sajjad, W.; Ahmad, F.; Ahmad, S.; Hanif, F.; Faheem, M.; Khalil, A.A.K. Quorum Quenchers from Reynoutria Japonica in the Battle against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules 2023, 28, 2635. [Google Scholar] [CrossRef] [PubMed]
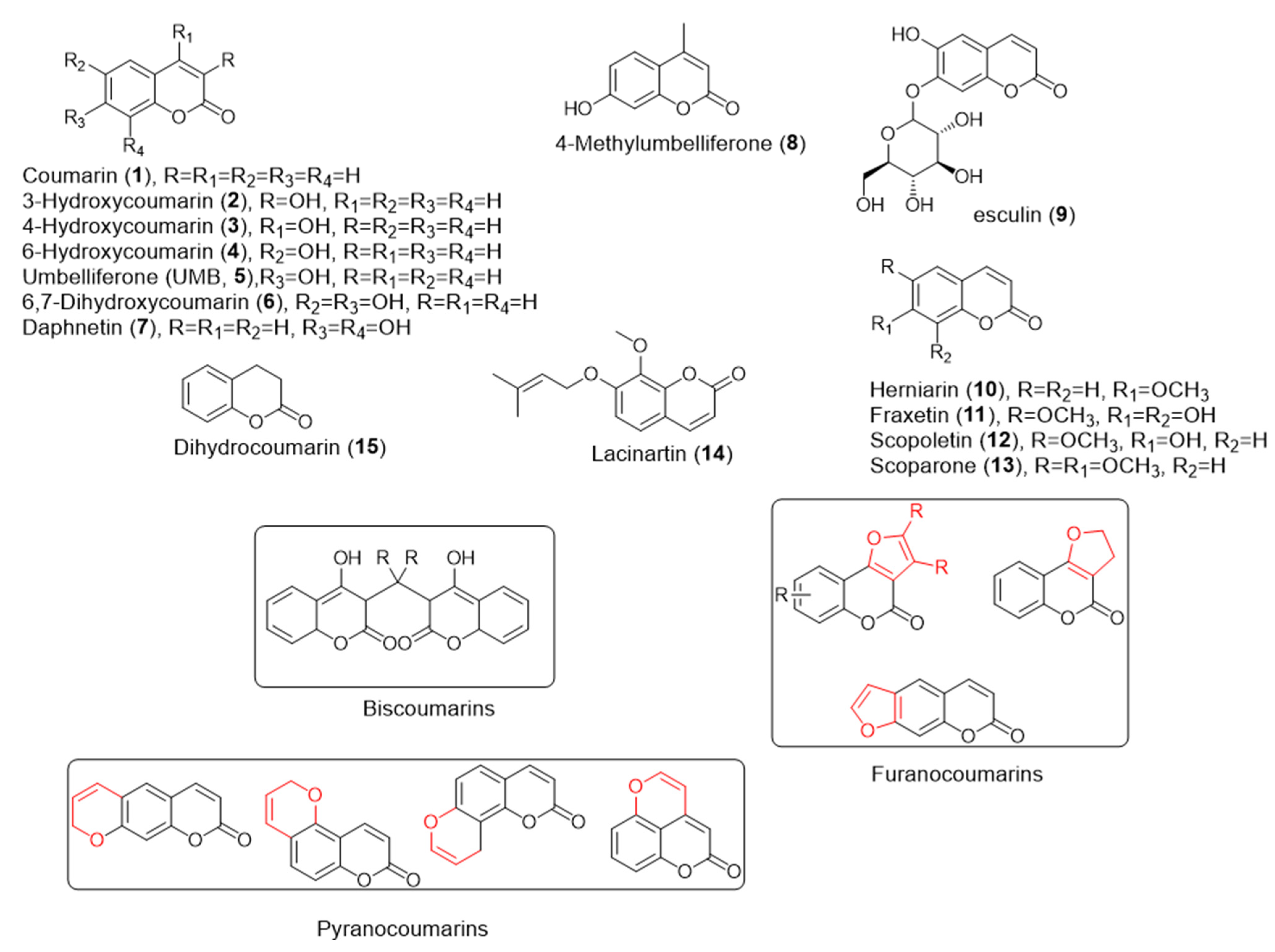

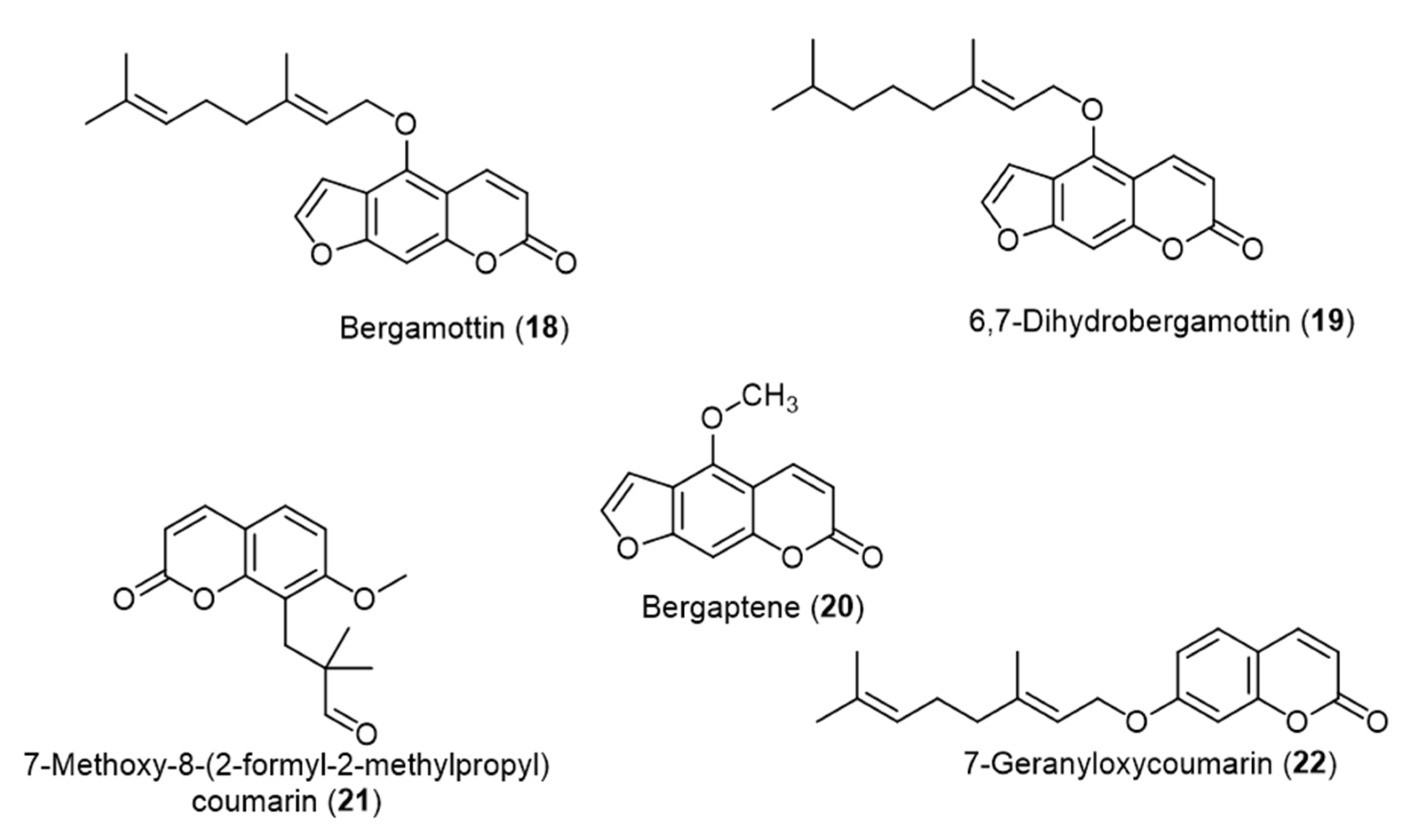

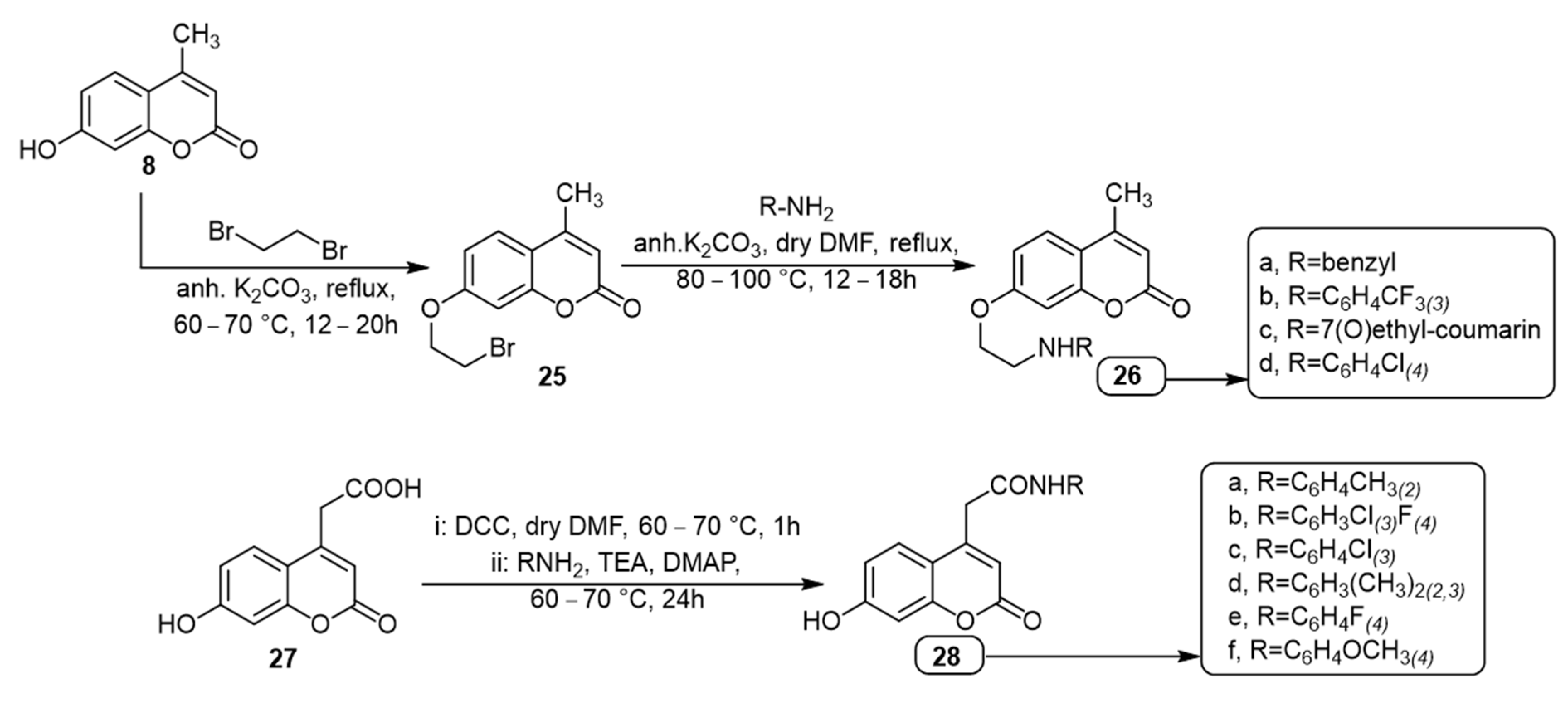
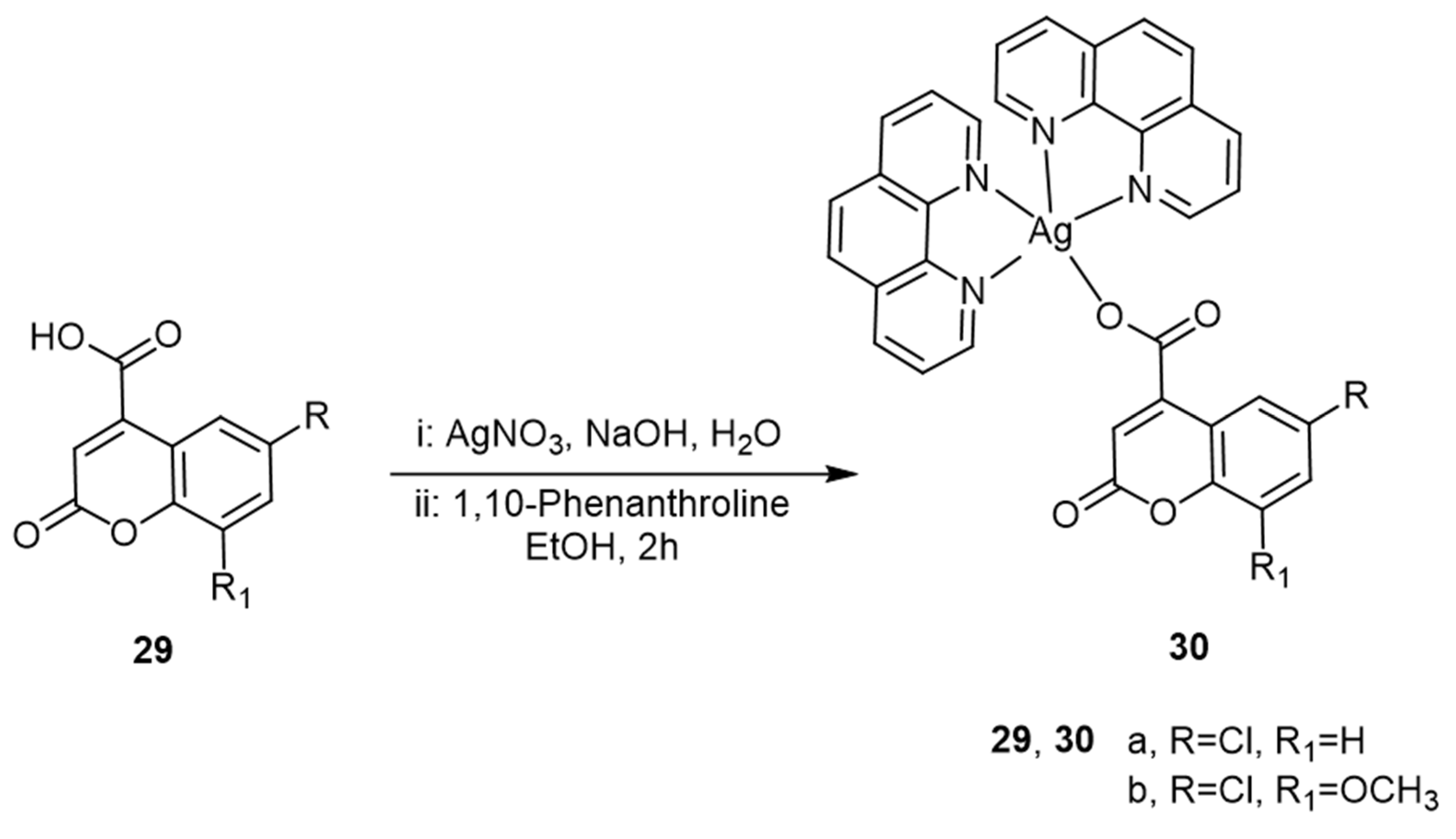
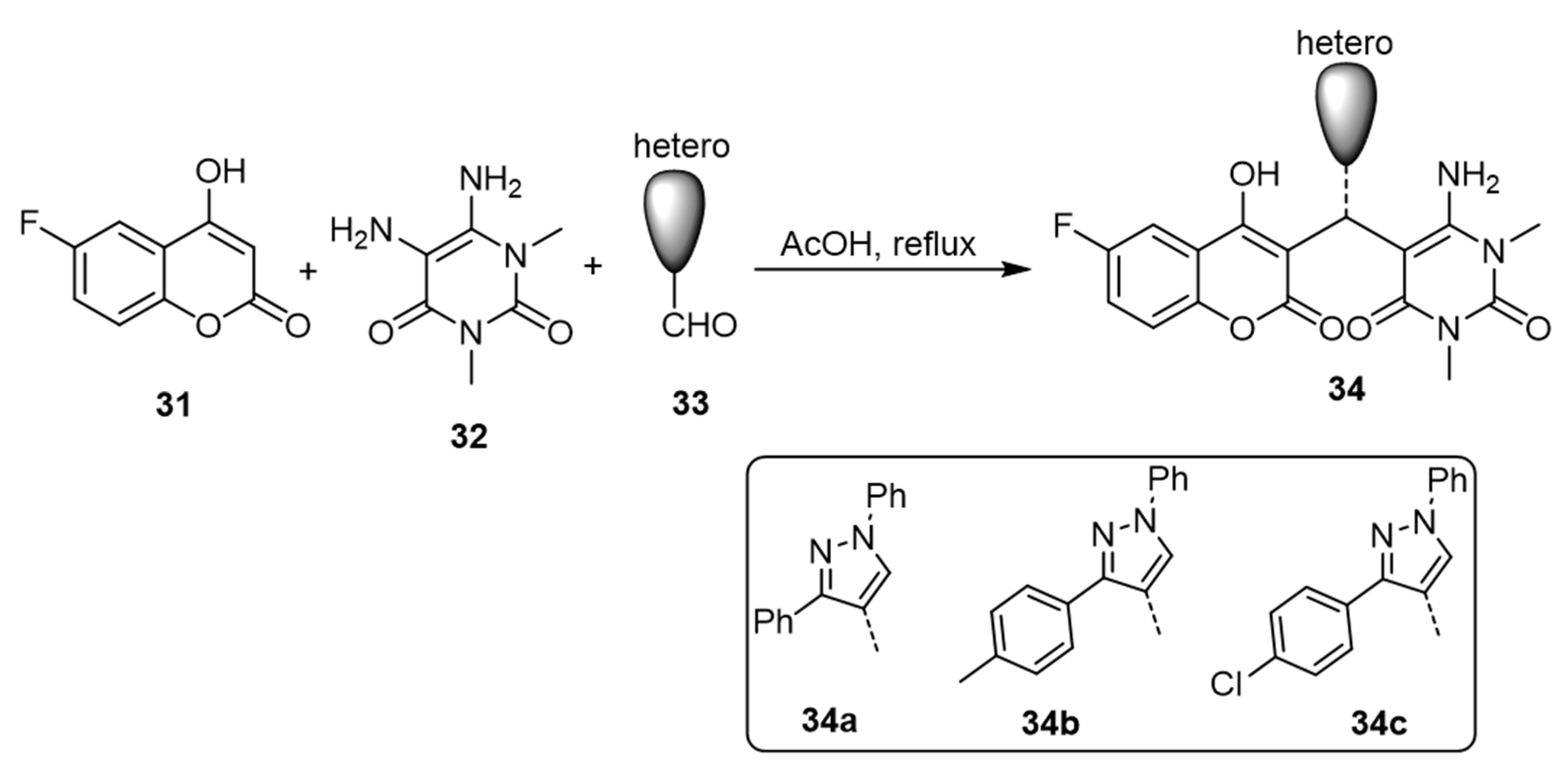

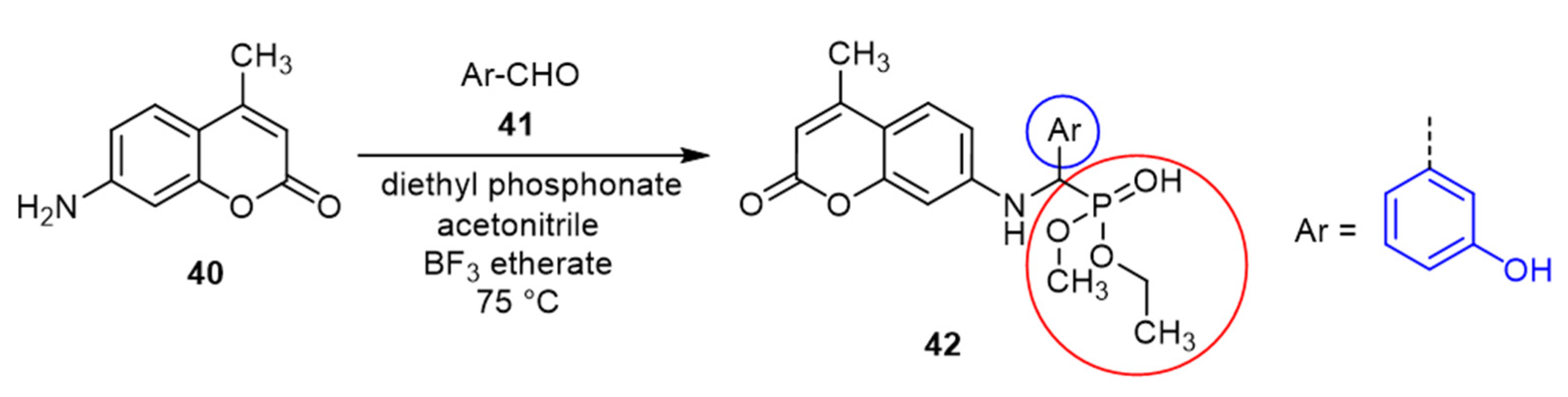


| Coumarin | Target Microorganism | Mode of Action | Ref |
|---|---|---|---|
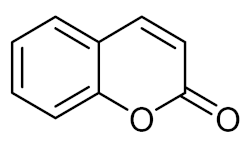 Coumarin (1) | E. coli O157:H7 |
| [56] |
| S. typhimurium SL1344 |
| [57] | |
| R. solanacearum |
| [58] | |
| C. albicans |
| [59] | |
| C. violaceum 12472 P. aeruginosa PAO1 Serratia marcescens MTCC 97 |
| [24] | |
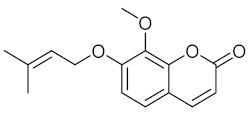 Lacinartin (14) | P. gingivalis |
| [60] |
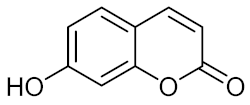 Umbelliferone (UMB, 5) | S. aureus CECT |
| [61] |
| Methicillin-Resistant S. epidermidis (MRSE) |
| [62] | |
| P. aeruginosa |
| [63] | |
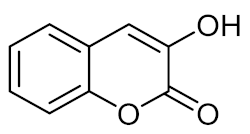 3-Hydroxycoumarin (2) | C. albicans 90028 |
| [64] |
 Daphnetin (7) | R. solanacearum |
| [65,66] |
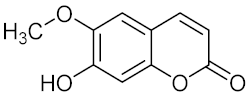 Scopoletin (12) | MDR Candida tropicalis |
| [67] |
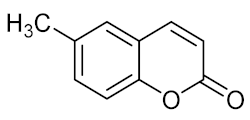 6-Methylcoumarin (16) | R. solanacearum |
| [68] |
| P. aeruginosa |
| [69] | |
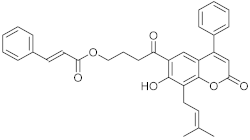 Cinnamoyloxy-mammeisin (CNM, 17) Cinnamoyloxy-mammeisin (CNM, 17) | S. aureus MRSA S. aureus |
| [70] |
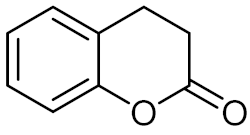 Dihydrocoumarin (15) | H. alvei H4 C. violaceum 026 |
| [71] |
| Target Gram-Negative Bacteria | Protein | PDB |
|---|---|---|
| P. aeruginosa | LasI | 1RO5 |
| Pantoea stewartii | EsaI | 1KZF |
| P. aeruginosa | LasR | 2UV0 |
| P. aeruginosa | LasA | 3IT7 |
| C. violaceum ATCC 12472 | CviR | 3QP5 |
| C. violaceum ATCC 12472 | CviR’ | 3QP1 |
| P. aeruginosa | PqsR | 4JVI |
| P. aeruginosa | Rh1R | SWISS-MODEL Repository (ID: P54292) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Sawy, E.R.; Abdel-Aziz, M.S.; Abdelmegeed, H.; Kirsch, G. Coumarins: Quorum Sensing and Biofilm Formation Inhibition. Molecules 2024, 29, 4534. https://doi.org/10.3390/molecules29194534
El-Sawy ER, Abdel-Aziz MS, Abdelmegeed H, Kirsch G. Coumarins: Quorum Sensing and Biofilm Formation Inhibition. Molecules. 2024; 29(19):4534. https://doi.org/10.3390/molecules29194534
Chicago/Turabian StyleEl-Sawy, Eslam R., Mohamed S. Abdel-Aziz, Heba Abdelmegeed, and Gilbert Kirsch. 2024. "Coumarins: Quorum Sensing and Biofilm Formation Inhibition" Molecules 29, no. 19: 4534. https://doi.org/10.3390/molecules29194534
APA StyleEl-Sawy, E. R., Abdel-Aziz, M. S., Abdelmegeed, H., & Kirsch, G. (2024). Coumarins: Quorum Sensing and Biofilm Formation Inhibition. Molecules, 29(19), 4534. https://doi.org/10.3390/molecules29194534








