Characterization and Crystal Structural Analysis of Novel Carvedilol Adipate and Succinate Ethanol-Solvated Salts
Abstract
:1. Introduction
2. Results and Discussion
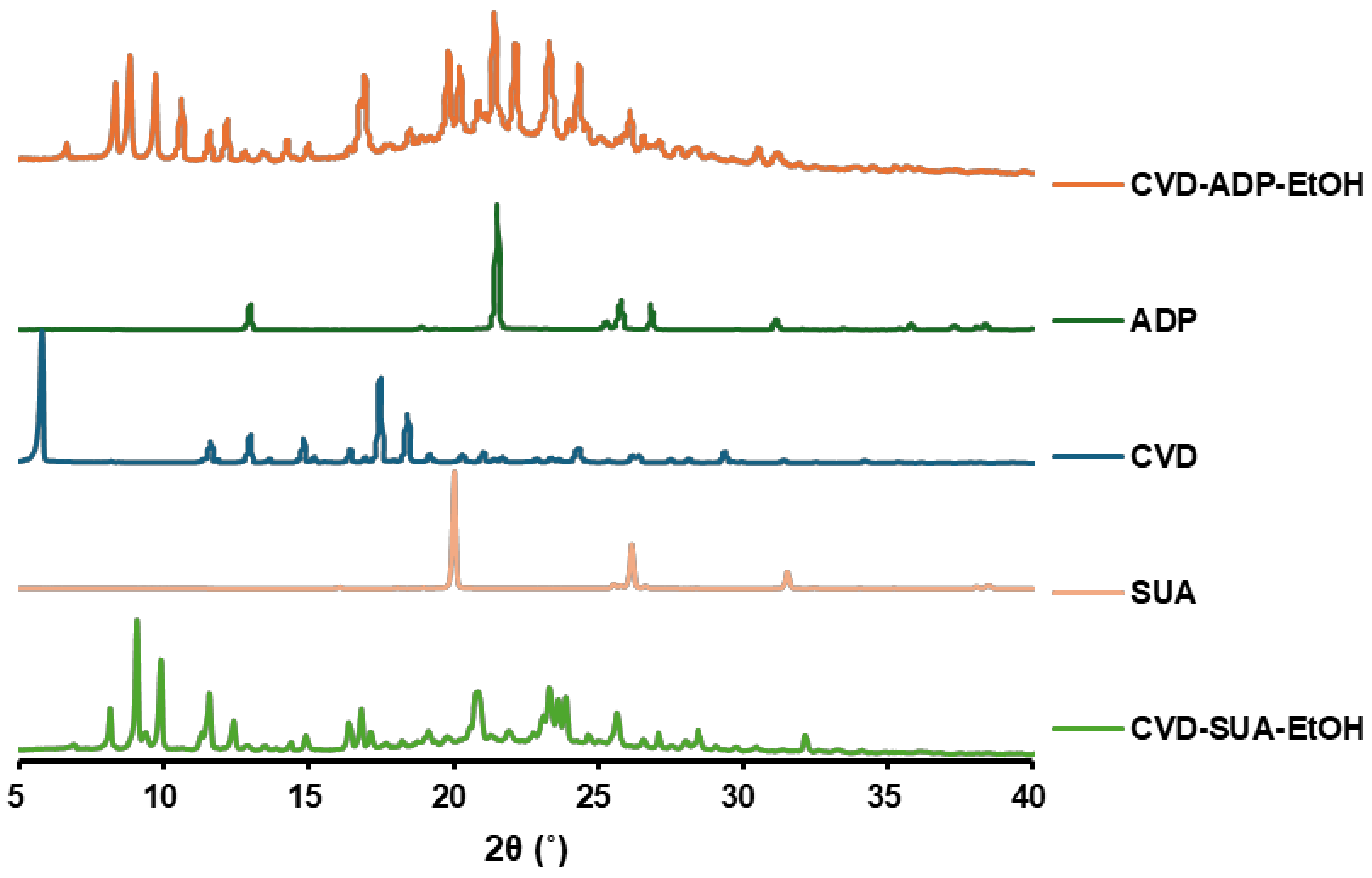
2.1. PXRD Measurements
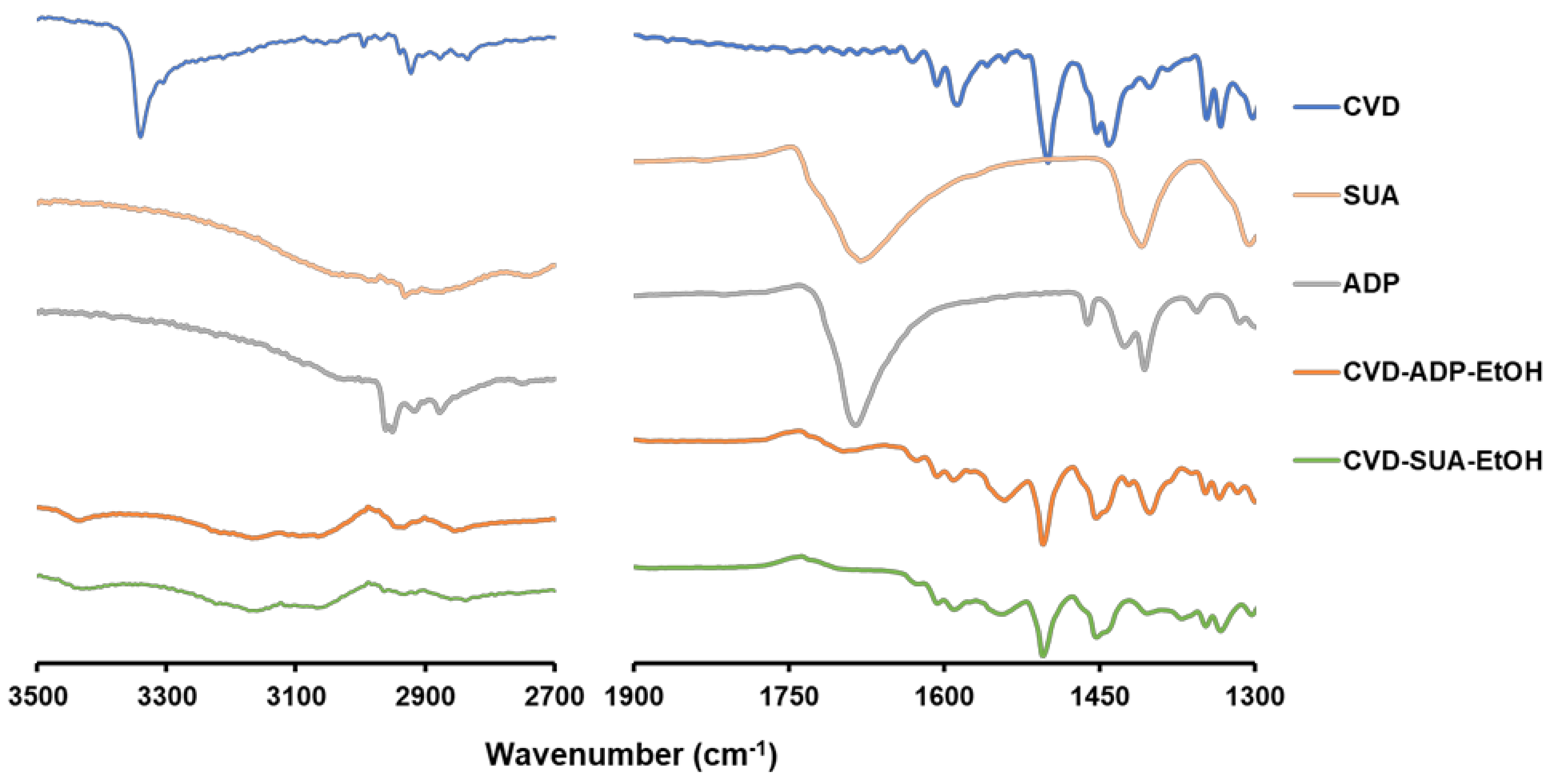
2.2. FT-IR Spectra of the Novel Salts
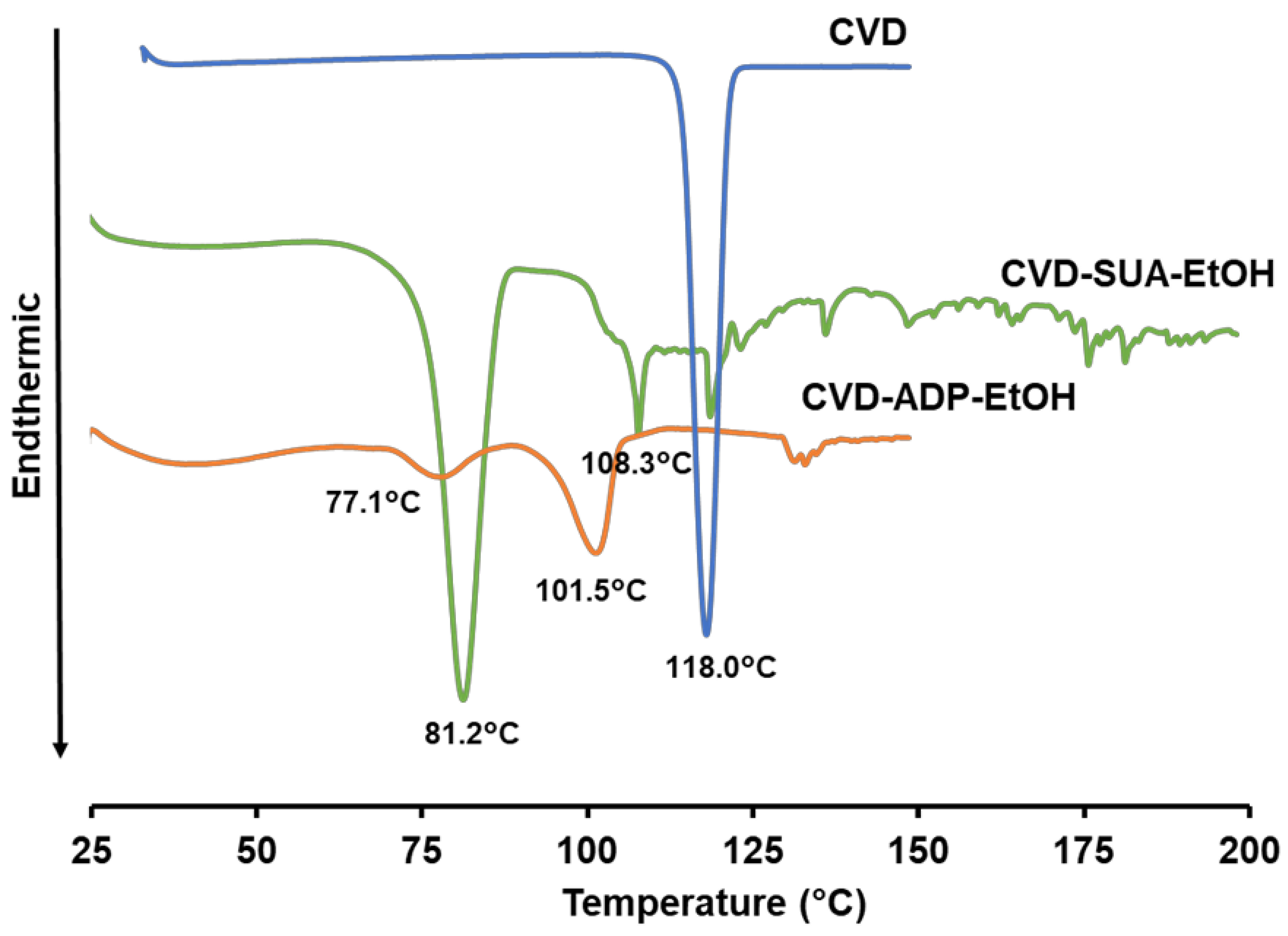
2.3. Thermal Properties of the Novel Salts
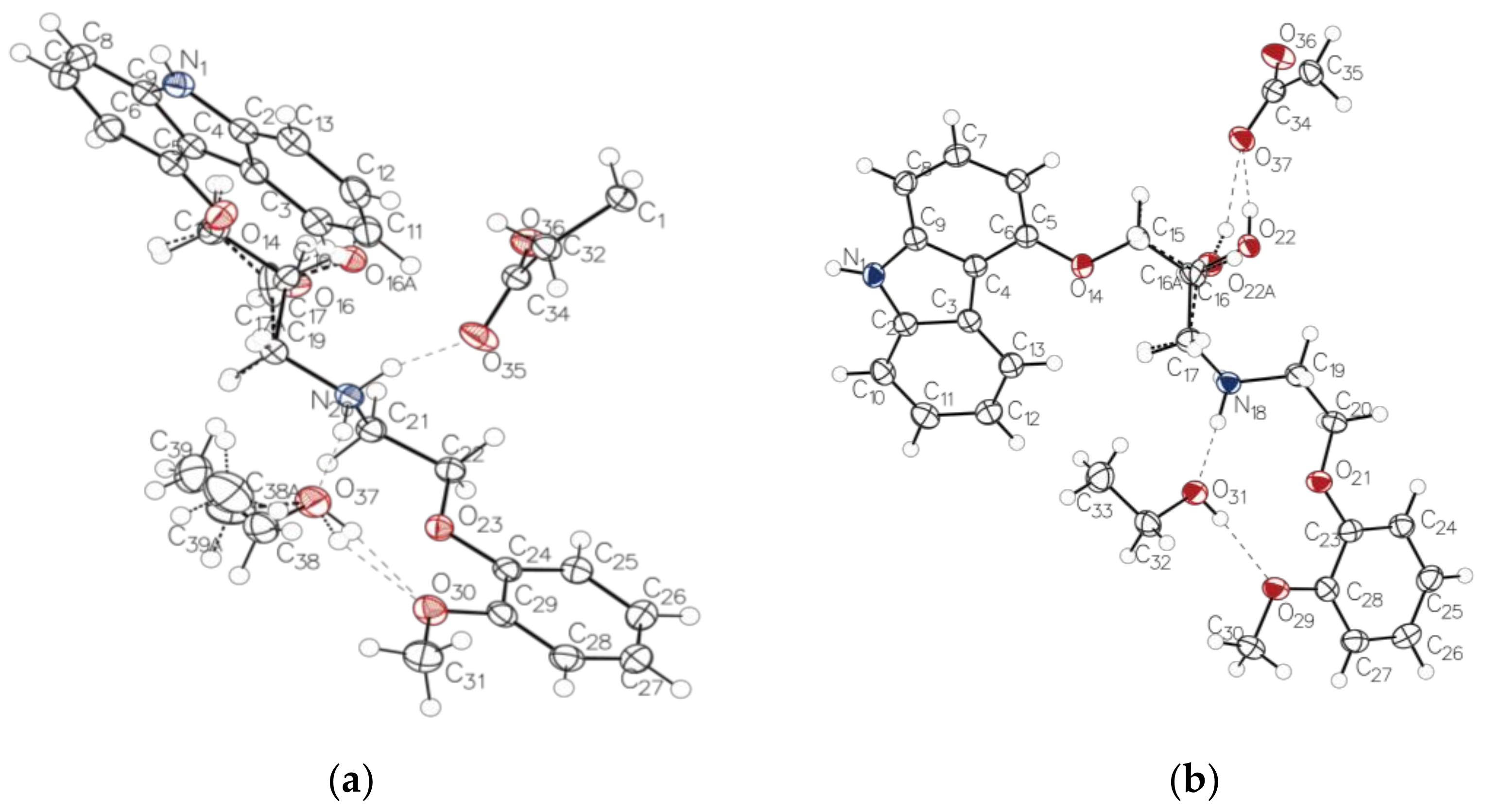
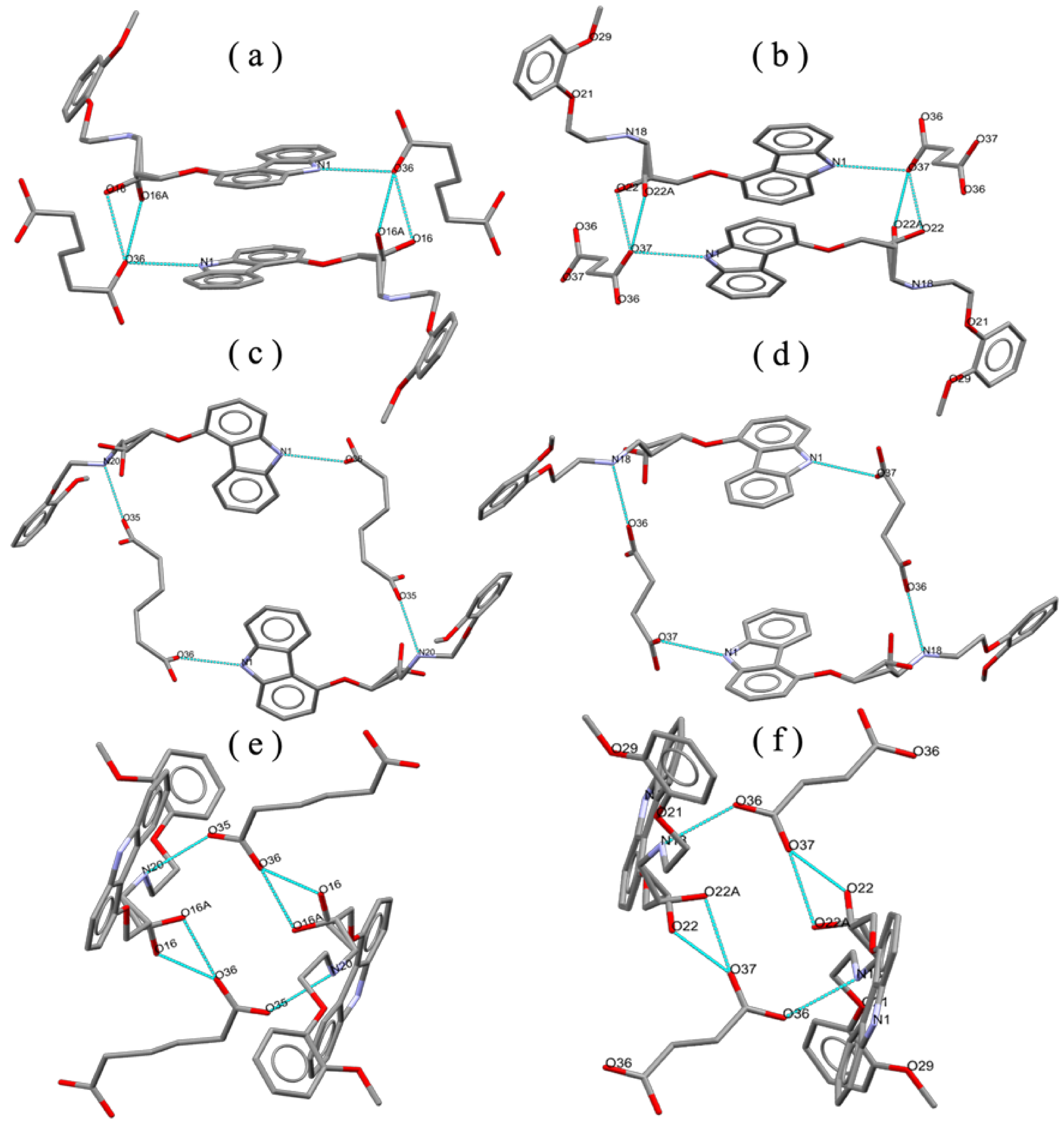
2.4. Crystal Structures of the Novel Salts
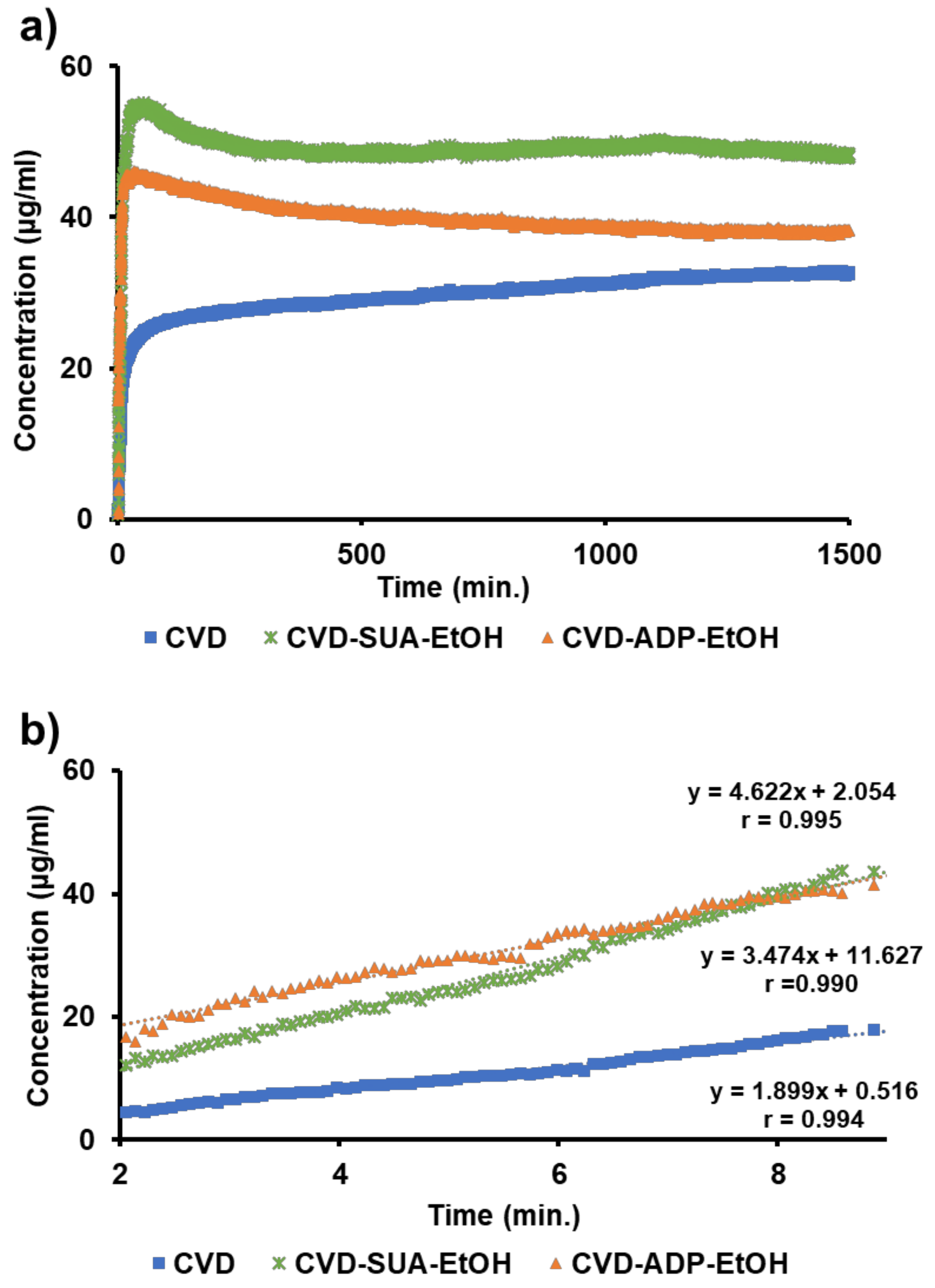
2.5. Dissolution Test
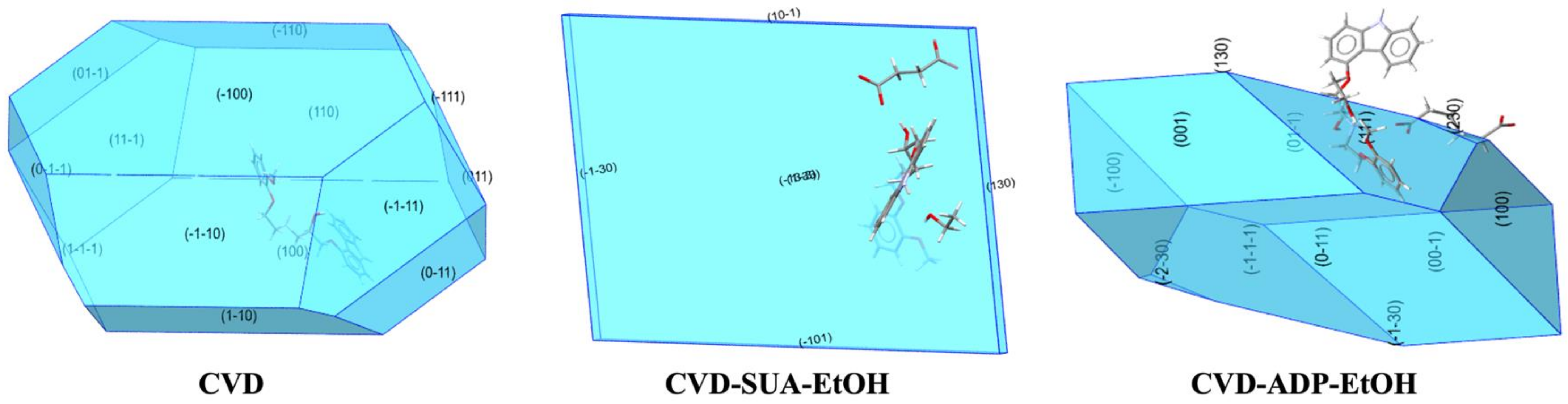
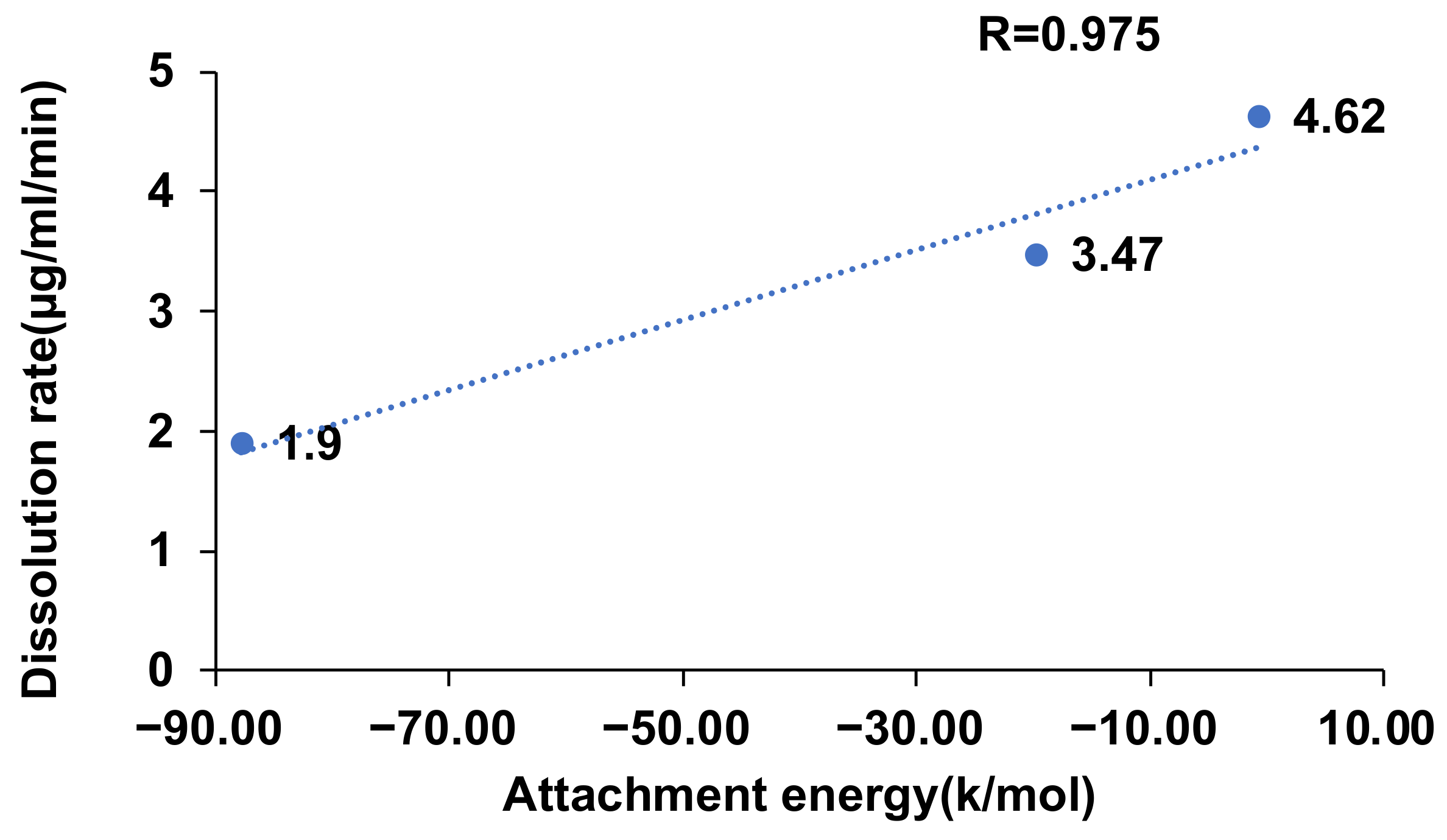
2.6. Effect of Attachment Energy on the Dissolution Rate of CVD-ADP-EtOH and CVD-SUA-EtOH
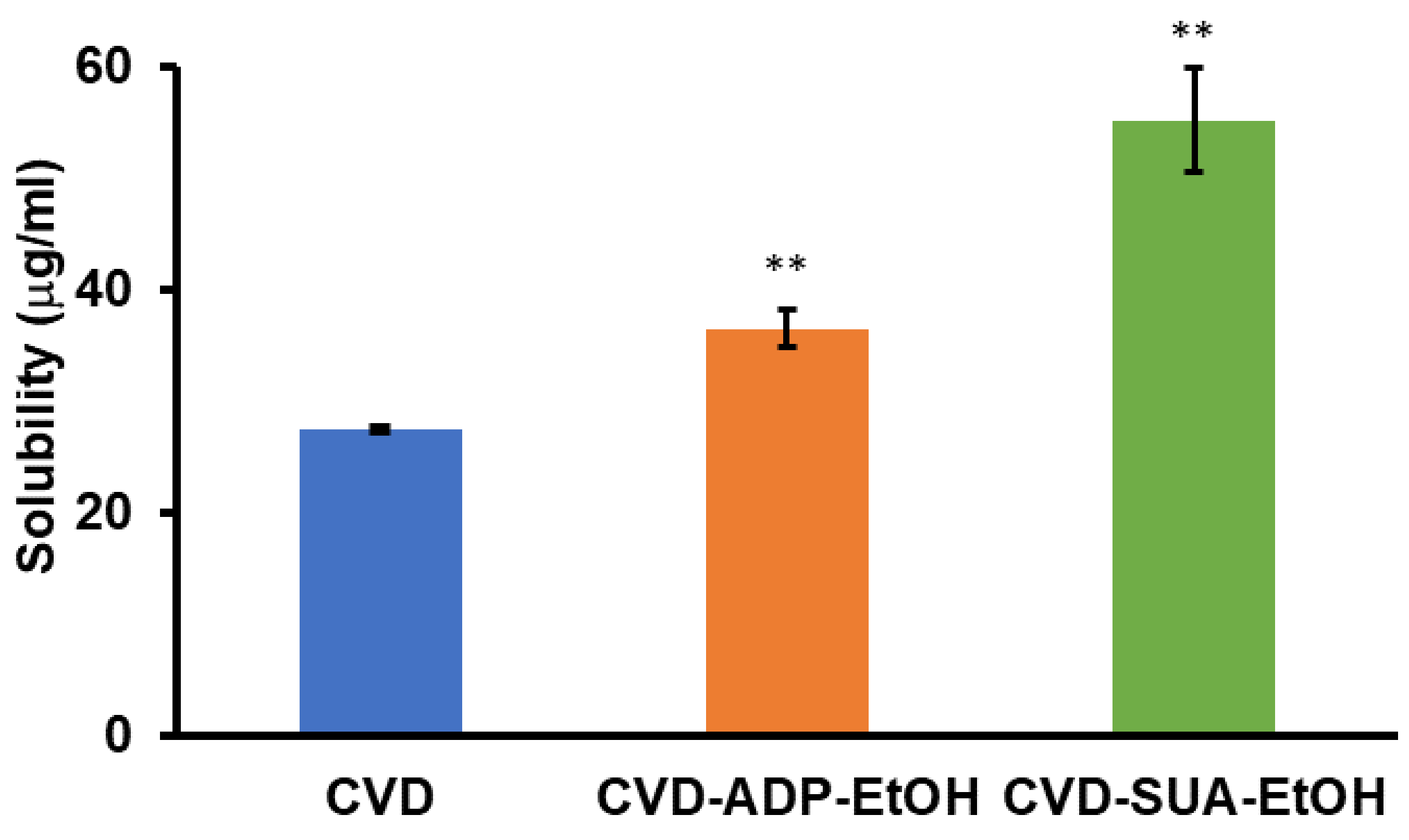
2.7. Equilibrium Solubility
3. Materials and Methods
3.1. Materials
3.2. Sample Preparation
3.3. Powder X-ray Diffraction (PXRD) Measurements
3.4. Fourier Transform Infrared Spectroscopy (FT-IR)
3.5. Differential Scanning Calorimetry (DSC) and Thermogravimetric (TG) Measurements
3.6. Single-Crystal X-ray Diffraction (SCXRD) Measurements
3.7. Dissolution Test
3.8. Computational Analysis
3.9. Equilibrium Solubility Experiments
3.10. High-Performance Liquid Chromatography (HPLC)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thakuria, R.; Delori, A.; Jones, W.; Lipert, M.P.; Roy, L.; Rodriguez-Hornedo, N. Pharmaceutical cocrystals and poorly soluble drugs. Int. J. Pharm. 2013, 453, 101–125. [Google Scholar] [CrossRef] [PubMed]
- Bolla, G.; Sarma, B.; Nangia, A.K. Crystal engineering of pharmaceutical cocrystals in the discovery and development of improved drugs. Chem. Rev. 2022, 122, 11514–11603. [Google Scholar] [CrossRef] [PubMed]
- Berge, S.M.; Bighley, L.D.; Monkhouse, D.C. Pharmaceutical salts. J. Pharm. Sci. 1977, 66, 1–19. [Google Scholar] [CrossRef]
- Aitipamula, S.; Banerjee, R.; Bansal, A.K.; Biradha, K.; Cheney, M.L.; Choudhury, A.R.; Desiraju, G.R.; Dikundwar, A.G.; Dubey, R.; Duggirala, N.; et al. Polymorphs, salts, and cocrystals: What’s in a name? Cryst. Growth Des. 2012, 12, 2147–2152. [Google Scholar] [CrossRef]
- Aakeröy, C.B.; Salmon, D.J. Building co-crystals with molecular sense and supramolecular sensibility. CrystEngComm 2005, 7, 439–448. [Google Scholar] [CrossRef]
- Griesser, U.J. The importance of solvates. In Polymorphism; Wiley-VCH Verlag GmbH & Co. KgaA: Weinheim, Germany, 2006; Chapter 8; pp. 211–233. [Google Scholar] [CrossRef]
- Khankari, R.K.; Grant, D.J.W. Pharmaceutical hydrates. Thermochim. Acta 1995, 248, 61–79. [Google Scholar] [CrossRef]
- Di, L.; Kerns, E.H.; Carter, G.T. Drug-like property concepts in pharmaceutical design. Curr. Pharm. Des. 2009, 15, 2184–2194. [Google Scholar] [CrossRef]
- Yadav, A.V.; Shete, A.S.; Dabke, A.P.; Kulkarni, P.V.; Sakhare, S.S. Co-crystals: A novel approach to modify physicochemical properties of active pharmaceutical ingredients. Indian J. Pharm. Sci. 2009, 71, 359–370. [Google Scholar] [CrossRef]
- Cao, F.; Amidon, G.L.; Rodriguez-Hornedo, N.; Amidon, G.E. Mechanistic basis of cocrystal dissolution advantage. J. Pharm. Sci. 2018, 107, 380–389. [Google Scholar] [CrossRef]
- Paulekuhn, G.S.; Dressman, J.B.; Saal, C. Trends in active pharmaceutical ingredient salt selection based on analysis of the Orange Book database. J. Med. Chem. 2007, 50, 6665–6672. [Google Scholar] [CrossRef]
- Cerreia Vioglio, P.; Chierotti, M.R.; Gobetto, R. Pharmaceutical aspects of salt and cocrystal forms of APIs and characterization challenges. Adv. Drug Deliv. Rev. 2017, 117, 86–110. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Bhatia, D.; Dave, V.; Sutariya, V.; Varghese Gupta, S. Salts of therapeutic agents: Chemical, physicochemical, and biological considerations. Molecules 2018, 23, 1719. [Google Scholar] [CrossRef] [PubMed]
- Black, S.N.; Collier, E.A.; Davey, R.J.; Roberts, R.J. Structure, solubility, screening, and synthesis of molecular salts. J. Pharm. Sci. 2007, 96, 1053–1068. [Google Scholar] [CrossRef] [PubMed]
- Bastin, R.J.; Bowker, M.J.; Slater, B.J. Salt selection and optimisation procedures for pharmaceutical new chemical entities. Org. Process Res. Dev. 2000, 4, 427–435. [Google Scholar] [CrossRef]
- Cruz-Cabeza, A.J. Acid–base crystalline complexes and the pKa rule. CrystEngComm 2012, 14, 6362–6365. [Google Scholar] [CrossRef]
- Hossain Mithu, M.S.; Economidou, S.; Trivedi, V.; Bhatt, S.; Douroumis, D. Advanced methodologies for pharmaceutical salt synthesis. Cryst. Growth Des. 2021, 21, 1358–1374. [Google Scholar] [CrossRef]
- Fisher, L.D. Carvedilol and the Food and Drug Administration (FDA) approval process: The FDA paradigm and reflections on hypothesis testing. Control. Clin. Trials 1999, 20, 16–39. [Google Scholar] [CrossRef]
- World Health Organization. The Selection and Use of Essential Medicines 2023: Web Annex A: World Health Organization Model List of Essential Medicines: 23rd List (2023); World Health Organization: Geneva, Switzerland, 2023. [Google Scholar]
- Fernandes, G.J.; Kumar, L.; Sharma, K.; Tunge, R.; Rathnanand, M. A review on solubility enhancement of carvedilol—A BCS Class II Drug. J. Pharm. Innov. 2018, 13, 197–212. [Google Scholar] [CrossRef]
- Liu, D.; Pan, H.; He, F.; Wang, X.; Li, J.; Yang, X.; Pan, W. Effect of particle size on oral absorption of carvedilol nanosuspensions: In vitro and in vivo evaluation. Int. J. Nanomed. 2015, 10, 6425–6434. [Google Scholar] [CrossRef]
- Krstic, M.; Manic, L.; Martic, N.; Vasiljevic, D.; Mracevic, S.D.; Vukmirovic, S.; Raskovic, A. Binary polymeric amorphous carvedilol solid dispersions: In vitro and in vivo characterization. Eur. J. Pharm. Sci. 2020, 150, 105343. [Google Scholar] [CrossRef]
- Savic-Gajic, I.; Savic, I.M.; Nikolic, V.D.; Nikolic, L.B.; Popsavin, M.M.; Kapor, A.J. Study of the solubility, photostability and structure of inclusion complexes of carvedilol with β-cyclodextrin and (2-hydroxypropyl)-β-cyclodextrin. J. Incl. Phenom. Macrocycl. Chem. 2016, 86, 7–17. [Google Scholar] [CrossRef]
- Mishra, J.; Lobmann, K.; Grohganz, H.; Rades, T. Influence of preparation technique on co-amorphization of carvedilol with acidic amino acids. Int. J. Pharm. 2018, 552, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Furuishi, T.; Sato-Hata, N.; Fukuzawa, K.; Yonemochi, E. Characterization of co-amorphous carvedilol-maleic acid system prepared by solvent evaporation. Pharm. Dev. Technol. 2023, 28, 309–317. [Google Scholar] [CrossRef]
- Diniz, L.F.; Carvalho, P.S.; da Nova Mussel, W.; Yoshida, M.I.; Diniz, R.; Fernandes, C. Racemic salts and solid solutions of enantiomers of the antihypertensive drug carvedilol. Cryst. Growth Des. 2019, 19, 4498–4509. [Google Scholar] [CrossRef]
- Vogt, F.G.; Copley, R.C.B.; Mueller, R.L.; Spoors, G.P.; Cacchio, T.N.; Carlton, R.A.; Katrincic, L.M.; Kennady, J.M.; Parsons, S.; Chetina, O.V. Isomorphism, disorder, and hydration in the crystal structures of racemic and single-enantiomer carvedilol phosphate. Cryst. Growth Des. 2010, 10, 2713–2733. [Google Scholar] [CrossRef]
- Sopyan, I.; Layyareza, R.T.; Megantara, S.; Marvita, S.S. Carvedilol solubility enhancement by multicomponent crystallization with coformers of benzoic acid, isonicotinamide, and saccharin. Pharmacia 2023, 70, 283–290. [Google Scholar] [CrossRef]
- Hiendrawan, S.; Widjojokusumo, E.; Veriansyah, B.; Tjandrawinata, R.R. Pharmaceutical salts of carvedilol: Polymorphism and physicochemical properties. AAPS PharmSciTech 2017, 18, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Hata, N.; Furuishi, T.; Tamboli, M.I.; Ishizaki, M.; Umeda, D.; Fukuzawa, K.; Yonemochi, E. Crystal structural analysis of dl-mandelate salt of carvedilol and its correlation with physicochemical properties. Crystals 2020, 10, 53. [Google Scholar] [CrossRef]
- Wolpert, M.; Hellwig, P. Infrared spectra and molar absorption coefficients of the 20 alpha amino acids in aqueous solutions in the spectral range from 1800 to 500 cm−1. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2006, 64, 987–1001. [Google Scholar] [CrossRef]
- Ibrahim, M.; Nada, A.; Kamal, D.E. Density functional theory and FTIR spectroscopic study of carboxyl group. Indian J. Pure Appl. Phys. 2005, 34, 911–917. [Google Scholar]
- Ahmed, K.; Masum, M.; Sultana, S.; Bhuiyan, M.; Reza, M. Impact analysis of inclusion complexation on micromeritic properties and dissolution behavior of carvedilol. J. Appl. Pharm. Sci. 2016, 6, 106–114. [Google Scholar] [CrossRef]
- Yathirajan, H.S.; Bindya, S.; Sreevidya, T.V.; Narayana, B.; Bolte, M. A second polymorph of carvedilol. Acta Crystallogr. Sect. E Struct. Rep. Online 2007, 63, o542–o544. [Google Scholar] [CrossRef]
- Hatanaka, T.; Yoshihashi, Y.; Terada, K.; Yonemochi, E. Understanding crystal cleavability and physical properties of crystal surfaces using in silico simulation. Chem. Pharm. Bull. 2021, 69, 185–198. [Google Scholar] [CrossRef]
- Lippold, B.C.; Ohm, A. Correlation between wettability and dissolution rate of pharmaceutical powders. Int. J. Pharm. 1986, 28, 67–74. [Google Scholar] [CrossRef]
- Greco, K.; Bogner, R. Solution-mediated phase transformation: Significance during dissolution and implications for bioavailability. J. Pharm. Sci. 2012, 101, 2996–3018. [Google Scholar] [CrossRef] [PubMed]
- Babu, N.J.; Nangia, A. Solubility advantage of amorphous drugs and pharmaceutical cocrystals. Cryst. Growth Des. 2011, 11, 2662–2679. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Donnay, J.D.H.; Harker, D. A new law of crystal morphology extending the Law of Bravais. Am. Mineral. 1937, 22, 446–467. [Google Scholar]
- Roberts, K.J.; Hammond, R.B.; Ramachandran, V.; Docherty, R. Synthonic engineering. In Computational Pharmaceutical Solid State Chemistry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; Chapter 8; pp. 175–210. [Google Scholar] [CrossRef]
- Song, L.; Zhao, F.Q.; Xu, S.Y.; Ju, X.H.; Ye, C.C. Crystal morphology prediction and anisotropic Evolution of 1,1-Diamino-2,2-dinitroethylene (FOX-7) by Temperature Tuning. Sci. Rep. 2020, 10, 2317. [Google Scholar] [CrossRef]
- Hartman, P.; Bennema, P. The attachment energy as a habit controlling factor. J. Cryst. Growth 1980, 49, 145–156. [Google Scholar] [CrossRef]
- Hartman, P. An approximate calculation of attachment energies for ionic crystals. Acta Crystallogr. 1956, 9, 569–572. [Google Scholar] [CrossRef]
- Docherty, R.; Clydesdale, G.; Roberts, K.J.; Bennema, P. Application of bravais-friedel-donnay-harker, attachment energy and ising models to predicting and understanding the morphology of molecular crystals. J. Phys. D Appl. Phys. 1991, 24, 89–99. [Google Scholar] [CrossRef]
- Coombes, D.S.; Catlow, C.R.A.; Gale, J.D.; Rohl, A.L.; Price, S.L. Calculation of attachment energies and relative volume growth rates as an aid to polymorph prediction. Cryst. Growth Des. 2005, 5, 879–885. [Google Scholar] [CrossRef]
- Clydesdale, G.; Roberts, K.J.; Docherty, R. HABIT95—A program for predicting the morphology of molecular crystals as a function of the growth environment. J. Cryst. Growth 1996, 166, 78–83. [Google Scholar] [CrossRef]
- Clydesdale, G.; Docherty, R.; Roberts, K.J. HABIT—A program for predicting the morphology of molecular crystals. Comput. Phys. Commun. 1991, 64, 311–328. [Google Scholar] [CrossRef]
- Beyer, T.; Day, G.M.; Price, S.L. The prediction, morphology, and mechanical properties of the polymorphs of paracetamol. J. Am. Chem. Soc. 2001, 123, 5086–5094. [Google Scholar] [CrossRef]



| Parameters | CVD (Form II) (a) | CVD-ADP-EtOH | CVD-SUA-EtOH |
|---|---|---|---|
| Empirical formula | C24H26N2O4 | C29H37N2O7 | C28H35N2O7 |
| Formula weight | 406.47 | 525.61 | 511.58 |
| Temperature [K] | 173(2) | 100.01 | 100.01 |
| Wavelength | 0.71073 | 1.54184 | 1.54184 |
| Crystal system | Monoclinic | Triclinic | Triclinic |
| Space group | P21/c | P-1 | P-1 |
| Unit cell dimensions | |||
| a [Å] | 15.5414 (14) | 9.9404(3) | 9.7393(3) |
| b [Å] | 15.2050 (12) | 10.5634(3) | 10.7950(3) |
| c [Å] | 9.1174 (8) | 13.0610(3) | 12.5360(3) |
| α [°] | 90° | 100.104(2) | 99.058(2) |
| β [°] | 100.730 (7) | 95.155(2) | 91.319(2) |
| γ [°] | 90° | 90.197(2) | 90.376(2) |
| Volume (Å3) | 2116.8 (3) | 1344.47 | 1300.94 |
| Molecules per cell (Z) | 4 | 2 | 2 |
| Molecules in the asymmetric unit (Z’) | 1 | 1 | 1 |
| Density (calculated) [g/cm3] | 1.275 | 1.298 | 1.306 |
| Absorption coefficient [mm⁻1] | 0.087 | 0.759 | 0.771 |
| F (000) | 864 | 562 | 546 |
| Crystal size [mm × mm × mm] | 0.36 × 0.33 × 0.32 | 0.8 × 0.5 × 0.4 | 0.46 × 0.08 × 0.04 |
| Theta range for data collection [°] | 3.51 to 25.68 | 3.452 to 68.241 | 3.571 to 68.319 |
| Index ranges [°] | −18 ≤ h ≤ 17, −18 ≤ k ≤ 18, −11 ≤ l ≤ 11 | −11 ≤ h ≤ 11, −12 ≤ k ≤ 12, −15 ≤ l ≤ 14 | −11 ≤ h ≤ 11, −12 ≤ k ≤ 12, −14 ≤ l ≤ 15 |
| Reflections collected | 3956 | 12,234 | 11,673 |
| Independent reflections | 3956 | 4877 | 4703 |
| Completeness to theta = 67.684° | 99.7% | 99.7% | 99.6% |
| µ (mm−1) | 0.09 | 0.76 | 0.77 |
| Absorption correction | ω scans | multi-scan | multi-scan |
| Max. and min. transmission | - | 1.00000 and 0.72819 | 1.00000 and 0.89934 |
| Refinement method | Fc = kFc [1 + 0.001xFc2λ3/sin(2θ)]^((−1)⁄4) | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 3956/0/285 | 4877/8/386 | 4703/5/358 |
| Goodness-of-fit on F2 | 1.029 | 1.058 | 1.115 |
| Final R indices [I > 2sigma(I)] | R1 = 0.0403, wR2 = 0.0986 | R1 = 0.0428, wR2 = 0.1096 | R1 = 0.0401, wR2 = 0.0975 |
| R indices (all data) | R1 = 0.0576, wR2 = 0.1056 | R1 = 0.0455, wR2 = 0.1115 | R1 = 0.0418, wR2 = 0.0985 |
| Largest diff peak and hole | 0.286 and −0.207 | 0.169 and −0.311 | 0.241 and −0.194 |
| CCDC number | 636656 | 2320421 | 2320422 |
| Donor–H…Acceptor | D-H (Å) | H…A (Å) | D…A (Å) | D-H...A (°) | Symmetry Codes |
|---|---|---|---|---|---|
| N1–H1…O36 | 0.88 | 2.03 | 2.8841 (18) | 163 | x, 1 + y, z |
| O16–H16…O36 | 0.84 | 1.90 | 2.7206 (17) | 164 | 1 − x, 1 − y, -z |
| N20–H20A…O23 | 0.91 | 2.53 | 2.8636 (16) | 102 | Intramolecular |
| N20–H20A…O37 | 0.91 | 1.92 | 2.8223 (17) | 171 | x, y, z |
| N20–H20B…O35 | 0.91 | 1.76 | 2.6372 (16) | 162 | x, y, z |
| O37–H37A…O23 | 0.84 | 2.53 | 3.0570 (16) | 122 | x, y, z |
| O37–H37A…O30 | 0.84 | 1.95 | 2.7816 (16) | 169 | x, y, z |
| C19–H19A…O14 | 0.99 | 2.44 | 2.8554 (19) | 105 | Intramolecular |
| C21–H21A…O16 | 0.99 | 2.42 | 3.092 (2) | 125 | Intramolecular |
| C21–H21B…O23 | 0.99 | 2.54 | 3.4637 (18) | 156 | 1 − x, 1 − y, 1 − z |
| C28–H28…O35 | 0.95 | 2.42 | 3.3390 (19) | 164 | 2 − x, 1 − y, 1 − z |
| C33–H33A…O16 | 0.99 | 2.5 | 3.386 (2) | 149 | 1 − x, 1 − y, −z |
| Donor–H…Acceptor | D-H (Å) | H…A (Å) | D…A (Å) | D-H…A (°) | Symmetry Codes |
|---|---|---|---|---|---|
| CVD-SUA-EtOH | |||||
| N1–H1…O37 | 0.88 | 2 | 2.8430 (18) | 161 | 1 − x, 2 − y, − z |
| N18–H18A…O36 | 0.91 | 1.82 | 2.6983 (17) | 160 | 1 − x, 1 − y, −z |
| N18–H18B…O21 | 0.91 | 2.54 | 2.8955 (17) | 104 | Intramolecular |
| N18–H18B…O31 | 0.91 | 1.94 | 2.8108 (17) | 160 | x, y, z |
| O22–H22…O37 | 0.84 | 1.84 | 2.6671 (18) | 169 | x, y, z |
| O31–H31…O21 | 0.84 | 2.46 | 2.9572 (15) | 119 | x, y, z |
| O31–H31…O29 | 0.84 | 1.97 | 2.7937 (16) | 168 | x, y, z |
| C17–H17B…O14 | 0.99 | 2.38 | 2.8076 (19) | 105 | Intramolecular |
| C19–H19A…O21 | 0.99 | 2.5 | 3.4554 (18) | 163 | 1 − x, 1 − y, 1 − z |
| C19–H19B…O22 | 0.99 | 2.41 | 3.025 (2) | 120 | Intramolecular |
| C27–H27…O36 | 0.95 | 2.46 | 3.391 (2) | 168 | −1 + x, y, 1 + z |
| C30–H30C…O22 | 0.98 | 2.58 | 3.238 (2) | 124 | 1 − x, 1 − y, 1 − z |
| CVD-ADP-EtOH | CVD-SUA-EtOH | CVD | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Surface | (01−1) | (001) | (100) | (111) | (230) | (1−33) | (10−1) | −130 | (1 0 0) | (1 1 0) | (1 1 −1) | (0 1 1) |
| Percentage surface area (%) | 26.63 | 10.26 | 7.05 | 5.85 | 0.22 | 47.79 | 1.22 | 1.09 | 20.34 | 7.16 | 5.28 | 2.39 |
| Electrostatic energy (kJ/mol) | 56.09 | 8.53 | 8.65 | 36.81 | 54.26 | 85.59 | 40.03 | 77.21 | −7.84 | −7.9 | −13.99 | −13.9 |
| van der Waals energy (kJ/mol) | −58.7 | −21.92 | −32.05 | −39.97 | −59.05 | −61.57 | −34.73 | −65.86 | −46.12 | −75.43 | −89.64 | −103.61 |
| Hydrogen bond energy (kJ/mol) | −9.39 | −10.63 | −12.18 | −24.89 | −25.04 | −24.26 | −15.33 | −21.63 | −0.01 | −10.3 | −19.18 | −19.19 |
| Attachment energy (kJ/mol) | −12 | −24.02 | −35.57 | −28.05 | −29.83 | −0.24 | −10.03 | −10.28 | −53.97 | −93.62 | −122.81 | −136.69 |
| Attachment energy of facet (kJ/mol) | −3.19 | −2.46 | −2.51 | −1.64 | −0.06 | −0.11 | −0.12 | −0.11 | −10.98 | −6.7 | −6.49 | −3.27 |
| Symmetry equivalent facets | (0 −1 1) | (0 0 −1) | (−1 0 0) | (−1 −1 −1) | (−2 −3 0) | (−13−3) | (−101) | (−1−30) | (−1 0 0) | (1 −1 0) (−1 1 0) (−1 −1 0) | (1 −1 −1) (−1 1 1) (−1 −1 1) | (0 1 −1) (0 −1 1) (0 −1 −1) |
| Total attachment energy (kJ/mol) | −19.74 | −0.69 | −87.78 | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, L.; Furuishi, T.; Yamashita, T.; Yonemochi, E. Characterization and Crystal Structural Analysis of Novel Carvedilol Adipate and Succinate Ethanol-Solvated Salts. Molecules 2024, 29, 4704. https://doi.org/10.3390/molecules29194704
Ye L, Furuishi T, Yamashita T, Yonemochi E. Characterization and Crystal Structural Analysis of Novel Carvedilol Adipate and Succinate Ethanol-Solvated Salts. Molecules. 2024; 29(19):4704. https://doi.org/10.3390/molecules29194704
Chicago/Turabian StyleYe, Li, Takayuki Furuishi, Takefumi Yamashita, and Etsuo Yonemochi. 2024. "Characterization and Crystal Structural Analysis of Novel Carvedilol Adipate and Succinate Ethanol-Solvated Salts" Molecules 29, no. 19: 4704. https://doi.org/10.3390/molecules29194704
APA StyleYe, L., Furuishi, T., Yamashita, T., & Yonemochi, E. (2024). Characterization and Crystal Structural Analysis of Novel Carvedilol Adipate and Succinate Ethanol-Solvated Salts. Molecules, 29(19), 4704. https://doi.org/10.3390/molecules29194704






