Ab Initio Neural Network Potential Energy Surface and Quantum Dynamics Calculations on Na(2S) + H2 → NaH + H Reaction
Abstract
:1. Introduction
2. Results and Discussion
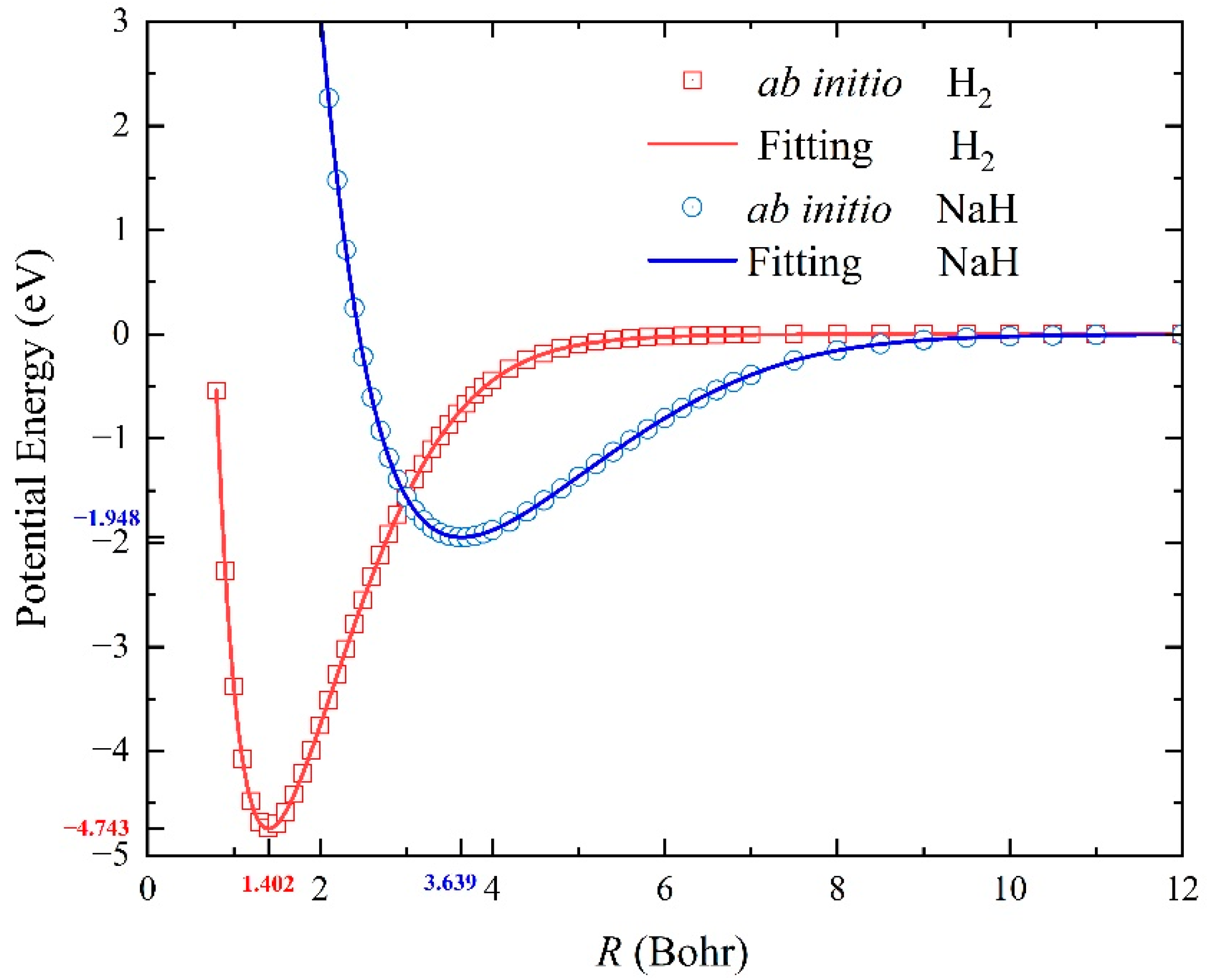
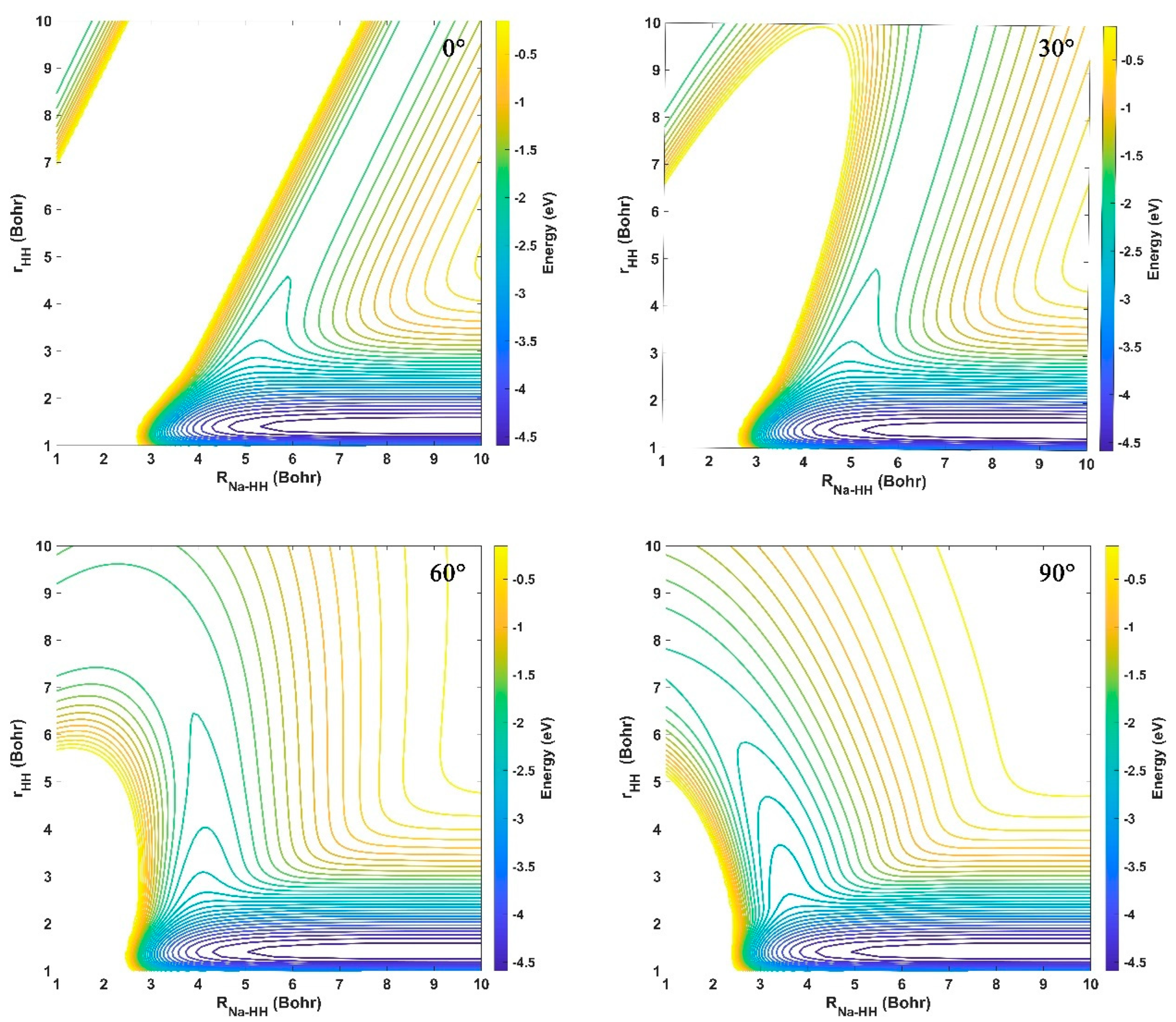
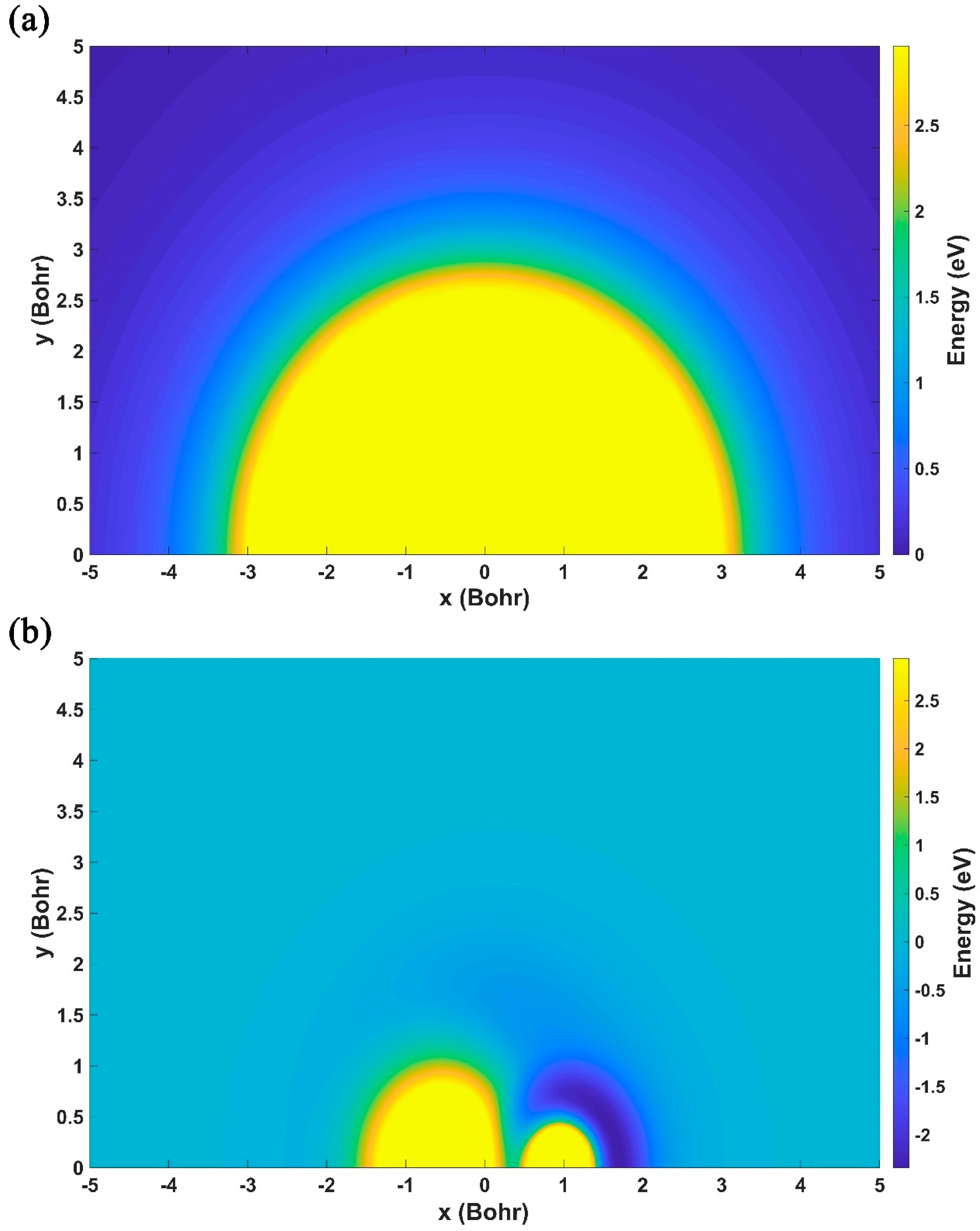
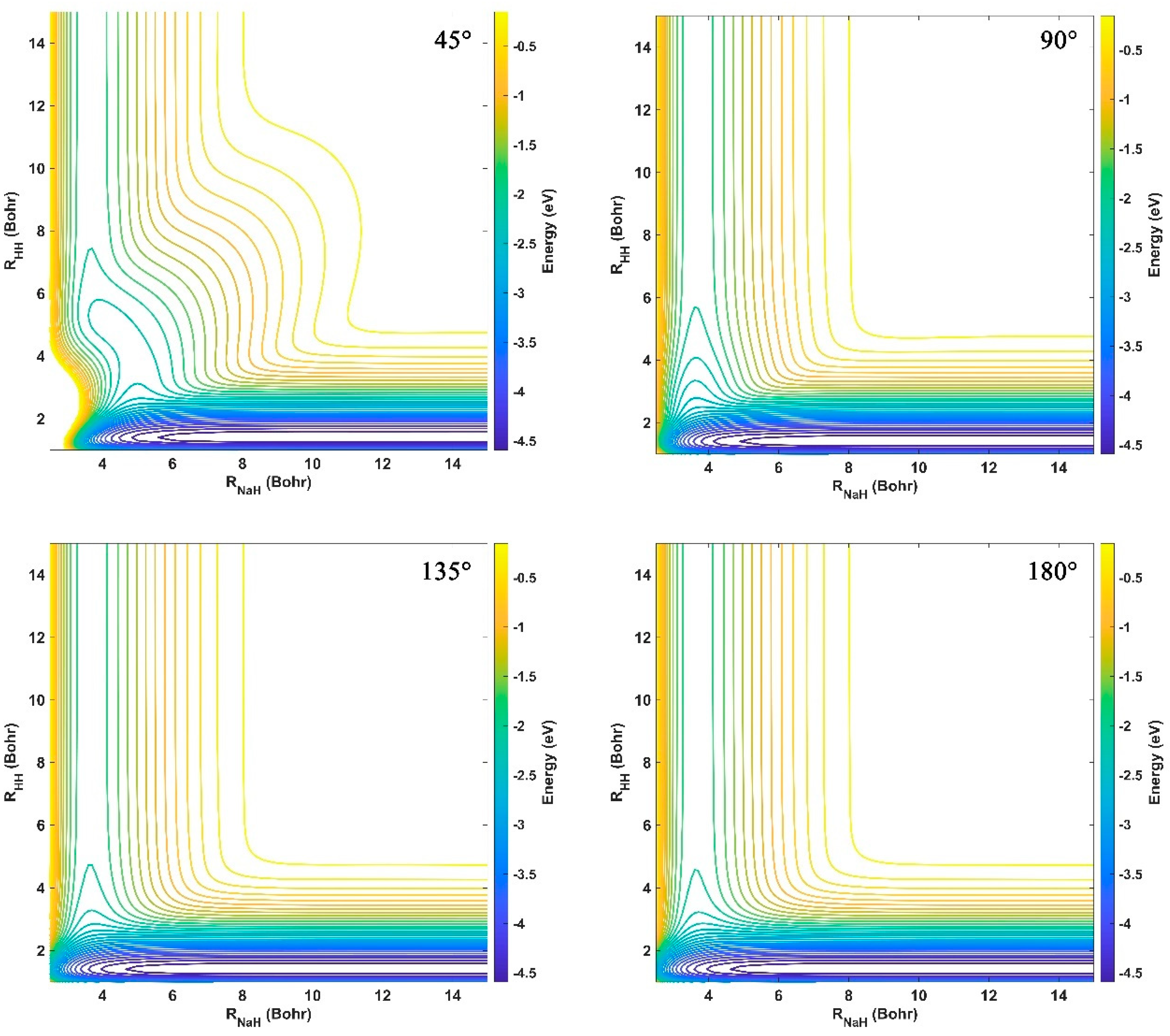
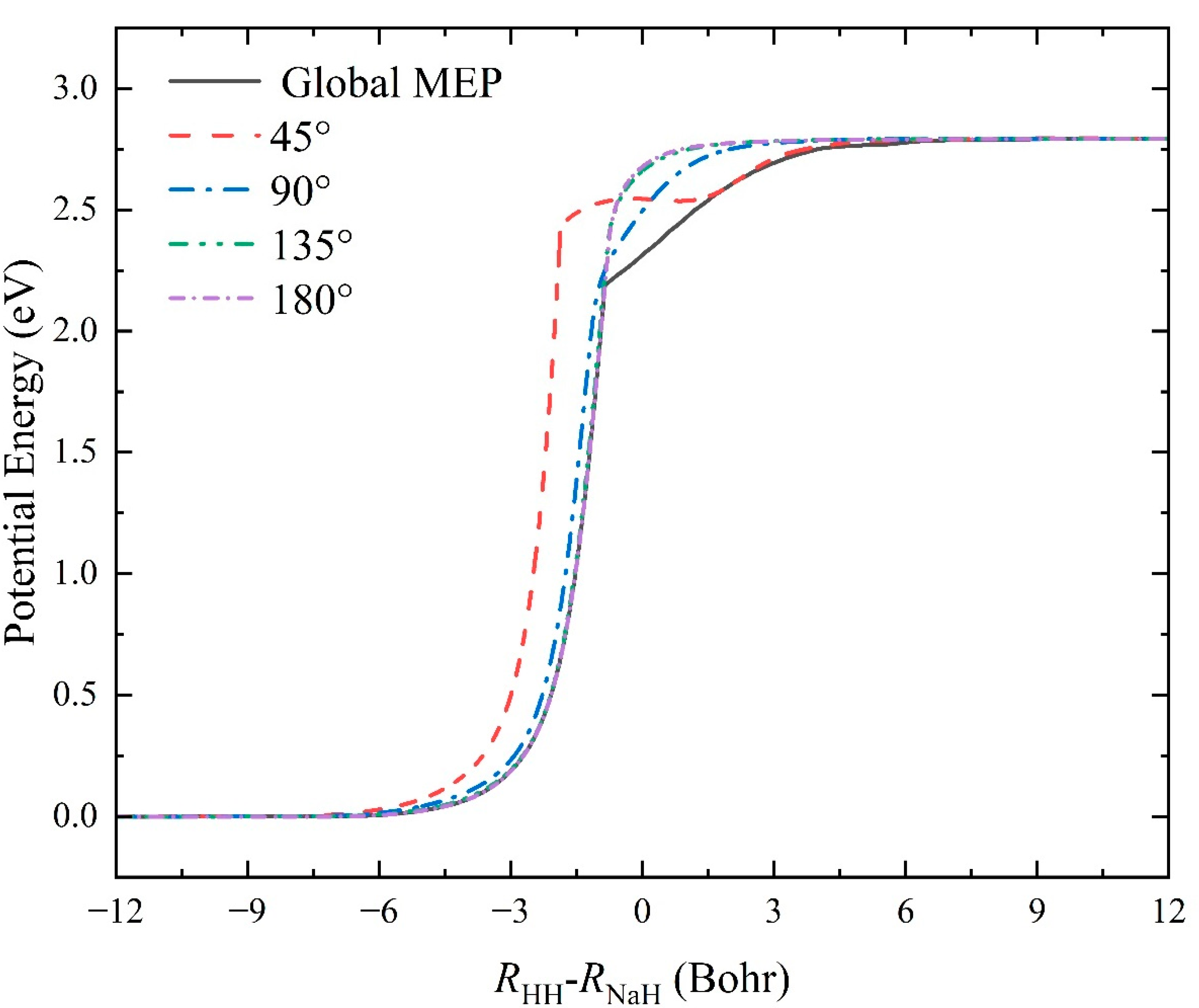
2.1. Characteristics of the Ground-State NaH2 PES
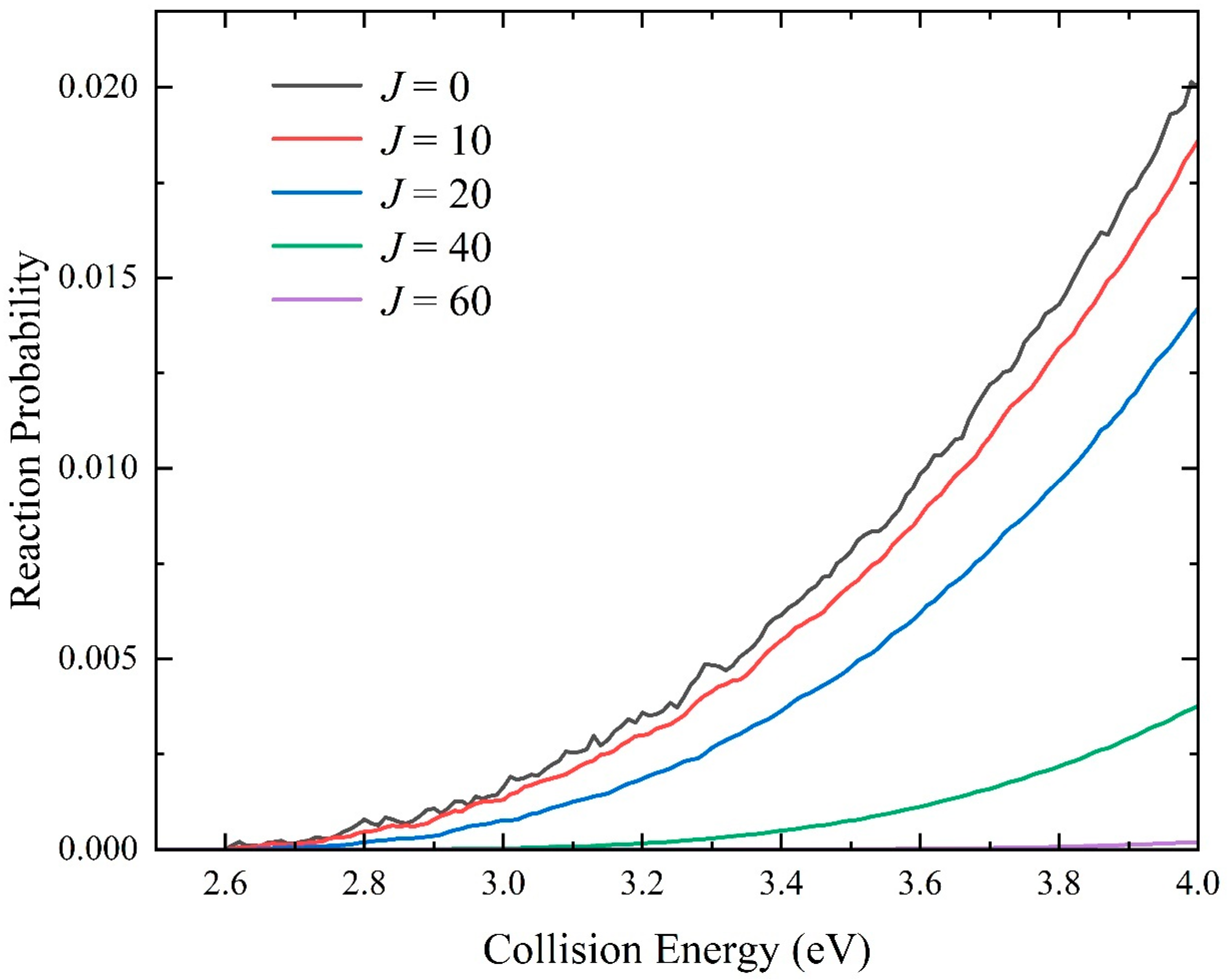
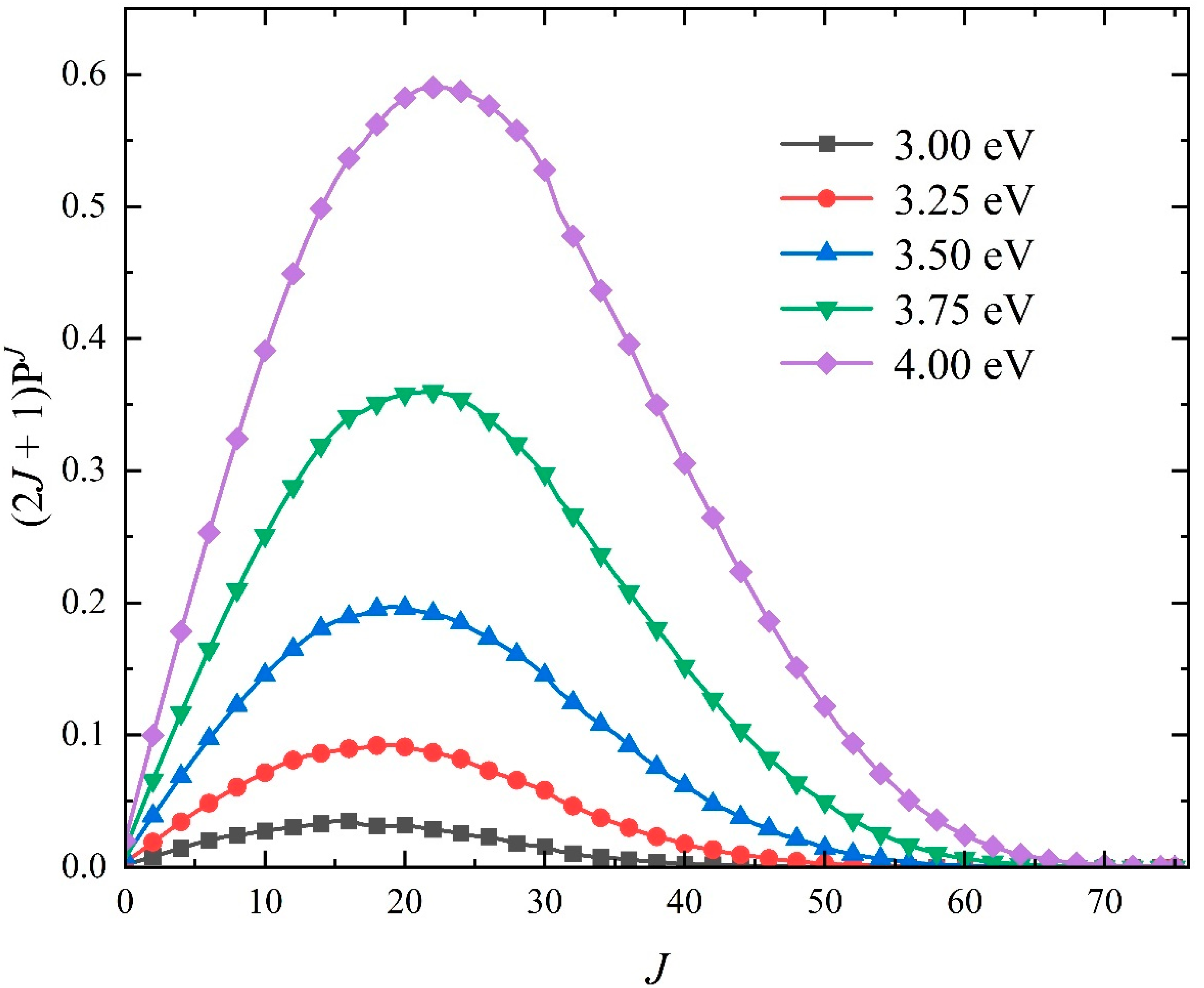
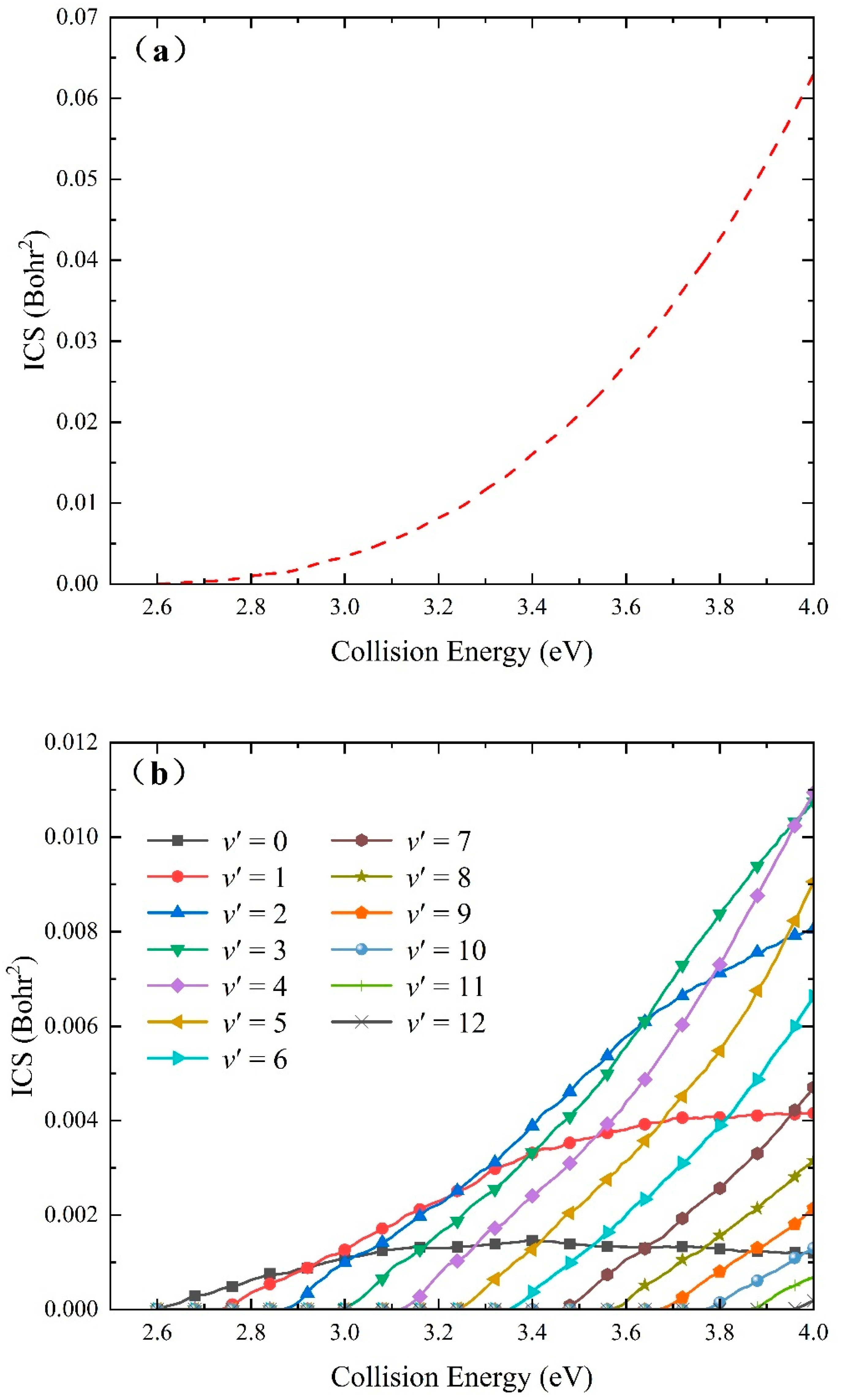
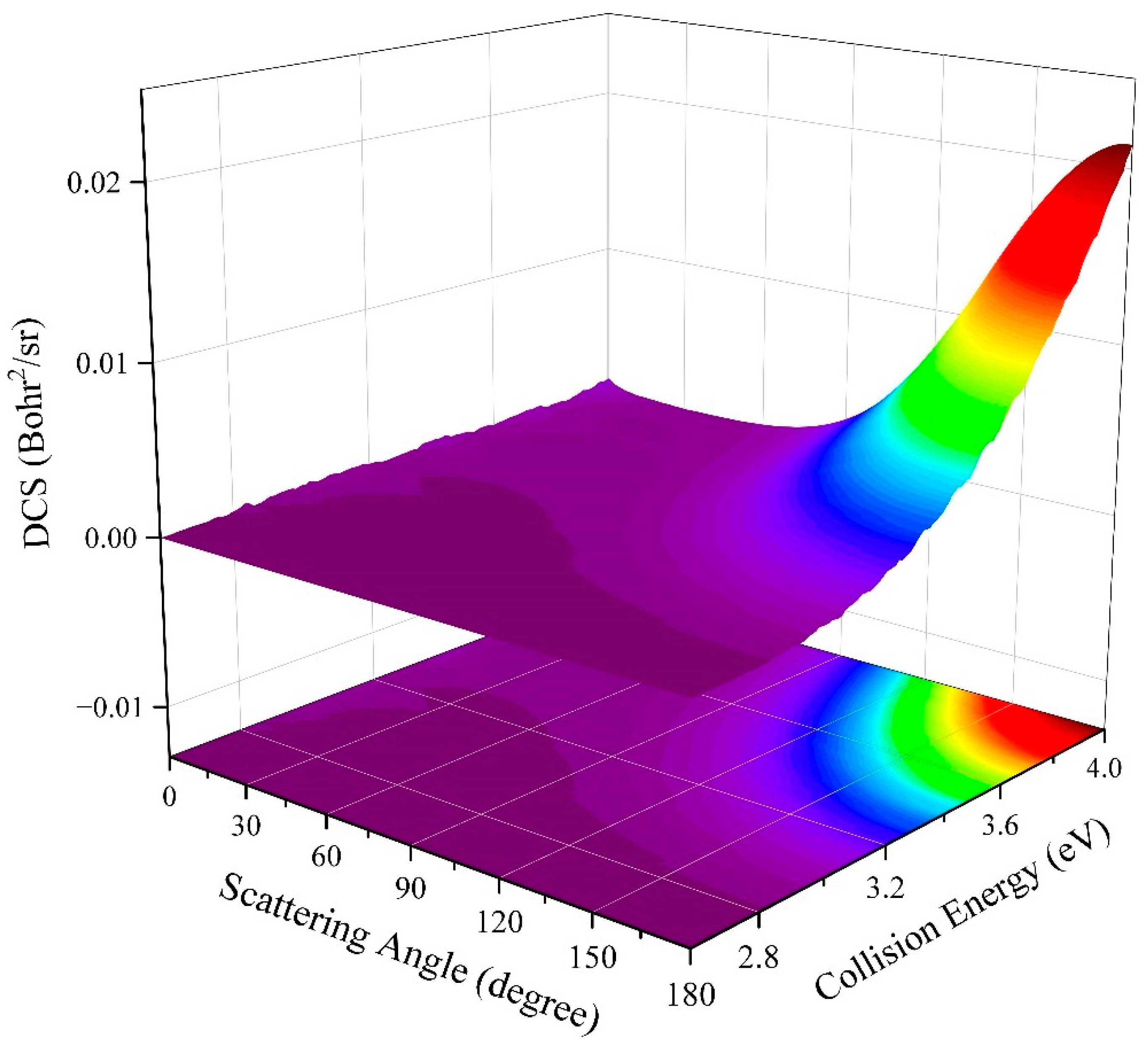
2.2. Quantum Dynamics Calculations
3. Theory and Method
3.1. Ab Initio Calculations
3.2. PES Fitting
3.3. TDWP Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bililign, S.; Kleiber, P.D.; Kearney, W.R.; Sando, K.M. Reactive Collision Dynamics of Na*(4 2P) + H2 and HD: Experiment and Theory. J. Chem. Phys. 1992, 96, 218–229. [Google Scholar] [CrossRef]
- Chen, J.J.; Hung, Y.M.; Liu, D.K.; Fung, H.S.; Lin, K.C. Reaction pathway, energy barrier, and rotational state distribution for Li(22PJ) + H2 → LiH(X1Σ+) + H. J. Chem. Phys. 2001, 114, 9395–9401. [Google Scholar] [CrossRef]
- Lee, H.S.; Lee, Y.S.; Jeung, G.H. Potential energy surfaces for LiH2 and photochemical reactions Li* + H2 ⇆ LiH + H. J. Phys. Chem. A 1999, 103, 11080–11088. [Google Scholar] [CrossRef]
- Martinez, T.J. Ab initio molecular dynamics around a conical intersection: Li(2p)+H2. Chem. Phys. Lett. 1997, 272, 139–147. [Google Scholar] [CrossRef]
- Wong, T.H.; Kleiber, P.D.; Yang, K.H. Chemical dynamics of the reaction K*(5p2P) + H2 → KH(v = 0; J) + H: Electronic orbital alignment effects. J. Chem. Phys. 1999, 110, 6743–6748. [Google Scholar] [CrossRef]
- Li, W.T.; Wang, X.M.; Zhao, H.L.; He, D. Non-adiabatic dynamics studies of the K(4p2P) + H2(X1Σ+g) reaction based on new diabatic potential energy surfaces. Phys. Chem. Chem. Phys. 2020, 22, 16203–16214. [Google Scholar] [CrossRef]
- Fan, L.H.; Chen, J.J.; Lin, Y.Y.; Luh, W.T. Reaction of Rb(52D, 72S) with H2. J. Phys. Chem. A 1999, 103, 1300–1305. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, J.Z.; Xing, G.Q.; Wang, X.B.; Bersohn, R. The reaction of Cs(82P) and Cs(92P) with hydrogen molecules. J. Chem. Phys. 1996, 104, 1338–1343. [Google Scholar] [CrossRef]
- Bililign, S.; Kleiber, P.D. Nascent Rotational Quantum State Distribution of NaH (NaD) from the Reaction of Na*(42P) with H2, D2, and HD. J. Chem. Phys. 1992, 96, 213–217. [Google Scholar] [CrossRef]
- Motzkus, M.; Pichler, G.; Kompa, K.L.; Hering, P. Comparison of the Na(4p) + H2 and Na(3p) + H2 reactive/quenching systems studied with CARS, resonance-enhanced CARS, and DFWM. J. Chem. Phys. 1997, 106, 9057–9066. [Google Scholar] [CrossRef]
- Chang, Y.P.; Hsiao, M.K.; Liu, D.K.; Lin, K.C. Rotational and vibrational state distributions of NaH in the reactions of Na (42S, 32D, and 62S) H2: Insertion versus harpoon-type mechanisms. J. Chem. Phys. 2008, 128, 234309. [Google Scholar] [CrossRef] [PubMed]
- Botschwina, P.; Meyer, W.; Hertel, I.V.; Reiland, W. Collisions of Excited Na Atoms with H2 Molecules. Ⅰ. Ab initio Potential-Energy Surfaces and Qualitative Discussion of the Quenching Process. J. Chem. Phys. 1981, 75, 5438–5448. [Google Scholar] [CrossRef]
- Truhlar, D.G.; Duff, J.W.; Blais, N.C.; Tully, J.C.; Garrett, B.C. The Quenching of Na(32P) by H2: Interactions and Dynamics. J. Chem. Phys. 1982, 77, 764–776. [Google Scholar] [CrossRef]
- Blais, N.C.; Truhlar, D.G.; Garrett, B.C. Improved Parametrization of Diatomics-in-Molecules Potential Energy Surface for Na(3p2P)+H2 → Na(3s2S) + H2. J. Chem. Phys. 1983, 78, 2956–2961. [Google Scholar] [CrossRef]
- Yarkony, D.R. On the Reaction Na(2P) + H2 → Na(2S)+ H2 Nonadiabatic Effects. J. Chem. Phys. 1986, 84, 3206–3211. [Google Scholar] [CrossRef]
- Halvick, P.; Truhlar, D.G. A New Diabatic Representation of the Coupled Potential Energy Surfaces for Na(3p2P) + H2 → Na(3s2S)+H2 or NaH + H. J. Chem. Phys. 1992, 96, 2895–2909. [Google Scholar] [CrossRef]
- Hack, M.D.; Truhlar, D.G. Analytic potential energy surfaces and their couplings for the electronically nonadiabatic chemical processes Na(3p) + H2 → Na(3s) + H2 Na(3p) + H2 → NaH + H. J. Chem. Phys. 1999, 110, 4315–4337. [Google Scholar] [CrossRef]
- Wang, S.F.; Yang, Z.J.; Yuan, J.C.; Chen, M.D. New diabatic potential energy surfaces of the NaH2 system and dynamics studies for the Na(3p) + H2 → NaH + H reaction. Sci. Rep. 2018, 8, 17960. [Google Scholar] [CrossRef]
- Buren, B.; Yang, Z.J.; Chen, M.D. Non-adiabatic state-to-state dynamic studies of Na(3p) + H2 (ν = 1, 2, 3; j = 0) → NaH + H reactions. Chem. Phys. Lett. 2019, 723, 128–132. [Google Scholar] [CrossRef]
- Wen, Y.P.; Buren, B.; Chen, M.D. Non-adiabatic quantum dynamical studies of Na(3p) + HD(ν = 1, j = 0)→NaH/NaD + D/H reaction. Chinese Phys. B 2019, 28, 063401. [Google Scholar] [CrossRef]
- Buren, B.; Yang, Z.J.; Chen, M.D. Dynamics study on the non-adiabatic Na(3p) + HD → NaH/NaD + D/H reaction: Insertion-abstraction mechanism. Phys. Chem. Chem. Phys. 2020, 22, 3633–3642. [Google Scholar] [CrossRef] [PubMed]
- Buren, B.; Chen, M.D. Stereodynamics-Controlled Product Branching in the Nonadiabatic H + NaD → Na(3s, 3p) + HD Reaction at Low Temperatures. J. Phys. Chem. A 2022, 126, 2453–2462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Zhu, Z.L.; Li, W.T. The quenching processes of the Na(3p) + H2 → Na(3s) + H2 reaction studied using the time-dependent wave packet method. Mol. Phys. 2022, 120, e2083713. [Google Scholar] [CrossRef]
- Bai, Y.W.; Buren, B.; Chen, M.D. A comparative study on adiabatic and nonadiabatic dynamics of the H(2S) + NaH(X1Σ+) reaction. Chem. Phys. 2024, 583, 112324. [Google Scholar] [CrossRef]
- Huber, K.P.; Herzberf, G. Constants of Diatomic Molecules; Springer: Boston, MA, USA, 1979. [Google Scholar]
- Stwalley, W.C.; Zemke, W.T.; Yang, S.C. Spectroscopy and Structure of the Alkali Hydride Diatomic-Molecules and Their Ions. J. Phys. Chem. Ref. Data 1991, 20, 153–187. [Google Scholar] [CrossRef]
- Knowles, P.J.; Werner, H.J. An Efficient Method for the Evaluation of Coupling-Coefficients in Configuration-Interaction Calculations. Chem. Phys. Lett. 1988, 145, 514–522. [Google Scholar] [CrossRef]
- Werner, H.J.; Knowles, P.J. An Efficient Internally Contracted Multiconfiguration Reference Configuration-Interaction Method. J. Chem. Phys. 1988, 89, 5803–5814. [Google Scholar] [CrossRef]
- Kendall, R.A.; Dunning, T.H.; Harrison, R.J. Electron Affinities of the First-row Atoms Revisited. Systematic Basis Sets and Wave Functions. J. Chem. Phys. 1992, 96, 6796–6806. [Google Scholar] [CrossRef]
- Werner, H.J.; Knowles, P.J. A Second Order Multiconfiguration SCF Procedure with Optimum Convergence. J. Chem. Phys. 1985, 82, 5053–5063. [Google Scholar] [CrossRef]
- Knowles, P.J.; Werner, H.J. An Efficient 2nd-Order Mc SCF Method for Long Configuration Expansions. Chem. Phys. Lett. 1985, 115, 259–267. [Google Scholar] [CrossRef]
- Werner, H.J.; Knowles, P.J.; Knizia, G.; Manby, F.R.; Schutz, M. Molpro: A general-purpose quantum chemistry program package. WIREs Comput. Mol. Sci. 2012, 2, 242–253. [Google Scholar] [CrossRef]
- Jiang, B.; Li, J.; Guo, H. High-Fidelity Potential Energy Surfaces for Gas-Phase and Gas-Surface Scattering Processes from Machine Learning. J. Phys. Chem. Lett. 2020, 11, 5120–5131. [Google Scholar] [CrossRef] [PubMed]
- Manzhos, S.; Carrington, T. Neural Network Potential Energy Surfaces for Small Molecules and Reactions. Chem. Rev. 2021, 121, 10187–10217. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.N.; Zhang, S.H.; Ranasinghe, K.D.; Isayev, O.; Roitberg, A.E. Machine Learning of Reactive Potentials. Annu. Rev. Phys. Chem. 2024, 75, 371–395. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.N.; Zhang, D.H. Accurate fundamental invariant-neural network representation of ab initio potential energy surfaces. Natl. Sci. Rev. 2023, 10, nwad321. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.L.; Lu, X.X.; Han, Y.C.; Fu, B.N.; Zhang, D.H.; Bowman, J.M. Collision-induced and complex-mediated roaming dynamics in the H + C2H4 → H2 + C2H3 reaction. Chem. Sci. 2020, 11, 2148–2154. [Google Scholar] [CrossRef]
- Qin, J.; Liu, Y.; Lu, D.D.; Li, J. Theoretical Study for the Ground Electronic State of the Reaction OH + SO → H + SO. J. Phys. Chem. A 2019, 123, 7218–7227. [Google Scholar] [CrossRef]
- Williams, D.M.G.; Eisfeld, W. Neural network diabatization: A new ansatz for accurate high-dimensional coupled potential energy surfaces. J. Chem. Phys. 2018, 149, 204106. [Google Scholar] [CrossRef]
- Xie, C.J.; Zhu, X.L.; Yarkony, D.R.; Guo, H. Permutation invariant polynomial neural network approach to fitting potential energy surfaces. IV. Coupled diabatic potential energy matrices. J. Chem. Phys. 2018, 149, 144107. [Google Scholar] [CrossRef]
- Lin, Q.D.; Zhang, L.; Zhang, Y.L.; Jiang, B. Searching Configurations in Uncertainty Space: Active Learning of High-Dimensional Neural Network Reactive Potentials. J. Chem. Theory Comput. 2021, 17, 2691–2701. [Google Scholar] [CrossRef]
- Xie, Y.R.; Zhao, H.L.; Wang, Y.F.; Huang, Y.; Wang, T.; Xu, X.; Xiao, C.L.; Sun, Z.G.; Zhang, D.H.; Yang, X.M. Quantum interference in H + HD → H2 + D between direct abstraction and roaming insertion pathways. Science 2020, 368, 767–771. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.J.; Wang, S.F.; Yuan, J.C.; Chen, M.D. Neural network potential energy surface and dynamical isotope effects for the N+(3p) + H2 → NH+ + H reaction. Phys. Chem. Chem. Phys. 2019, 21, 22203–22214. [Google Scholar] [CrossRef] [PubMed]
- Biswas, R.; Gianturco, F.A.; Giri, K.; González-Sánchez, L.; Lourderaj, U.; Sathyamurthy, N.; Yurtsever, E. An improved artificial neural network fit of the ab initio potential energy surface points for HeH+ + H2 and its ensuing rigid rotors quantum dynamics. Artif. Intell. Chem. 2023, 1, 10007. [Google Scholar] [CrossRef]
- Song, K.S.; Song, H.W.; Li, J. Validating experiments for the reaction H2 + NH2− by dynamical calculations on an accurate full-dimensional potential energy surface. Phys. Chem. Chem. Phys. 2022, 24, 10160–10167. [Google Scholar] [CrossRef]
- Yang, Z.J.; Cao, F.R.; Cheng, H.Y.; Liu, S.W.; Sun, J.C. A Globally Accurate Neural Network Potential Energy Surface and Quantum Dynamics Studies on Be+(2S) + H2/D2 → BeH+/BeD+ + H/D Reactions. Molecules 2024, 29, 3436. [Google Scholar] [CrossRef]
- Buren, B.; Zhang, J.P.; Li, Y.Q. Quantum Dynamics Studies of the Li + Na(v = 0, j = 0) → Na + NaLi Reaction on a New Neural Network Potential Energy Surface. J. Phys. Chem. A 2024, 128, 5115–5127. [Google Scholar] [CrossRef]
- Goodlett, S.M.; Turney, J.M.; Schaefer, H.F. Comparison of multifidelity machine learning models for potential energy surfaces. J. Chem. Phys. 2023, 159, 044111. [Google Scholar] [CrossRef]
- Tao, C.; Yang, J.W.; Hong, Q.Z.; Sun, Q.H.; Li, J. Global and Full-Dimensional Potential Energy Surfaces of the N2+O2 Reaction for Hyperthermal Collisions. J. Phys. Chem. A 2023, 127, 4027–4042. [Google Scholar] [CrossRef]
- Braams, B.J.; Bowman, J.M. Permutationally invariant potential energy surfaces in high dimensionality. Int. Rev. Phys. Chem. 2009, 28, 577–606. [Google Scholar] [CrossRef]
- Jiang, B.; Guo, H. Permutation invariant polynomial neural network approach to fitting potential energy surfaces. J. Chem. Phys. 2013, 139, 054112. [Google Scholar] [CrossRef]
- Qu, C.; Yu, Q.; Bowman, J.M. Permutationally Invariant Potential Energy Surfaces. Annu. Rev. Phys. Chem. 2018, 69, 151–175. [Google Scholar] [CrossRef] [PubMed]
- Hagan, M.T.; Menhaj, M.B. Training Feedforward Networks with the Marquardt Algorithm. IEEE Trans. Neural Netw. 1994, 5, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.G.; Lee, S.Y.; Guo, H.; Zhang, D.H. Comparison of second-order split operator and Chebyshev propagator in wave packet based state-to-state reactive scattering calculations. J. Chem. Phys. 2009, 130, 174102. [Google Scholar] [CrossRef]
- Sun, Z.G.; Guo, H.; Zhang, D.H. Extraction of state-to-state reactive scattering attributes from wave packet in reactant Jacobi coordinates. J. Chem. Phys. 2010, 132, 084112. [Google Scholar] [CrossRef] [PubMed]
- Buren, B.; Chen, M.D.; Sun, Z.G.; Guo, H. Quantum Wave Packet Treatment of Cold Nonadiabatic Reactive Scattering at the State-To-State Level. J. Phys. Chem. A 2021, 125, 10111–10120. [Google Scholar] [CrossRef]
- Huang, J.Y.; Liu, S.; Zhang, D.H.; Krems, R.V. Time-Dependent Wave Packet Dynamics Calculations of Cross Sections for Ultracold Scattering of Molecules. Phys. Rev. Lett. 2018, 120, 143401. [Google Scholar] [CrossRef]
- Sun, Z.G.; Lin, X.; Lee, S.Y.; Zhang, D.H. A Reactant-Coordinate-Based Time-Dependent Wave Packet Method for Triatomic State-to-State Reaction Dynamics: Application to the H + O2 Reaction. J. Phys. Chem. A 2009, 113, 4145–4154. [Google Scholar] [CrossRef]
- Gómez-Carrasco, S.; Roncero, O. Coordinate transformation methods to calculate state-to-state reaction probabilities with wave packet treatments. J. Chem. Phys. 2006, 125, 054102. [Google Scholar] [CrossRef]
- Feit, M.D.; Fleck, J.A.; Steiger, A. Solution of the Schrödinger-equation by a spectral method. J. Comput. Phys. 1982, 47, 412–433. [Google Scholar] [CrossRef]

| NN PES | Experimental Data | ||
|---|---|---|---|
| H2(X1Σg+) | Re (Bohr) | 1.402 | 1.401 |
| De (eV) | 4.743 | 4.747 | |
| ωe (cm−1) | 4400.1 | 4401.2 | |
| ωexe (cm−1) | 120.3 | 121.3 | |
| NaH(X1Σ+) | Re (Bohr) | 3.639 | 3.566 |
| De (eV) | 1.948 | 1.971 | |
| ωe (cm−1) | 1170.4 | 1171.9 | |
| ωexe (cm−1) | 19.3 | 19.7 | |
| Na(2S) + H2(v0 = 0, j0 = 0) → NaH + H | |
|---|---|
| Grid ranges and sizes | R∈[0.01,25.0], = 299, = 199 |
| r∈[0.1,25.0], = 249, = 11 | |
| = 109, = 19 | |
| Initial wave packet | Rc = 14.0 ∆R = 0.6 E0 = 0.5 eV |
| Propagation time | 15,000 |
| Highest J value | 75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Cheng, H.; Cao, F.; Sun, J.; Yang, Z. Ab Initio Neural Network Potential Energy Surface and Quantum Dynamics Calculations on Na(2S) + H2 → NaH + H Reaction. Molecules 2024, 29, 4871. https://doi.org/10.3390/molecules29204871
Liu S, Cheng H, Cao F, Sun J, Yang Z. Ab Initio Neural Network Potential Energy Surface and Quantum Dynamics Calculations on Na(2S) + H2 → NaH + H Reaction. Molecules. 2024; 29(20):4871. https://doi.org/10.3390/molecules29204871
Chicago/Turabian StyleLiu, Siwen, Huiying Cheng, Furong Cao, Jingchang Sun, and Zijiang Yang. 2024. "Ab Initio Neural Network Potential Energy Surface and Quantum Dynamics Calculations on Na(2S) + H2 → NaH + H Reaction" Molecules 29, no. 20: 4871. https://doi.org/10.3390/molecules29204871
APA StyleLiu, S., Cheng, H., Cao, F., Sun, J., & Yang, Z. (2024). Ab Initio Neural Network Potential Energy Surface and Quantum Dynamics Calculations on Na(2S) + H2 → NaH + H Reaction. Molecules, 29(20), 4871. https://doi.org/10.3390/molecules29204871







