Ruthenium Complexes with Pyridazine Carboxylic Acid: Synthesis, Characterization, and Anti-Biofilm Activity
Abstract
:1. Introduction
2. Results and Discussion
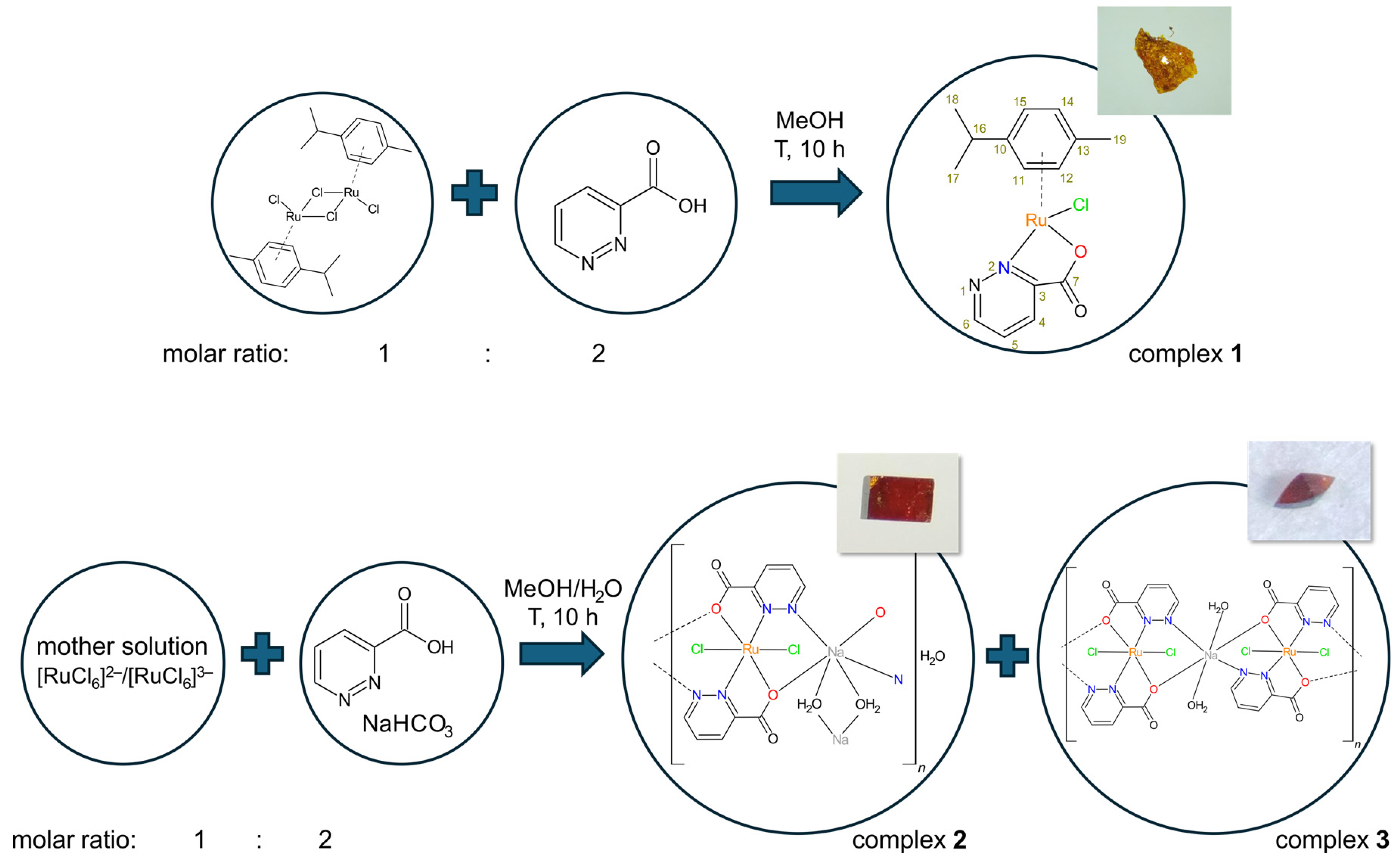
2.1. Preparation of the Complexes
2.2. Spectroscopic Characterization and Magnetic Measurements
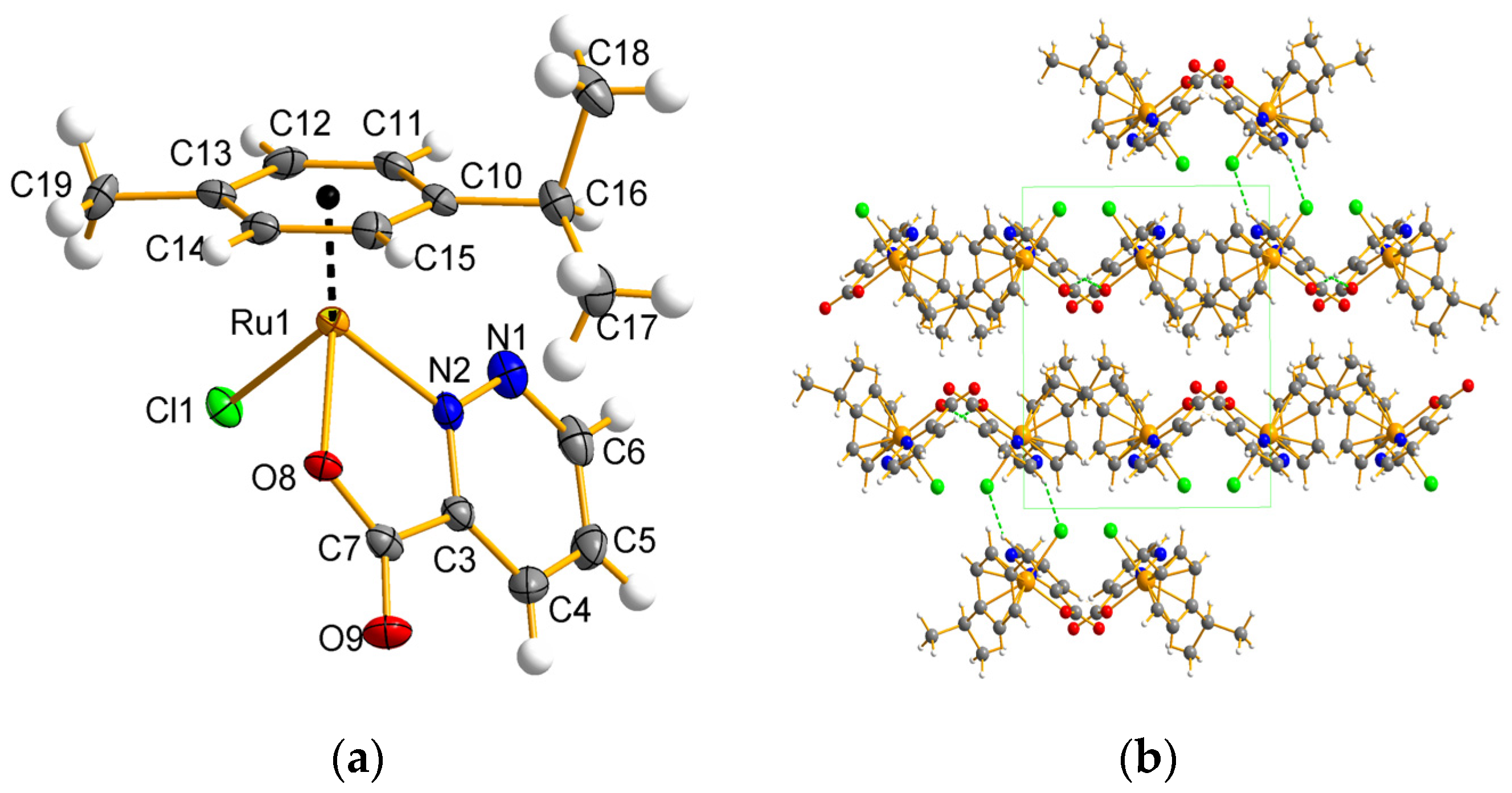
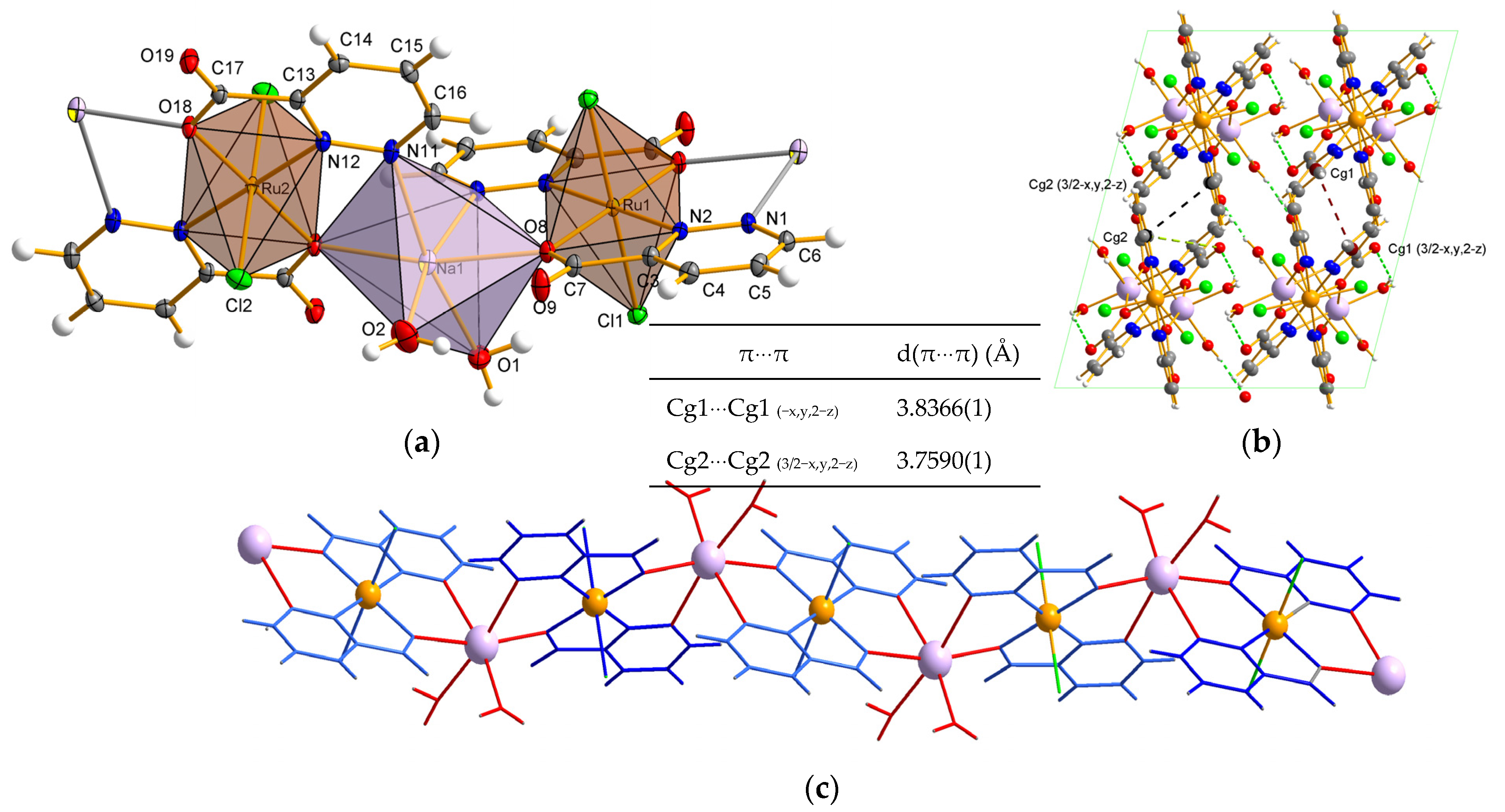
2.3. Molecular and Crystal Structure Description for Ru Complexes in Different Oxidation States
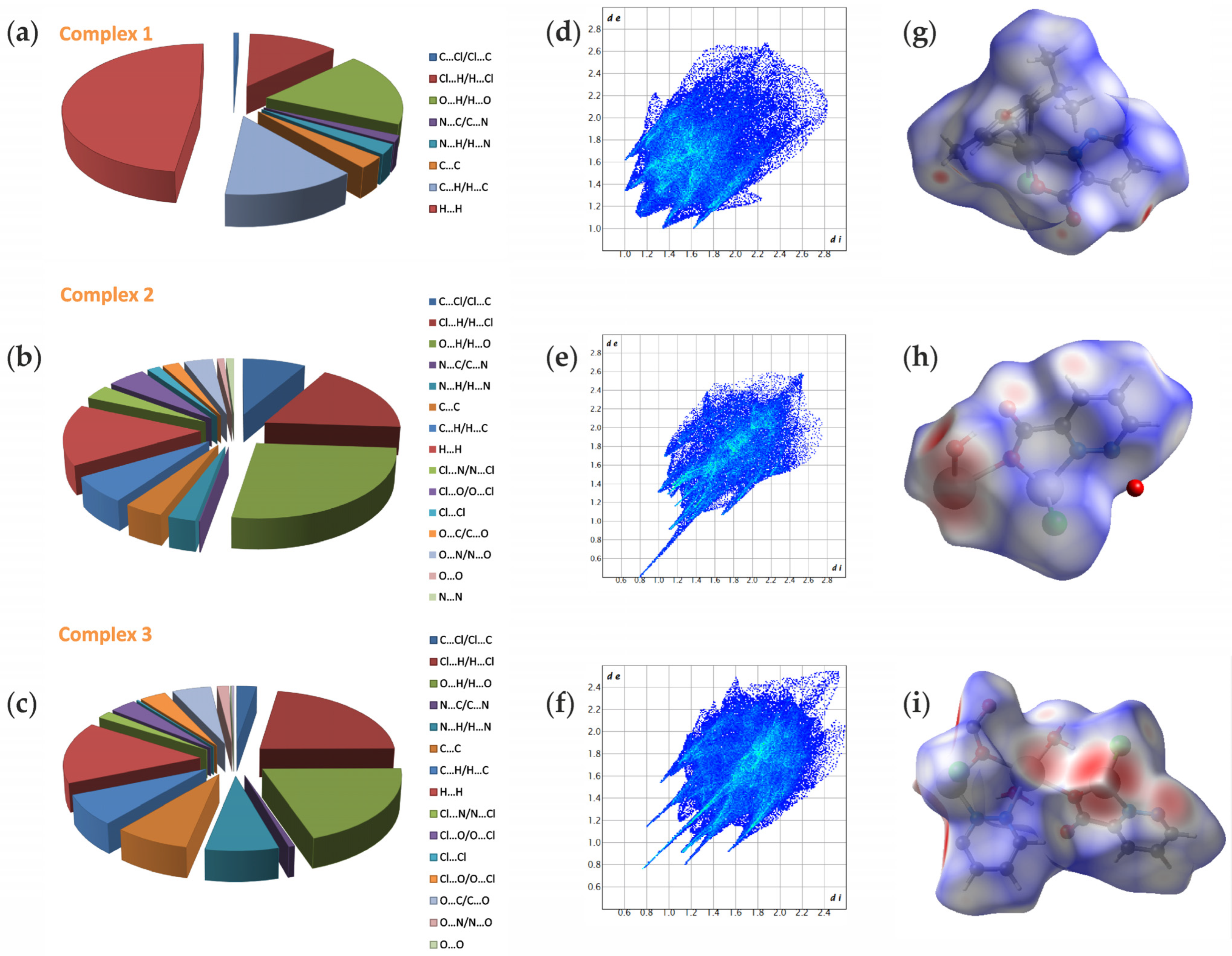
2.4. Hirshfeld Surface Analysis (HS)
2.5. Electrochemical Studies
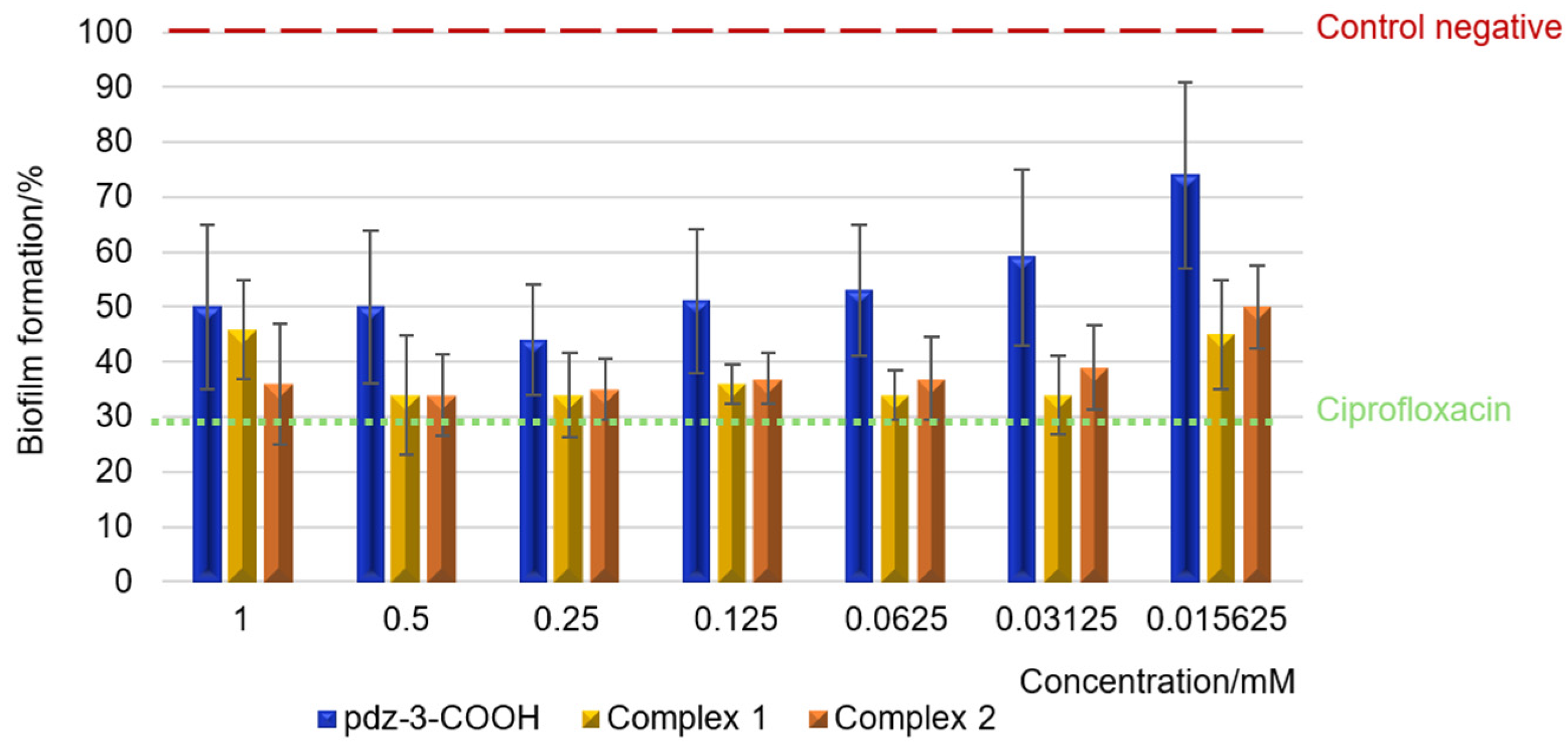
2.6. Minimum Inhibitory Concentration and Biofilm Biomass Quantification
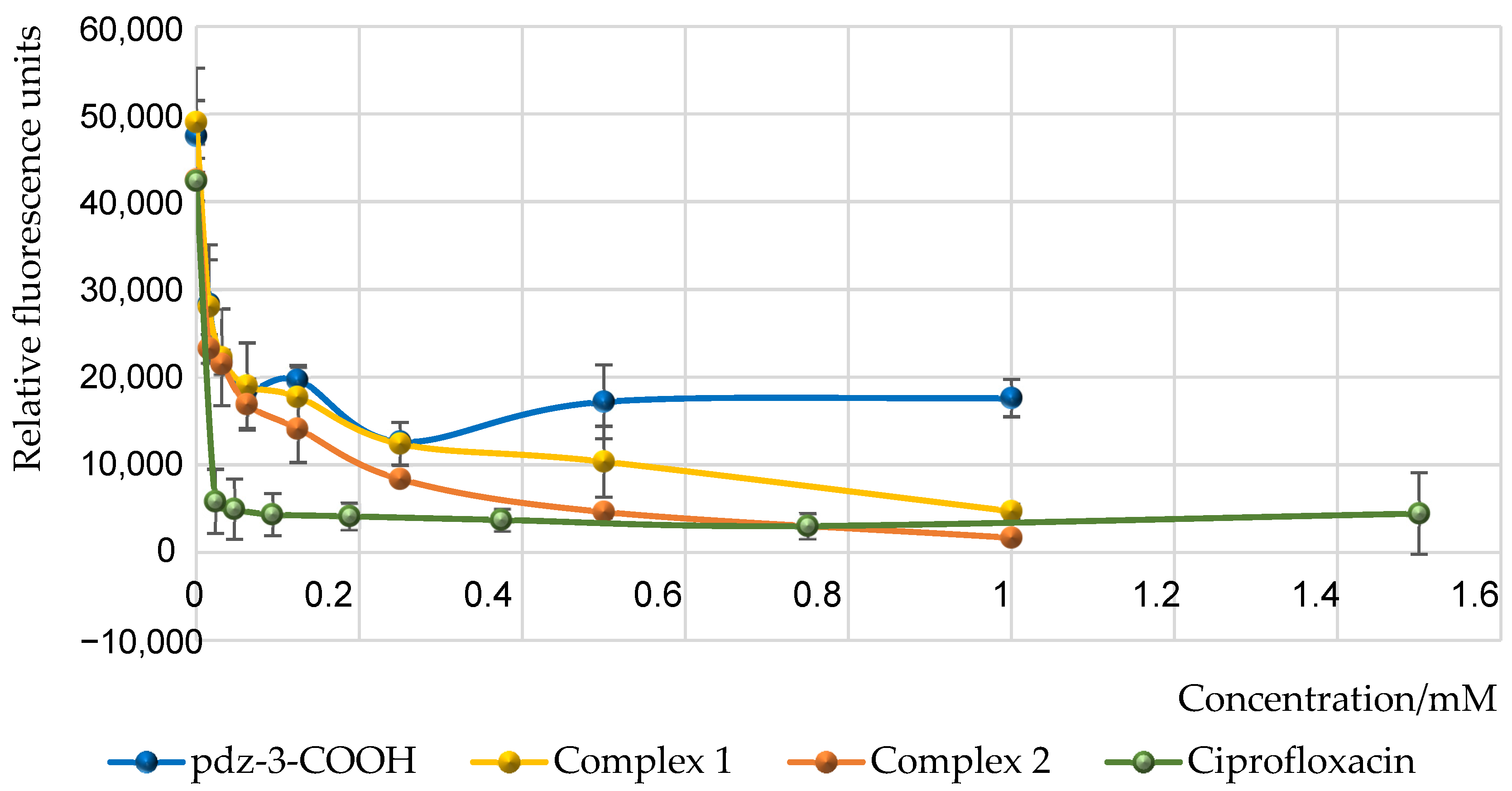
2.7. Pyoverdine Inhibition Assay
2.8. Cytotoxicity Activity
2.9. HSA Binding Studies
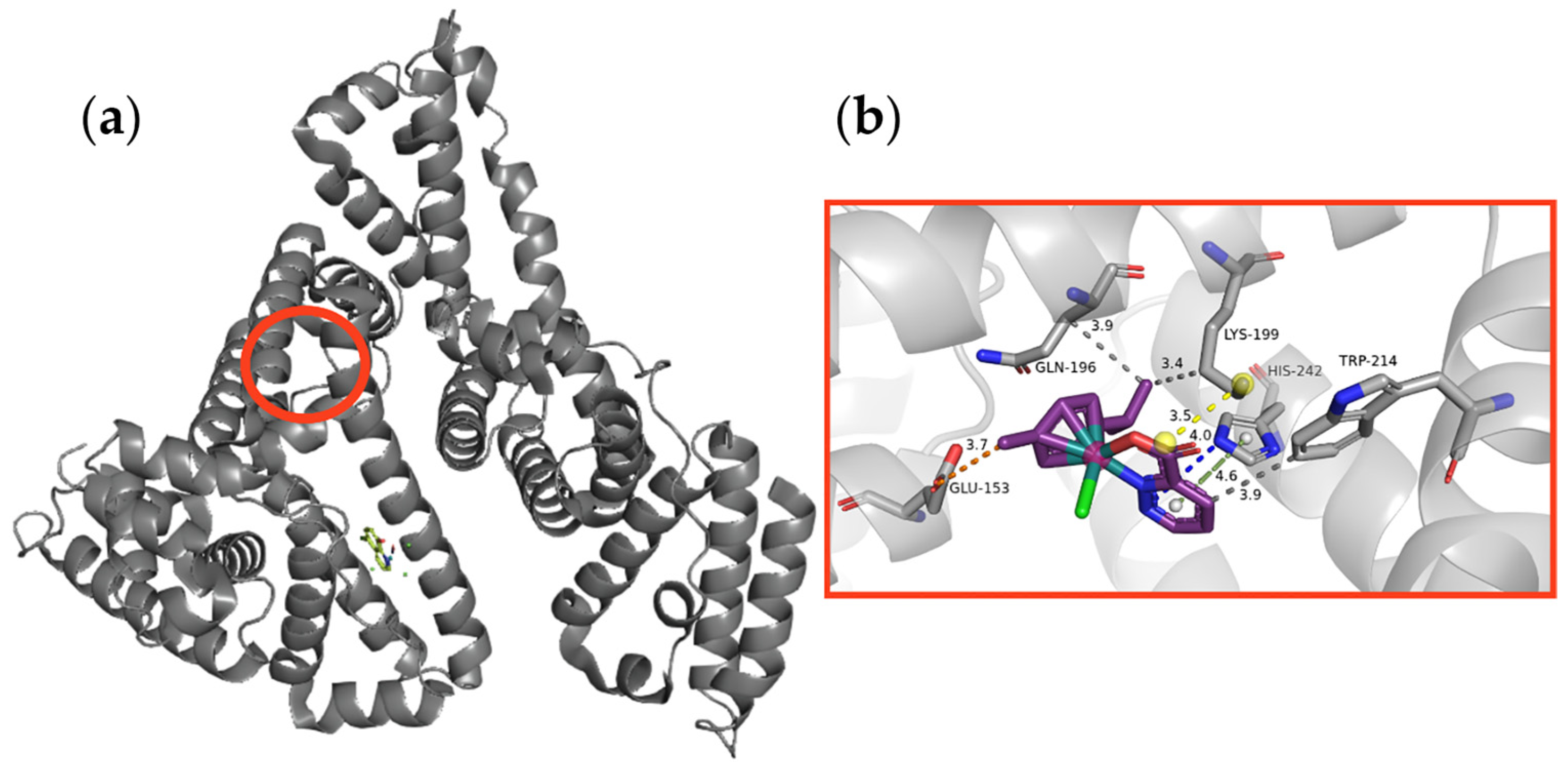
2.9.1. Molecular Docking of the Studied Ruthenium Complexes to HSA
2.9.2. Human Serum Albumin Fluorescence Quenching Assay
2.10. In Silico Pharmacokinetic and Drug-Likeness Predictions (ADME) for Ruthenium Complexes
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jabłońska-Wawrzycka, A.; Rogala, P.; Michałkiewicz, S.; Hodorowicz, M.; Barszcz, B. Ruthenium complexes in different oxidation states: Synthesis, crystal structure, spectra and redox properties. Dalton Trans. 2013, 42, 6092–6101. [Google Scholar] [CrossRef] [PubMed]
- Rogala, P.; Jabłońska-Wawrzycka, A.; Kazimierczuk, K.; Borek, A.; Błażejczyk, A.; Wietrzyk, J.; Barszcz, B. Synthesis, crystal structure and cytotoxic activity of ruthenium(II) piano-stool complex with N,N-chelating ligand. J. Mol. Struct. 2016, 1126, 74–82. [Google Scholar] [CrossRef]
- Rogala, P.; Czerwonka, G.; Michałkiewicz, S.; Hodorowicz, M.; Barszcz, B.; Jabłońska-Wawrzycka, A. Synthesis, structural characterization and antimicrobial evaluation of ruthenium complexes with heteroaromatic carboxylic acids. Chem. Biodivers. 2019, 16, e1900403. [Google Scholar] [CrossRef] [PubMed]
- Czerwonka, G.; Gmiter, D.; Guzy, A.; Rogala, P.; Jabłońska-Wawrzycka, A.; Borkowski, A.; Cłapa, T.; Narożna, D.; Kowalczyk, P.; Syczewski, M.; et al. A benzimidazole-based ruthenium(IV) complex inhibits Pseudomonas aeruginosa biofilm formation by interacting with siderophores and the cell envelope, and inducing oxidative stress. Biofouling 2019, 35, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Jabłońska-Wawrzycka, A.; Rogala, P.; Czerwonka, G.; Michałkiewicz, S.; Hodorowicz, M.; Kowalczyk, P. Ruthenium(IV) Complexes as Potential Inhibitors of Bacterial Biofilm Formation. Molecules 2020, 25, 4938. [Google Scholar] [CrossRef] [PubMed]
- Jabłońska-Wawrzycka, A.; Rogala, P.; Czerwonka, G.; Gałczyńska, K.; Drabik, M.; Dańczuk, M. Ruthenium Complexes with 2-Pyridin-2-yl-1H-benzimidazole as Potential Antimicrobial Agents: Correlation between Chemical Properties and Anti-Biofilm Effects. Int. J. Mol. Sci. 2021, 22, 10113. [Google Scholar] [CrossRef] [PubMed]
- Rogala, P.; Jabłońska-Wawrzycka, A.; Czerwonka, G.; Kazimierczuk, K.; Gałczyńska, K.; Michałkiewicz, S.; Kalinowska-Tłuścik, J.; Karpiel, M.; Klika, K.D. Synthesis, Characterization and Biological Investigations of Half-Sandwich Ruthenium(II) Complexes Containing Benzimidazole Moiety. Molecules 2023, 28, 40. [Google Scholar] [CrossRef] [PubMed]
- Figgis, B.N.; Nyholm, R.S. A convenient solid for calibration of the Gouy magnetic susceptibility apparatus. J. Chem. Soc. 1958, 4190, 1. [Google Scholar]
- Munteanu, A.-C.; Uivarosi, V. Ruthenium complexes in the fight against pathogenic microorganisms. An extensive review. Pharmaceutics 2021, 13, 874. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, C.Y.K.; Nam, T.-G. Ruthenium Complexes as Anticancer Agents: A Brief History and Perspectives. Drug Des. Devel. Ther. 2020, 14, 5375–5392. [Google Scholar] [CrossRef] [PubMed]
- Kanaoujiya, R.; Meenakshi, R.; Srivastava, S.; Singh, R.; Mustafa, G. Recent advances and application of ruthenium complexes in tumor malignancy. Mater. Today Proc. 2023, 72, 2822–2827. [Google Scholar] [CrossRef]
- Ardiwlnata, E.S.; Craig, D.C.; Phillips, D.J. Metal complexes of 3-pyridazinecarboxylic acid (Hpdc). The X-ray crystal structure of [Mn(pdc)2(H2O)2]·2H2O and a structural comparison between equivalent pyridine and pyridazine systems. Inorg. Chim. Acta 1989, 166, 233–238. [Google Scholar] [CrossRef]
- Gryz, M.; Starosta, W.; Ptasiewicz-Bąk, H.; Leciejewicz, J. Crystal and molecular structure of pyridazine-3-carboxylic acid hydrochloride and zinc(II) pyridazine-3-carboxylate tetrahydrate. J. Coord. Chem. 2003, 56, 1505–1511. [Google Scholar] [CrossRef]
- Gryz, M.; Starosta, W.; Leciejewicz, J. Diaqua-trans-bis(pyridazine-3-carboxylato-κ2O,N)zinc(II). Acta Crystallogr. E Struct. Rep. Online 2004, 60, m1481–m1483. [Google Scholar] [CrossRef]
- Gryz, M.; Starosta, W.; Leciejewicz, J. trans-Diaquabis(pyridazine-3-carboxylato-κ2N,O)magnesium(II) dihydrate. Acta Crystallogr. E Struct. Rep. Online 2006, 62, m123–m124. [Google Scholar] [CrossRef]
- Starosta, W.; Leciejewicz, J. trans-Diaqua(pyridazine-3-carboxylato-κ2N2,O)lithium. Acta Crystallogr. E Struct. Rep. Online 2011, 67, m202. [Google Scholar] [CrossRef] [PubMed]
- Artetxe, B.; Reinoso, S.; San Felices, L.; Martín-Caballero, J.; Gutiérrez-Zorrilla, J.M. trans-Diaquabis(pyridazine-3-carboxylato-κ2N2,O)cobalt(II) dihydrate. Acta Crystallogr. E Struct. Rep. Online 2013, 69, m420–m421. [Google Scholar] [CrossRef]
- Pache, A.; Iturrospe, A.; San Felices, L.; Reinoso, S.; Gutiérrez-Zorrilla, J.M. trans-Diaquabis(pyridazine-3-carboxylato-κ2N2,O)copper(II). Acta Crystallogr. E Struct. Rep. Online 2014, 70, m114–m115. [Google Scholar] [CrossRef] [PubMed]
- Świderski, G.; Łyszczek, R.; Wojtulewski, S.; Kalinowska, M.; Świsłocka, R.; Lewandowski, W. Comparison of structural, spectroscopic, theoretical and thermal properties of metal complexes (Zn(II), Mn(II), Cu(II), Ni(II) and Co(II)) of pyridazine-3-carboxylic acid and pyridazine-4-carboxylic acids. Inorg. Chim. Acta 2020, 512, 119865. [Google Scholar] [CrossRef]
- Starosta, W.; Leciejewicz, J. Bis(μ-pyridazine-3-carboxylato)-κ3N,O:O;κ3O:N,O.-bis[triaquacalcium(II)]. Acta Crystallogr. E Struct. Rep. Online 2007, 63, m1662–m1663. [Google Scholar] [CrossRef]
- Leciejewicz, J.; Starosta, W. Bis(μ-pyridazine-3-carboxylato-κ2O:O′)-bis[aquadioxido(pyridazine-3-carboxylato-κ2N2,O)uranium(VI)] dihydrate. Acta Crystallogr. E Struct. Rep. Online 2009, 65, m94. [Google Scholar] [CrossRef]
- Starosta, W.; Leciejewicz, J. catena-Poly[lead(II)-bis(μ2-pyridazine-3-carboxylato-κ3N2,O:O)]. Acta Crystallogr. E Struct. Rep. Online 2010, 66, m192. [Google Scholar] [CrossRef] [PubMed]
- Mangalagiu, I.I. Recent Achievements in the Chemistry of 1,2-Diazines. Curr. Org. Chem. 2011, 15, 730–752. [Google Scholar] [CrossRef]
- Malik, A.; Mishra, R.; Mazumder, R.; Mazumder, A.; Mishra, P.S. A comprehensive study on synthesis and biological activities of Pyridazine Derivatives. Res. J. Pharm. Technol. 2021, 14, 3423–3429. [Google Scholar] [CrossRef]
- Asif, M. Various Chemical and Biological Activities of Pyridazinone Derivatives. Cent. Eur. J. Exp. Biol. 2017, 5, 1–19. [Google Scholar]
- Ibrahim, M.A.; Elmenoufy, A.H.; Elagawany, M.; Ghoneim, M.M.; Moawad, A. “Pyridopyridazine”: A Versatile Nucleus in Pharmaceutical Field. J. Biosci. Med. 2015, 3, 59–66. [Google Scholar] [CrossRef]
- Asif, M.; Abida; Imran, M. Study of various fused heterocyclic pyridazine derivatives as potent anticancer agents: A brief overview. Acta Sci. Pharm. Sci. 2019, 3, 43–49. [Google Scholar]
- Vigil-De Gracia, P.; Lasso, M.; Ruiz, E.; Vega-Malek, J.C.; de Mena, F.T.; López, J.C. Severe hypertension in pregnancy: Hydralazine or labetalol: A randomized clinical trial. Eur. J. Obstet. Gynecol. Reprod. Biol. 2006, 128, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Bizière, K.; Worms, P.; Kan, J.P.; Mandel, P.; Garattini, S.; Roncucci, R. Minaprine, a new drug with antidepressant properties. Drugs Exp. Clin. Res. 1985, 11, 831–840. [Google Scholar] [PubMed]
- Ikawa, K.; Morikawa, N.; Ikeda, K.; Nomura, K.; Taniwaki, M.; Ohge, H.; Sueda, T. Pharmacokinetic-pharmacodynamic target attainment analysis of cefozopran in Japanese adult patients. J. Infect. Chemother. 2008, 14, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Świderski, G.; Kalinowska, M.; Jabłońska-Trypuć, A.; Wołejko, E.; Wydro, U.; Łyszczek, R.; Rusinek, I.; Lewandowski, W. Studies on the relationship between the structure of pyrimidinecarboxylic, pyridazinecarboxylic and pyrazinecarboxylic acids and their antimicrobial and cytotoxic activity. J. Mol. Struct. 2021, 1231, 129903. [Google Scholar] [CrossRef]
- Deacon, G.B.; Phillips, R.J. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord. Chem. Rev. 1980, 33, 227–250. [Google Scholar] [CrossRef]
- Lever, A.B.P. Inorganic Electronic Spectroscopy, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 1984. [Google Scholar]
- Rardin, R.L.; Tolman, W.B.; Lippard, S.J. Monodentate carboxylate complexes and the carboxylate shift: Implications for polymetalloprotein structure and function. New J. Chem. 1991, 15, 417–430. [Google Scholar]
- Browning, K.; Abboud, K.A.; Palenik, G.J. Sodium hydrogen dipicolinate dipicolinic acid trihydrate: Synthesis, structure, and valence bond sums. J. Chem. Crystallogr. 1995, 25, 851–855. [Google Scholar] [CrossRef]
- Lainé, P.; Gourdon, A.; Launay, J.-P. Chemistry of Iron with Dipicolinic Acid. 2. Bridging Role of Carboxylate Groups in Solid State Structures. Inorg. Chem. 1995, 34, 5138–5149. [Google Scholar] [CrossRef]
- Marken, F.; Neudeck, A.; Bond, A.M.; Stojek, Z. Cyclic Voltammetry; Pulse Voltammetry. In Electroanalytical Methods. Guide to Experiments and Applications; Scholz, F., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 57–119. ISBN 978-3-642-02914-1. [Google Scholar]
- Meyer, J.-M.; Neely, A.; Stintzi, A.; Georges, C.; Holder, I.A. Pyoverdin is Essential for Virulence of Pseudomonas aeruginosa. Infect. Immun. 1996, 64, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Visca, P.; Imperi, F.; Lamont, I.L. Pyoverdine siderophores: From biogenesis to biosignificance. Trends Microbiol. 2007, 15, 22–30. [Google Scholar] [CrossRef] [PubMed]
- FDA. The Drug Development Process. Available online: https://www.fda.gov/ForPatients/Approvals/Drugs/default.htm (accessed on 10 October 2024).
- Larsen, M.T.; Kuhlmann, M.; Hvam, M.L.; Howard, K.A. Albumin-based drug delivery: Harnessing nature to cure disease. Mol. Cell. Ther. 2016, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Chuang, V.T.G.; Maruyama, T.; Otagiri, M. Albumin–drug interaction and its clinical implication. Biochim. Biophys. Acta-Gen. Subj. 2013, 1830, 5435–5443. [Google Scholar] [CrossRef] [PubMed]
- Sathyadevi, P.; Krishnamoorthy, P.; Bhuvanesh, N.S.P.; Kalaiselvi, P.; Vijaya Padma, V.; Dharmaraj, N. Organometallic ruthenium(II) complexes: Synthesis, structure and influence of substitution at azomethine carbon towards DNA/BSA binding, radical scavenging and cytotoxicity. Eur. J. Med. Chem. 2012, 55, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R.; Weber, G. Quenching of protein fluorescence by oxygen. Detection of structural fluctuations in proteins on the nanosecond time scale. Biochemistry 1973, 12, 4171–4179. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Weljie, A.M.; Vogel, H.J. Tryptophan Fluorescence Quenching by Methionine and Selenomethionine Residues of Calmodulin: Orientation of Peptide and Protein Binding. Biochemistry 1998, 37, 3187–3195. [Google Scholar] [CrossRef] [PubMed]
- Eftink, M.R.; Ghiron, C.A. Fluorescence quenching of indole and model micelle systems. J. Phys. Chem. 1976, 80, 486–493. [Google Scholar] [CrossRef]
- Eftink, M.R.; Ghiron, C.A. Fluorescence quenching studies with proteins. Anal. Biochem. 1981, 114, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Kandagal, P.B.; Ashoka, S.; Seetharamappa, J.; Shaikh, S.M.T.; Jadegoud, Y.; Ijare, O.B. Study of the interaction of an anticancer drug with human and bovine serum albumin: Spectroscopic approach. J. Pharm. Biomed. Anal. 2006, 41, 393–399. [Google Scholar] [CrossRef]
- Gong, A.; Zhu, X.; Hu, Y.; Yu, S. A fluorescence spectroscopic study of the interaction between epristeride and bovin serum albumine and its analytical application. Talanta 2007, 73, 668–673. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Ertl, P.; Rohde, B.; Selzer, P. Fast Calculation of Molecular Polar Surface Area as a Sum of Fragment-Based Contributions and Its Application to the Prediction of Drug Transport Properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef] [PubMed]
- Velders, A.H.; Pazderski, L.; Ugozzoli, F.; Biagini-Cingi, M.; Manotti-Lanfredi, A.M.; Haasnoot, J.G.; Reedijk, J. Synthesis, characterization and crystal structure of trans-aquatrichlorobis (5,7-dimethyl[1,2,4]triazolo[1,5-a]pyrimidine-N3)ruthenium(III) monohydrate. Inorg. Chim. Acta 1998, 273, 259–265. [Google Scholar] [CrossRef]
- Blanc, R.; González-Casado, A.; Navalón, A.; Vílchez, J.L. On the estimate of blanks in differential pulse voltammetric techniques: Application to detection limits evaluation as recommended by IUPAC. Anal. Chim. Acta 2000, 403, 117–123. [Google Scholar] [CrossRef]
- Rigaku Oxford Diffraction. CrysAlisPro, Version 1.171.36.20; Rigaku Oxford Diffraction: Oxfordshire, England, 2015.
- Sheldrick, G.M. SHELXT – Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, C71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Putz, K.H. Brandenburg, Diamond - Crystal and Molecular Structure Visualization Crystal Impact, GbR. Kreuzherrenstr 2014, 102, 53227. [Google Scholar]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- Cullen, L.; Weiser, R.; Olszak, T.; Maldonado, R.F.; Moreira, A.S.; Slachmuylders, L.; Brackman, G.; Paunova-Krasteva, T.S.; Żarnowiec, P.; Czerwonka, G.; et al. Phenotypic characterization of an international Pseudomonas aeruginosa reference panel: Strains of cystic fibrosis (CF) origin show less in vivo virulence than non-CF strains. Microbiology 2015, 161, 1961–1977. [Google Scholar] [CrossRef] [PubMed]
- Borkowski, A.; Gutowski, Ł.; Syczewski, M.; Cłapa, T.; Czerwonka, G. Adaptation of bacteria Escherichia coli in presence of quaternary ammonium ionic liquids. Ecotoxicol. Environ. Saf. 2018, 164, 370–378. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Schrödinger LLC. Schrödinger Release 2024-1: Maestro; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein–ligand docking using GOLD. Proteins Struct. Funct. Bioinform. 2003, 52, 609–623. [Google Scholar] [CrossRef]
- Sapundzhi, F.; Prodanova, K.; Lazarova, M. Survey of the scoring functions for protein-ligand docking. AIP Conf. Proc. 2019, 2172, 100008. [Google Scholar] [CrossRef]
- Schrödinger LLC. The PyMOL Molecular Graphics System, Version 2.5; Schrödinger LLC: New York, NY, USA, 2021.
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer US: Boston, MA, USA, 2006; ISBN 978-0-387-31278-1. [Google Scholar]
- Lineweaver, H.; Burk, D. The Determination of Enzyme Dissociation Constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]



| Bond Lengths (Å) | |||
|---|---|---|---|
| Ru1–N2 | 2.053(5) | Ru1–Cl1 | 2.4126(2) |
| Ru1–O8 | 2.083(4) | Ru1–Ct1 | 1.6567(5) |
| Valence Angles (°) | |||
| N2–Ru1–O8 | 78.10(2) | N2–Ru1–Ct1 | 133.09(1) |
| N2–Ru1–Cl1 | 83.08(2) | O8–Ru1–Ct1 | 129.80(1) |
| O8–Ru1–Cl1 | 86.30(1) | Cl1–Ru1–Ct1 | 128.42(4) |
| Bond Lengths (Å) | |||
|---|---|---|---|
| Ru1–N2 | 2.0304(9) | Na1–O2 | 2.3827(1) |
| Ru1–O8 | 2.0327(8) | Na1–O8 | 2.4897(8) |
| Ru1–Cl1 | 2.3352(3) | Na1–N1(−x+1/2,−y+1/2,−z+1) | 2.5385(1) |
| Na1–Na1(−x+1,−y+1/2,z) | 3.6636(2) | ||
| Valence Angles (°) | |||
| N2(−x+1,−y+1/2,z)–Ru1–N2 | 180.0 | ||
| N2–Ru1–O8(−x+1,−y+1/2,z) | 99.97(3) | O2–Na1–O2(−x+1,−y+1/2,z) | 79.51(5) |
| N2–Ru1–O8 | 80.03(3) | O2–Na1–O8 | 80.29(2) |
| O8(−x+1,−y+1/2,z)–Ru1–O8 | 180.0 | O2–Na1–O8(x,−y+1/2,−z+3/2) | 114.31(3) |
| N2–Ru1–Cl1 | 92.92(3) | O8–Na1–O8(x,−y+1/2,−z+3/2) | 161.81(5) |
| O8–Ru1–Cl1 | 90.15(3) | O2–Na1–N1(−x+1/2,−y+1/2,−z+1) | 158.86(2) |
| N2–Ru1–Cl1(−x+1,−y+1/2,z) | 87.08(3) | O8–Na1–N1(−x+1/2,−y+1/2,−z+1) | 80.18(3) |
| O8–Ru1–Cl1(−x+1,−y+1/2,z) | 89.85(3) | O2–Na1–N1(−x+1/2,y,z+1/2) | 101.38(3) |
| Cl1–Ru1–Cl1(−x+3/2,−y+1/2,−z+3/2) | 180.0 | O8–Na1–N1(−x+1/2,y,z+1/2) | 86.46(3) |
| N1(−x+1/2,−y+1/2,−z+1)–Na1–N1(−x+1/2,y,z+1/2) | 85.39(5) | ||
| Bond Lengths (Å) | |||
|---|---|---|---|
| Ru1–N2 | 2.0311(1) | Na1–O2 | 2.2996(2) |
| Ru1–O8 | 2.0324(1) | Na1–O1 | 2.3507(2) |
| Ru1–Cl1 | 2.3402(4) | Na1–O18 | 2.3833(1) |
| Ru2–N12 | 2.0316(1) | Na1–O8 | 2.4498(1) |
| Ru2–O18 | 2.0388(1) | Na1–N1(−x+3/2,−y+1/2,−z+3/2) | 2.5726(2) |
| Ru2–Cl2 | 2.3285(5) | Na1–N11 | 2.6265(2) |
| Valence Angles (°) | |||
| N2(−x+3/2,−y+1/2,−z+3/2)–Ru1–N2 | 180.00(9) | Cl2(−x+3/2,−y+1/2,−z+3/2)–Ru2–Cl2 | 180.0 |
| N2–Ru1–O8(−x+3/2,−y+1/2,−z+3/2) | 100.00(5) | O2–Na1–O1 | 88.52(6) |
| N2–Ru1–O8 | 80.00(5) | O2–Na1–O18 | 101.28(6) |
| O8(−x+3/2,−y+1/2,−z+3/2)–Ru1–O8 | 180.00(5) | O1–Na1–O18 | 108.92(5) |
| N2–Ru1–Cl1 | 89.26(4) | O2–Na1–O8 | 89.81(6) |
| O8–Ru1–Cl1 | 92.55(3) | O1–Na1–O8 | 78.77(5) |
| N2–Ru1–Cl1(−x+3/2,−y+1/2,−z+3/2) | 90.74(4) | O18–Na1–O8 | 166.48(5) |
| O8–Ru1–Cl1(−x+3/2,−y+1/2,−z+3/2) | 87.45(3) | O2–Na1–N1(−x+3/2,−y+1/2,−z+3/2) | 168.31(6) |
| Cl1–Ru1–Cl1(−x+3/2,−y+1/2,−z+3/2) | 180.0 | O1–Na1–N1(−x+3/2,−y+1/2,−z+3/2) | 83.39(5) |
| N12–Ru2–N12(−x+3/2,−y+1/2,−z+3/2) | 180.00(6) | O18–Na1–N1(−x+3/2,−y+1/2,−z+3/2) | 89.35(5) |
| N12–Ru2–O18(−x+3/2,−y+1/2,−z+3/2) | 79.83(5) | O8–Na1–N1(−x+3/2,−y+1/2,−z+3/2) | 80.37(4) |
| N12–Ru2–O18 | 100.17(5) | O2–Na1–N11 | 92.19(6) |
| O18(−x+3/2,−y+1/2,−z+3/2)–Ru2–O18 | 180.0 | O1–Na1–N11 | 170.16(5) |
| N12–Ru2–Cl2(−x+3/2,−y+1/2,−z+3/2) | 90.63(4) | O18–Na1–N11 | 80.57(5) |
| O18–Ru2–Cl2(−x+3/2,−y+1/2,−z+3/2) | 90.95(4) | O8–Na1–N11 | 91.41(5) |
| N12–Ru2–Cl2 | 89.37(4) | N1(−x+3/2,−y+1/2,−z+3/2)–Na1–N11 | 94.36(5) |
| O18–Ru2–Cl2 | 89.05(4) |
| Complex | Scan Rate mV/s | Ru(IV) ↔ Ru(III) | Ru(III) ↔ Ru(II) | Ru(II) ↔ Ru(I) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Epa | Epc | ΔEp | E1/2 | Epa | Epc | ΔEp | E1/2 | Epa | Epc | ΔEp | E1/2 | ||
| 1 | 50 | 0.780 | 0.720 | 0.060 | 0.750 | ||||||||
| 100 | 0.780 | 0.720 | 0.060 | 0.750 | |||||||||
| 200 | 0.790 | 0.710 | 0.080 | 0.750 | |||||||||
| 500 | 0.795 | 0.705 | 0.090 | 0.750 | |||||||||
| 2 | 50 | 1.435 | 1.345 | 0.090 | 1.390 | 0.225 | 0.160 | 0.065 | 0.193 | −0.240 | −0.300 | 0.060 | −0.270 |
| 100 | 1.440 | 1.350 | 0.090 | 1.395 | 0.230 | 0.160 | 0.070 | 0.195 | −0.245 | −0.310 | 0.065 | −0.278 | |
| 200 | 1.450 | 1.360 | 0.090 | 1.405 | 0.240 | 0.165 | 0.075 | 0.203 | −0.245 | −0.315 | 0.070 | −0.280 | |
| 500 | 1.465 | 1.375 | 0.090 | 1.420 | 0.245 | 0.170 | 0.075 | 0.208 | −0.250 | −0.320 | 0.070 | −0.285 | |
| 3 | 50 | 0.165 | 0.115 | 0.050 | 0.140 | −0.300 | −0.425 | 0.125 | −0.363 | ||||
| 100 | 0.170 | 0.110 | 0.060 | 0.140 | −0.290 | −0.430 | 0.140 | −0.360 | |||||
| 200 | 0.180 | 0.105 | 0.075 | 0.143 | −0.285 | −0.440 | 0.155 | −0.363 | |||||
| 500 | 0.190 | 0.100 | 0.090 | 0.145 | −0.280 | −0.450 | 0.170 | −0.365 | |||||
| Compound | Quenching | Binding | Thermodynamic | |||
|---|---|---|---|---|---|---|
| KSV [M−1] | Kq [M−1 s−1] | Kb * [M−1] | Kb ** [M−1] | n | ΔG [kJ mol−1] | |
| 1 | 6.97 × 103 | 5.4 × 1011 | 4.23 × 105 | 7.03 × 103 | 1.46 | −31.45 |
| 2 | 5.03 × 103 | 4.4 × 1011 | 2.78 × 104 | 5.88 × 103 | 1.18 | −24.84 |
| Descriptor | Compound | ||
|---|---|---|---|
| 1 | 2 | ||
| Pharmacokinetics | GI absorption | High | High |
| BBB permeability | Yes | No | |
| P-gp substrate | Yes | Yes | |
| Water solubility (ESOL) | moderately soluble | moderately soluble | |
| Drug-Likeness | Lipiński | Yes, 0 violation | Yes, 0 violation |
| Veber | Yes | Yes | |
| Bioavability score | 0.55 | 0.55 | |
| TPSA [Å2] | 44.12 | 88.24 | |
| Physicochemical Properties (Ro5) | Number of H-Bond Acceptors | 3 | 6 |
| Number of H-Bond Donors | 0 | 0 | |
| Log Po/w (MLOGP) | 2.61 | 0.15 | |
| MW [g/mol] | 389.80 | 418.16 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rogala, P.; Jabłońska-Wawrzycka, A.; Czerwonka, G.; Hodorowicz, M.; Michałkiewicz, S.; Kalinowska-Tłuścik, J.; Karpiel, M.; Gałczyńska, K. Ruthenium Complexes with Pyridazine Carboxylic Acid: Synthesis, Characterization, and Anti-Biofilm Activity. Molecules 2024, 29, 5694. https://doi.org/10.3390/molecules29235694
Rogala P, Jabłońska-Wawrzycka A, Czerwonka G, Hodorowicz M, Michałkiewicz S, Kalinowska-Tłuścik J, Karpiel M, Gałczyńska K. Ruthenium Complexes with Pyridazine Carboxylic Acid: Synthesis, Characterization, and Anti-Biofilm Activity. Molecules. 2024; 29(23):5694. https://doi.org/10.3390/molecules29235694
Chicago/Turabian StyleRogala, Patrycja, Agnieszka Jabłońska-Wawrzycka, Grzegorz Czerwonka, Maciej Hodorowicz, Sławomir Michałkiewicz, Justyna Kalinowska-Tłuścik, Marta Karpiel, and Katarzyna Gałczyńska. 2024. "Ruthenium Complexes with Pyridazine Carboxylic Acid: Synthesis, Characterization, and Anti-Biofilm Activity" Molecules 29, no. 23: 5694. https://doi.org/10.3390/molecules29235694
APA StyleRogala, P., Jabłońska-Wawrzycka, A., Czerwonka, G., Hodorowicz, M., Michałkiewicz, S., Kalinowska-Tłuścik, J., Karpiel, M., & Gałczyńska, K. (2024). Ruthenium Complexes with Pyridazine Carboxylic Acid: Synthesis, Characterization, and Anti-Biofilm Activity. Molecules, 29(23), 5694. https://doi.org/10.3390/molecules29235694


.jpg)





