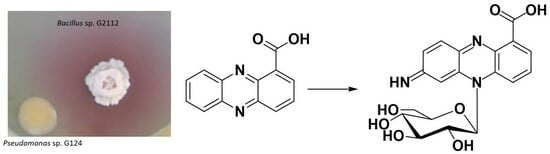
Bacillus sp. G2112 Detoxifies Phenazine-1-carboxylic Acid by N5 Glucosylation
Abstract
:1. Introduction
2. Results
2.1. Co-Cultivation of Bacillus sp. G2112 and Pseudomonas sp. G124
2.2. Identification of the Antibiotic Phenazine-1-carboxylic Acid as Precursor of Red Pigments
2.3. Identification of the Red Pigments
2.4. Detoxification Products Did Not Inhibit Bacillus sp. G2112
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glick, B.R. The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 1995, 41, 109–117. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Christophe, C.; Barka, A.E. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.A. Biological control of plant diseases. Australas. Plant Pathol. 2017, 46, 293–304. [Google Scholar] [CrossRef]
- Popp, J.; Pető, K.; Nagy, J. Pesticide productivity and food security. A review. Agron. Sustain. Dev. 2013, 33, 243–255. [Google Scholar] [CrossRef]
- Gerhardson, B. Biological substitutes for pesticides. Trends Biotechnol. 2002, 20, 338–343. [Google Scholar] [CrossRef]
- De Silva, N.I.; Brooks, S.; Lumyong, S.; Hyde, K.D. Use of endophytes as biocontrol agents. Fungal Biol. Rev. 2019, 33, 133–148. [Google Scholar] [CrossRef]
- Card, S.D.; Walter, M.; Jaspers, M.V.; Sztejnberg, A.; Stewart, A. Targeted selection of antagonistic microorganisms for control of Botrytis cinerea of strawberry in New Zealand. Australas. Plant Pathol. 2009, 38, 183–192. [Google Scholar] [CrossRef]
- Bejarano, A.; Puopolo, G. Bioformulation of microbial biocontrol agents for a sustainable agriculture. In How Research Can Stimulate the Development of Commercial Biological Control Against Plant Diseases. Progress in Biological Control; De Cal, A., Melgarejo, P., Magan, N., Eds.; Springer: Cham, Switzerland, 2020; Volume 21, pp. 275–293. [Google Scholar] [CrossRef]
- Velivelli, S.L.S.; De Vos, P.; Kromann, P.; Declerck, S.; Prestwich, B.D. Biological control agents: From field to market, problems, and challenges. Trends Biotechnol. 2014, 32, 493–496. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van Wees, S.C.M.; Van Pelt, J.A.; Knoester, M.; Laan, R.; Gerrits, H.; Weisbeek, P.J.; Van Loon, L.C. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 1998, 10, 1571–1580. [Google Scholar] [CrossRef]
- Montesinos, E. Development, registration and commercialization of microbial pesticides for plant protection. Int. Microbiol. 2003, 6, 245–252. [Google Scholar] [CrossRef]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Thomashow, L.S.; Weller, D.M.; Bonsall, R.F.; Pierson, L.S. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl. Environ. Microbiol. 1990, 56, 908–912. [Google Scholar] [CrossRef]
- Höfte, M. The use of Pseudomonas spp. as bacterial biocontrol agents to control plant diseases. In Microbial Bioprotectants for Plant Disease Management; Köhl, J., Ravensberg, W.J., Eds.; Burleigh Dodds Science Publishing: Cambridge, UK, 2021; pp. 1–74. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Bais, H.P.; Fall, R.; Vivanco, J.M. Biocontrol of Bacillus subtilis against infection of arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004, 134, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Anckaert, A.; Arias, A.A.; Hoff, G.; Calonne-Salmon, M.; Declerck, S.; Ongena, M. The use of Bacillus spp. as bacterial biocontrol agents to control plant diseases. In Microbial Bioprotectants for Plant Disease Management; Köhl, J., Ravensberg, W.J., Eds.; Burleigh Dodds Science Publishing: Cambridge, UK, 2021; pp. 1–54. [Google Scholar] [CrossRef]
- Arguelles-Arias, A.; Ongena, M.; Halimi, B.; Lara, Y.; Brans, A.; Joris, B.; Fickers, P. Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Fact. 2009, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, T.N.; Veselov, S.U.; Melentiev, A.I.; Martynenko, E.V.; Kudoyarova, G.R. Ability of bacterium Bacillus subtilis to produce cytokinins and to influence the growth and endogenous hormone content of lettuce plants. Plant Soil 2005, 272, 201–209. [Google Scholar] [CrossRef]
- Santoyo, G.; Orozco-Mosqueda, M.C.; Govindappa, M. Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: A review. Biocontrol Sci. Technol. 2012, 22, 855–872. [Google Scholar] [CrossRef]
- Lyng, M.; Kovács, Á.T. Frenemies of the soil: Bacillus and Pseudomonas interspecies interactions. Trends Microbiol. 2023, 31, 845–857. [Google Scholar] [CrossRef]
- Molina-Santiago, C.; Vela-Corcía, D.; Petras, D.; Díaz-Martínez, L.; Pérez-Lorente, A.I.; Sopeña-Torres, S.; Pearson, J.; Caraballo-Rodríguez, A.M.; Dorrestein, P.C.; de Vicente, A.; et al. Chemical interplay and complementary adaptative strategies toggle bacterial antagonism and co-existence. Cell Rep. 2021, 36, 109449. [Google Scholar] [CrossRef]
- Andrić, S.; Rigolet, A.; Argüelles Arias, A.; Steels, S.; Hoff, G.; Balleux, G.; Ongena, L.; Höfte, M.; Meyer, T.; Ongena, M. Plant-associated Bacillus mobilizes its secondary metabolites upon perception of the siderophore pyochelin produced by a Pseudomonas competitor. ISME J. 2023, 17, 263–275. [Google Scholar] [CrossRef]
- Powers, M.J.; Sanabria-Valentín, E.; Bowers, A.A.; Shank, E.A. Inhibition of cell differentiation in Bacillus subtilis by Pseudomonas protegens. J. Bacteriol. 2015, 197, 2129–2138. [Google Scholar] [CrossRef]
- Andrić, S.; Meyer, T.; Rigolet, A.; Prigent-Combaret, C.; Höfte, M.; Balleux, G.; Steels, S.; Hoff, G.; De Mot, R.; McCann, A.; et al. Lipopeptide interplay mediates molecular interactions between soil bacilli and pseudomonads. Microbiol. Spectr. 2021, 9, e02038-21. [Google Scholar] [CrossRef]
- Rojas-Ruiz, N.E.; Sansinenea-Royano, E.; Cedillo-Ramirez, M.L.; Marsch-Moreno, R.; Sanchez-Alonso, P.; Vazquez-Cruz, C. Analysis of Bacillus thuringiensis population dynamics and its interaction with Pseudomonas fluorescens in soil. Jundishapur J. Microbiol. 2015, 8, e27953. [Google Scholar] [CrossRef]
- Vick, S.H.W.; Fabian, B.K.; Dawson, C.J.; Foster, C.; Asher, A.; Hassan, K.A.; Midgley, D.J.; Paulsen, I.T.; Tetu, S.G. Delving into defence: Identifying the Pseudomonas protegens Pf-5 gene suite involved in defence against secreted products of fungal, oomycete and bacterial rhizosphere competitors. Microb. Genom. 2021, 7, 000671. [Google Scholar] [CrossRef]
- Chakraborty, B.; Mallick, A.; Annagiri, S.; Sengupta, S.; Sengupta, T.K. Deciphering a survival strategy during the interspecific competition between Bacillus cereus MSM-S1 and Pseudomonas sp. MSM-M1. R. Soc. Open Sci. 2016, 3, 160438. [Google Scholar] [CrossRef]
- Garbeva, P.; Silby, M.W.; Raaijmakers, J.M.; Levy, S.B.; Boer, W.D. Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors. ISME J. 2011, 5, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Chevrette, M.G.; Thomas, C.S.; Hurley, A.; Rosario-Melendez, N.; Sankaran, K.; Tu, Y.; Hall, A.; Magesh, S.; Handelsman, J. Microbiome composition modulates secondary metabolism in a multispecies bacterial community. Proc. Natl. Acad. Sci. USA 2022, 119, e2212930119. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xu, Z.; Xie, J.; Hesselberg-Thomsen, V.; Tan, T.; Zheng, D.; Strube, M.L.; Dragoš, A.; Shen, Q.; Zhang, R.; et al. Bacillus velezensis stimulates resident rhizosphere Pseudomonas stutzeri for plant health through metabolic interactions. ISME J. 2022, 16, 774–787. [Google Scholar] [CrossRef] [PubMed]
- Ghazy, N.; El-Nahrawy, S. Siderophore production by Bacillus subtilis MF497446 and Pseudomonas koreensis MG209738 and their efficacy in controlling Cephalosporium maydis in maize plant. Arch. Microbiol. 2021, 203, 1195–1209. [Google Scholar] [CrossRef]
- Ansari, F.A.; Ahmad, I. Fluorescent Pseudomonas -FAP2 and Bacillus licheniformis interact positively in biofilm mode enhancing plant growth and photosynthetic attributes. Sci. Rep. 2019, 9, 4547. [Google Scholar] [CrossRef]
- Durairaj, K.; Velmurugan, P.; Park, J.H.; Chang, W.S.; Park, Y.J.; Senthilkumar, P.; Choi, K.M.; Lee, J.H.; Oh, B.T. Potential for plant biocontrol activity of isolated Pseudomonas aeruginosa and Bacillus stratosphericus strains against bacterial pathogens acting through both induced plant resistance and direct antagonism. FEMS Microbiol. Lett. 2017, 364, fnx225. [Google Scholar] [CrossRef] [PubMed]
- Comeau, D.; Balthazar, C.; Novinscak, A.; Bouhamdani, N.; Joly, D.L.; Filion, M. Interactions between Bacillus spp., Pseudomonas spp. and Cannabis sativa promote plant growth. Front. Microbiol. 2021, 12, 715758. [Google Scholar] [CrossRef]
- Lozano, G.L.; Bravo, J.I.; Garavito Diago, M.F.; Park, H.B.; Hurley, A.; Peterson, S.B.; Stabb, E.V.; Crawford, J.M.; Broderick, N.A.; Handelsman, J. Introducing THOR, a model microbiome for genetic dissection of community behavior. MBio 2019, 10, 10-1128. [Google Scholar] [CrossRef]
- Baliarda, A.; Winkler, M.; Tournier, L.; Tinsley, C.R.; Aymerich, S. Dynamic interspecies interactions and robustness in a four-species model biofilm. Microbiol. Open 2021, 10, e1254. [Google Scholar] [CrossRef]
- Monmeyran, A.; Benyoussef, W.; Thomen, P.; Dahmane, N.; Baliarda, A.; Jules, M.; Aymerich, S.; Henry, N. Four species of bacteria deterministically assemble to form a stable biofilm in a millifluidic channel. NPJ Biofilms Microbiomes 2021, 7, 64. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Kieser, T.E.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Colney, UK, 2000. [Google Scholar]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Guthke, R.; Nüske, J.; Schorcht, R.; Fritsche, W.; Knorre, W.A. Dynamic model of discontinuous and continuous phaseolotoxin production of Pseudomonas syringae pv. phaseolicola. Z. Allg. Mikrobiol. 1984, 24, 427–435. [Google Scholar]
- Wensing, A.; Braun, S.D.; Büttner, P.; Expert, D.; Völksch, B.; Ullrich, M.S.; Weingart, H. Impact of siderophore production by Pseudomonas syringae pv. syringae 22d/93 on epiphytic fitness and biocontrol activity against Pseudomonas syringae pv. glycinea 1a/96. Appl. Environ. Microbiol. 2010, 76, 2704–2711. [Google Scholar] [CrossRef]
- Lee, J.Y.; Moon, S.S.; Hwang, B.K. Isolation and in vitro and in vivo activity against Phytophthora capsici and Colletotrichum orbiculare of phenazine-1-carboxylic acid from Pseudomonas aeruginosa strain GC-B26. Pest Manag. Sci. 2003, 59, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Hu, H.; Peng, H.; Zhang, X.; Wang, W. Isolation and structural identification of two bioactive phenazines from Streptomyces griseoluteus P510. Chin. J. Chem. Eng. 2015, 23, 699–703. [Google Scholar] [CrossRef]
- Raio, A.; Puopolo, G.; Cimmino, A.; Danti, R.; Della Rocca, G.; Evidente, A. Biocontrol of cypress canker by the phenazine producer Pseudomonas chlororaphis subsp. aureofaciens strain M71. Biol. Control 2011, 58, 133–138. [Google Scholar] [CrossRef]
- Kunigami, T.; Shin-Ya, K.; Furihata, K.; Furihata, K.; Hayakawa, Y.; Seto, H. A novel neuronal cell protecting substance, aestivophoenin C produced by Streptomyces purpeofuscus. J. Antibiot. 1998, 51, 880–882. [Google Scholar] [CrossRef] [PubMed]
- Fujimatu, E.; Ishikawa, T.; Kitajima, J. Aromatic compound glucosides, alkyl glucoside and glucide from the fruit of anise. Phytochemistry 2003, 63, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Nishibe, S.; Tsukamoto, H.; Hisada, S. Effects of O-methylation and O-glucosylation on carbon-13 nuclear magnetic resonance chemical shifts of matairesinol,(+)-pinoresinol and (+)-epipinoresinol. Chem. Pharm. Bull. 1984, 32, 4653–4657. [Google Scholar] [CrossRef]
- Tanaka, T.; Nakashima, T.; Ueda, T.; Tomii, K.; Kouno, I. Facile discrimination of aldose enantiomers by reversed-phase HPLC. Chem. Pharm. Bull. 2007, 55, 899–901. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, G.W.; Dawson, B.A. Aromatic imine stereochemistry as studied by 13C and 1H NMR of 15N-enriched materials. Org. Magn. Reson. 1980, 13, 293–298. [Google Scholar] [CrossRef]
- Abu, E.A.; Su, S.; Sallans, L.; Boissy, R.E.; Greatens, A.; Heineman, W.R.; Hassett, D.J. Cyclic voltammetric, fluorescence and biological analysis of purified aeruginosin A, a secreted red pigment of Pseudomonas aeruginosa PAO1. Microbiology 2013, 159, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Holliman, F.G. Pigments of Pseudomonas species. part 1. Structure and synthesis of aeruginosin A. J. Chem. Soc. C 1969, 19, 2514–2516. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Parejko, J.A.; Mavrodi, O.V.; Kwak, Y.S.; Weller, D.M.; Blankenfeldt, W.; Thomashow, L.S. Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ. Microbiol. 2013, 15, 675–686. [Google Scholar] [CrossRef]
- Zendah, I.; Riaz, N.; Nasr, H.; Frauendorf, H.; Schüffler, A.; Raies, A.; Laatsch, H. Chromophenazines from the terrestrial Streptomyces sp. ANK 315. J. Nat. Prod. 2012, 75, 2–8. [Google Scholar] [CrossRef]
- Wu, C.; Van Wezel, G.P.; Hae Choi, Y. Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66. J. Antibiot. 2015, 68, 445–452. [Google Scholar] [CrossRef]
- Heine, D.; Martin, K.; Hertweck, C. Genomics-guided discovery of endophenazines from Kitasatospora sp. HKI 714. J. Nat. Prod. 2014, 77, 1083–1087. [Google Scholar] [CrossRef]
- Laursen, J.B.; Nielsen, J. Phenazine natural products: Biosynthesis, synthetic analogues, and biological activity. Chem. Rev. 2004, 104, 1663–1685. [Google Scholar] [CrossRef] [PubMed]
- Saleh, O.; Bonitz, T.; Flinspach, K.; Kulik, A.; Burkard, N.; Mühlenweg, A.; Vente, A.; Polnick, S.; Lämmerhofer, M.; Gust, B.; et al. Activation of a silent phenazine biosynthetic gene cluster reveals a novel natural product and a new resistance mechanism against phenazines. Medchemcomm 2012, 3, 1009–1019. [Google Scholar] [CrossRef]
- Abdelfattah, M.S.; Toume, K.; Ishibashi, M. Izumiphenazine D, a new phenazoquinoline N-oxide from Streptomyces sp. IFM 11204. Chem. Pharm. Bull. 2011, 59, 508–510. [Google Scholar] [CrossRef] [PubMed]
- Rusman, Y.; Oppegard, L.M.; Hiasa, H.; Gelbmann, C.; Salomon, C.E. Solphenazines A-F, glycosylated phenazines from Streptomyces sp. strain DL-93. J. Nat. Prod. 2013, 76, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Shin-Ya, K.; Shimizu, S.; Kunigami, T.; Hayakawa, Y.; Seto, H.; Furihata, K. Novel neuronal cell protecting substances, aestivophoenins A and B, produced by Streptomyces purpeofuscus. J. Antibiot. 1995, 48, 1378–1381. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Medema, M.H.; Läkamp, R.M.; Zhang, L.; Dorrestein, P.C.; Choi, Y.H.; Van Wezel, G.P. Leucanicidin and endophenasides result from methyl-rhamnosylation by the same tailoring enzymes in Kitasatospora sp. MBT66. ACS Chem. Biol. 2016, 11, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Letourneau, M.K.; Marshall, M.J.; Grant, M.; Freeze, P.M.; Strawn, D.G.; Lai, B.; Dohnalkova, A.C.; Harsh, J.B.; Weller, D.M.; Thomashow, L.S. Phenazine-1-carboxylic acid-producing bacteria enhance the reactivity of iron minerals in dryland and irrigated wheat rhizospheres. Environ. Sci. Technol. 2019, 53, 14273–14284. [Google Scholar] [CrossRef]
- Wang, Y.; Wilks, J.C.; Danhorn, T.; Ramos, I.; Croal, L.; Newman, D.K. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J. Bacteriol. 2011, 193, 3606–3617. [Google Scholar] [CrossRef]
- Tsypin, L.M.; Newman, D.K. Nitrate reduction stimulates and is stimulated by phenazine-1carboxylic acid oxidation by Citrobacter portucalensis MBL. MBio 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Bonsall, R.F.; Delaney, S.M.; Soule, M.J.; Phillips, G.; Thomashow, L.S. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001, 183, 6454–6465. [Google Scholar] [CrossRef]
- Thierbach, S.; Birmes, F.S.; Letzel, M.C.; Hennecke, U.; Fetzner, S. Chemical modification and detoxification of the Pseudomonas aeruginosa toxin 2-heptyl-4-hydroxyquinoline N-oxide by environmental and pathogenic bacteria. ACS Chem. Biol. 2017, 12, 2305–2312. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Tan, Y.; Liu, N.; Yan, Z.; Liao, Y.; Chen, J.; De Saeger, S.; Yang, H.; Zhang, Q.; Wu, A. Detoxification of deoxynivalenol via glycosylation represents novel insights on antagonistic activities of Trichoderma when confronted with Fusarium graminearum. Toxins 2016, 8, 335. [Google Scholar] [CrossRef]
- Pedras, M.S.C.; Zaharia, I.L.; Gai, Y.; Zhou, Y.; Ward, D.E. In planta sequential hydroxylation and glycosylation of a fungal phytotoxin: Avoiding cell death and overcoming the fungal invader. Proc. Natl. Acad. Sci. USA 2001, 98, 747–752. [Google Scholar] [CrossRef]
- Salminen, J.P.; Lahtinen, M.; Lempa, K.; Kapari, L.; Haukioja, E.; Pihlaja, K. Metabolic modifications of birch leaf phenolics by an herbivorous insect: Detoxification of flavonoid aglycones via glycosylation. Z. Naturforsch. Sect. C J. Biosci. 2004, 59, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, N.; Teramoto, T.; Kasai, F.; Sano, T.; Tamaoki, M.; Aono, M.; Kubo, A.; Kamada, H.; Azumi, Y.; Saji, H. Glycosylation of bisphenol a by freshwater microalgae. Chemosphere 2007, 69, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Malouin, F.; Bryan, L.E. Modification of penicillin-binding proteins of beta-lactam resistance. Antimicrob. Agents Chemother. 1986, 30, 1–5. [Google Scholar] [CrossRef]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Luthra, S.; Rominski, A.; Sander, P. The role of antibiotic-target-modifying and antibiotic-modifying enzymes in Mycobacterium abscessus drug resistance. Front. Microbiol. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Molina-Santiago, C.; Pearson, J.R.; Navarro, Y.; Berlanga-Clavero, M.V.; Caraballo-Rodriguez, A.M.; Petras, D.; García-Martín, M.L.; Lamon, G.; Haberstein, B.; Cazorla, F.M.; et al. The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat. Commun. 2019, 10, 1919. [Google Scholar] [CrossRef] [PubMed]
- Boutigny, A.L.; Richard-Forget, F.; Barreau, C. Natural mechanisms for cereal resistance to the accumulation of Fusarium trichothecenes. Eur. J. Plant Pathol. 2008, 121, 411–423. [Google Scholar] [CrossRef]
- Stupp, G.S.; Von Reuss, S.H.; Izrayelit, Y.; Ajredini, R.; Schroeder, F.C.; Edison, A.S. Chemical detoxification of small molecules by Caenorhabditis elegans. ACS Chem. Biol. 2013, 8, 309–313. [Google Scholar] [CrossRef]
- Krastel, P.; Zeeck, A. Endophenazines A-D, new phenazine antibiotics from the athropod associated endosymbiont Streptomyces anulatus. J. Antibiot. 2002, 55, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Shindo, K.; Yamagishi, Y.; Matsuoka, M.; Kawai, H.; Mochizuki, J. Phenazoviridin, a novel free radical scavenger from Streptomyces sp. Taxonomy, fermentation, isolation, structure elucidation and biological properties. J. Antibiot. 1993, 46, 1485–1493. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, F.X.; Liu, C.; Wang, L.; Qi, Y.; Cao, M.; Guo, X.; Li, J.; Huang, X.; Yang, J.; et al. Isolation and biosynthesis of phenazine-polyketide hybrids from Streptomyces sp. KIB-H483. J. Nat. Prod. 2022, 85, 1324–1331. [Google Scholar] [CrossRef]
- Laursen, J.B.; Petersen, L.; Jensen, K.J.; Nielsen, J. Efficient synthesis of glycosylated phenazine natural products and analogs with DISAL (methyl 3,5-dinitrosalicylate) glycosyl donors. Org. Biomol. Chem. 2003, 1, 3147–3153. [Google Scholar] [CrossRef]
- Pathirana, C.; Jensen, P.R.; Dwight, R.; Fenical, W. Rare phenazine l-quinovose esters from a marine actinomycete. J. Org. Chem. 1992, 57, 740–742. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Mavrodi, O.V.; Parejko, J.A.; Bonsall, R.F.; Kwak, Y.S.; Paulitz, T.C.; Thomashow, L.S.; Weller, D.M. Accumulation of the antibiotic phenazine-1-carboxylic acid in the rhizosphere of dryland cereals. Appl. Environ. Microbiol. 2012, 78, 804–812. [Google Scholar] [CrossRef]
- Briard, B.; Bomme, P.; Lechner, B.E.; Mislin, G.L.A.; Lair, V.; Prévost, M.C.; Latgé, J.P.; Haas, H.; Beauvais, A. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci. Rep. 2015, 5, 8220. [Google Scholar] [CrossRef]
- Meirelles, L.A.; Newman, D.K. Phenazines and toxoflavin act as interspecies modulators of resilience to diverse antibiotics. Mol. Microbiol. 2022, 117, 1384–1404. [Google Scholar] [CrossRef]
- Chen, K.; Hu, H.; Wang, W.; Zhang, X.; Xu, Y. Metabolic degradation of phenazine-1-carboxylic acid by the strain Sphingomonas sp. DP58: The identification of two metabolites. Biodegradation 2008, 19, 659–667. [Google Scholar] [CrossRef]
- Costa, K.C.; Moskatel, L.S.; Meirelles, L.A.; Newman, D.K. PhdA catalyzes the first step of phenazine-1-carboxylic acid degradation in Mycobacterium fortuitum. J. Bacteriol. 2018, 200, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Hu, H.-B.; Wei, W.; Huang, X.-Q.; Zhang, X.-H. Novel three-component phenazine-1-carboxylic acid 1,2-dioxygenase in Sphingomonas wittichii DP58. Appl. Environ. Microbiol. 2017, 83, e00133-17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Bilal, M.; Yue, S.; Hu, H.; Wang, W.; Zhang, X. Identification of biphenyl 2, 3-dioxygenase and its catabolic role for phenazine degradation in Sphingobium yanoikuyae B1. J. Environ. Manage. 2017, 204, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Moree, W.J.; Phelan, V.V.; Wu, C.H.; Bandeira, N.; Cornett, D.S.; Duggan, B.M.; Dorrestein, P.C. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. USA 2012, 109, 13811–13816. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.C.; Johnson, G.T. Microbial transformation of phenazines by Aspergillus sclerotiorum. Mycologia 1969, 61, 452–467. [Google Scholar] [CrossRef]
- Gottlieb, H.E.; Kotlyar, V.; Nudelman, A. NMR chemical shifts of common laboratory solvents as trace impurities. J. Org. Chem. 1997, 62, 7512–7515. [Google Scholar] [CrossRef]
- Desai, C.; Madamwar, D. Extraction of inhibitor-free metagenomic DNA from polluted sediments, compatible with molecular diversity analysis using adsorption and ion-exchange treatments. Bioresour. Technol. 2007, 98, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production megablast searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Dasgupta, F.; Singh, P.P.; Srivastava, H.C. Acetylation of carbohydrates using ferric chloride in acetic anhydride. Carbohydr. Res. 1980, 80, 346–349. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iloabuchi, K.; Spiteller, D. Bacillus sp. G2112 Detoxifies Phenazine-1-carboxylic Acid by N5 Glucosylation. Molecules 2024, 29, 589. https://doi.org/10.3390/molecules29030589
Iloabuchi K, Spiteller D. Bacillus sp. G2112 Detoxifies Phenazine-1-carboxylic Acid by N5 Glucosylation. Molecules. 2024; 29(3):589. https://doi.org/10.3390/molecules29030589
Chicago/Turabian StyleIloabuchi, Kenechukwu, and Dieter Spiteller. 2024. "Bacillus sp. G2112 Detoxifies Phenazine-1-carboxylic Acid by N5 Glucosylation" Molecules 29, no. 3: 589. https://doi.org/10.3390/molecules29030589
APA StyleIloabuchi, K., & Spiteller, D. (2024). Bacillus sp. G2112 Detoxifies Phenazine-1-carboxylic Acid by N5 Glucosylation. Molecules, 29(3), 589. https://doi.org/10.3390/molecules29030589