Chemical Epigenetic Regulation Secondary Metabolites Derived from Aspergillus sydowii DL1045 with Inhibitory Activities for Protein Tyrosine Phosphatases
Abstract
:1. Introduction
2. Results
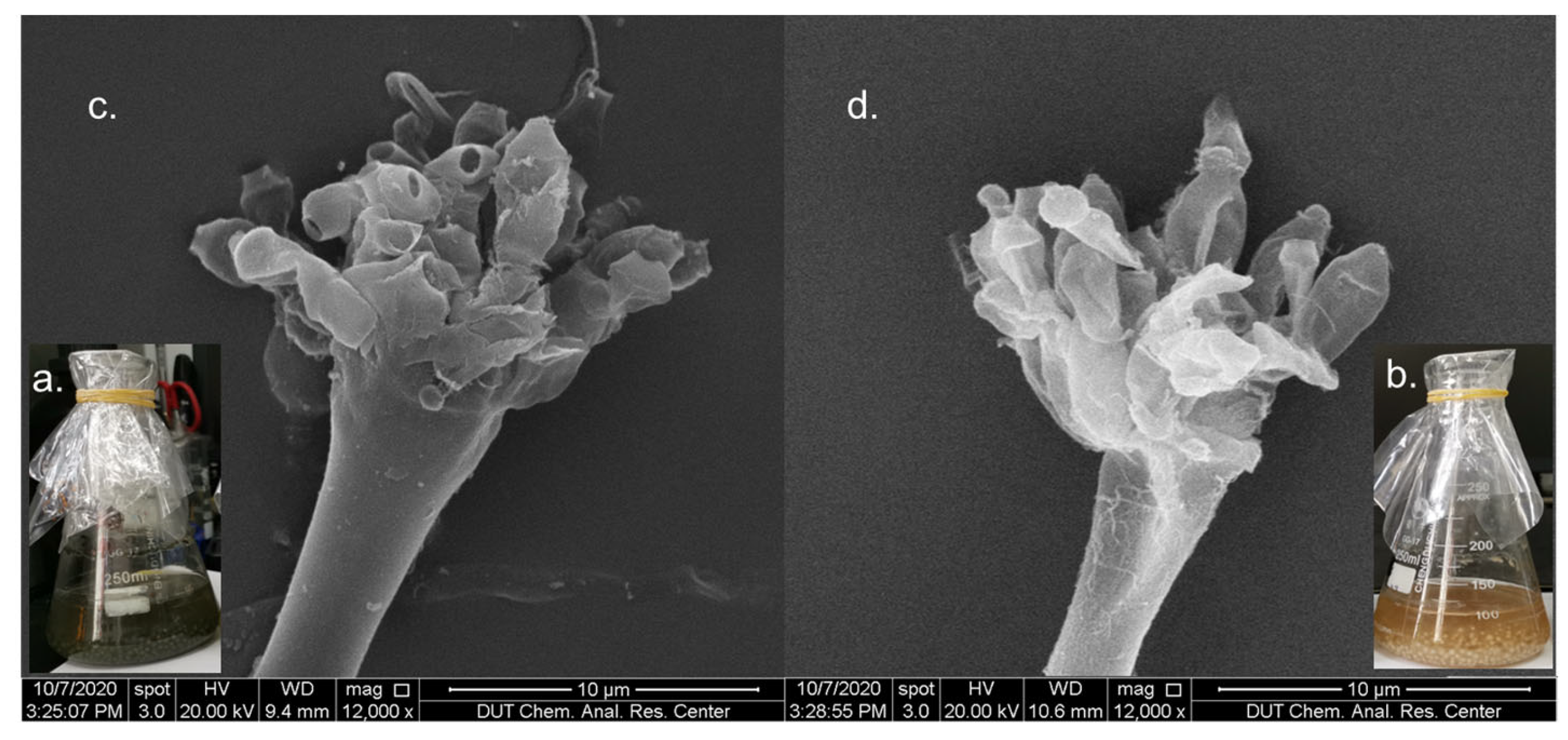
2.1. Effects of CER on Growth of A. sydowii DL1045
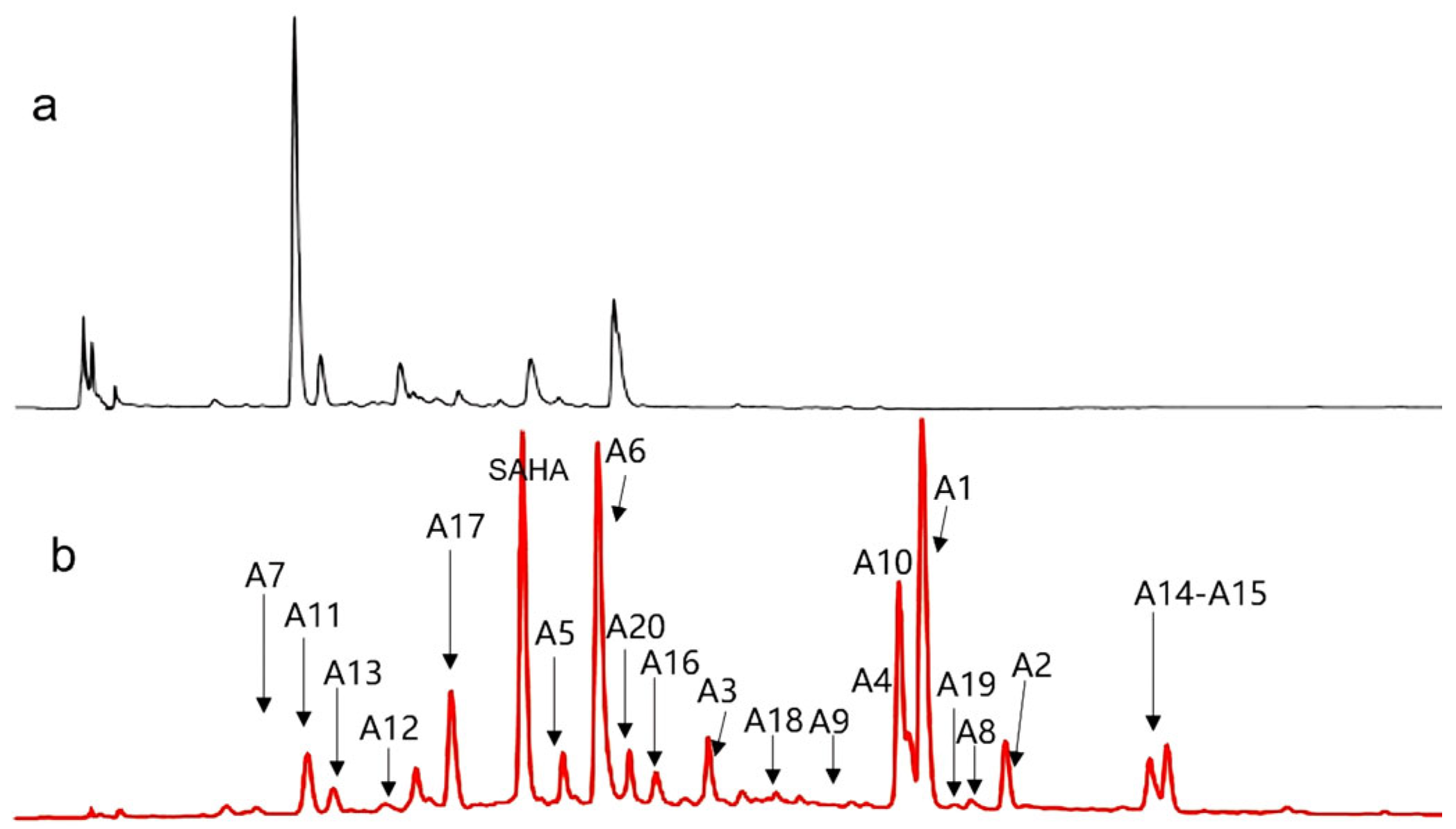
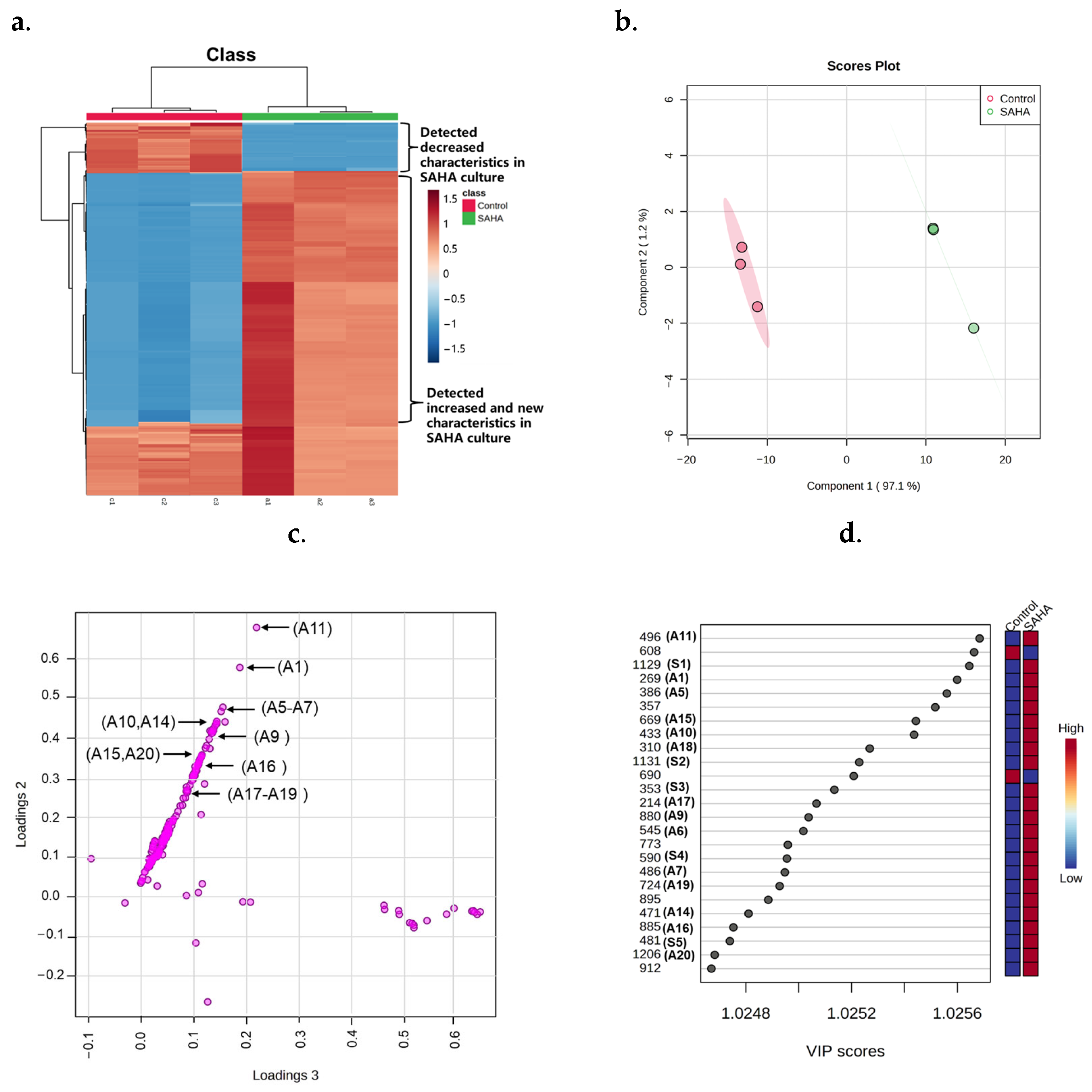
2.2. Metabolites Profile Induced by Adding SAHA into A. sydowii DL1045
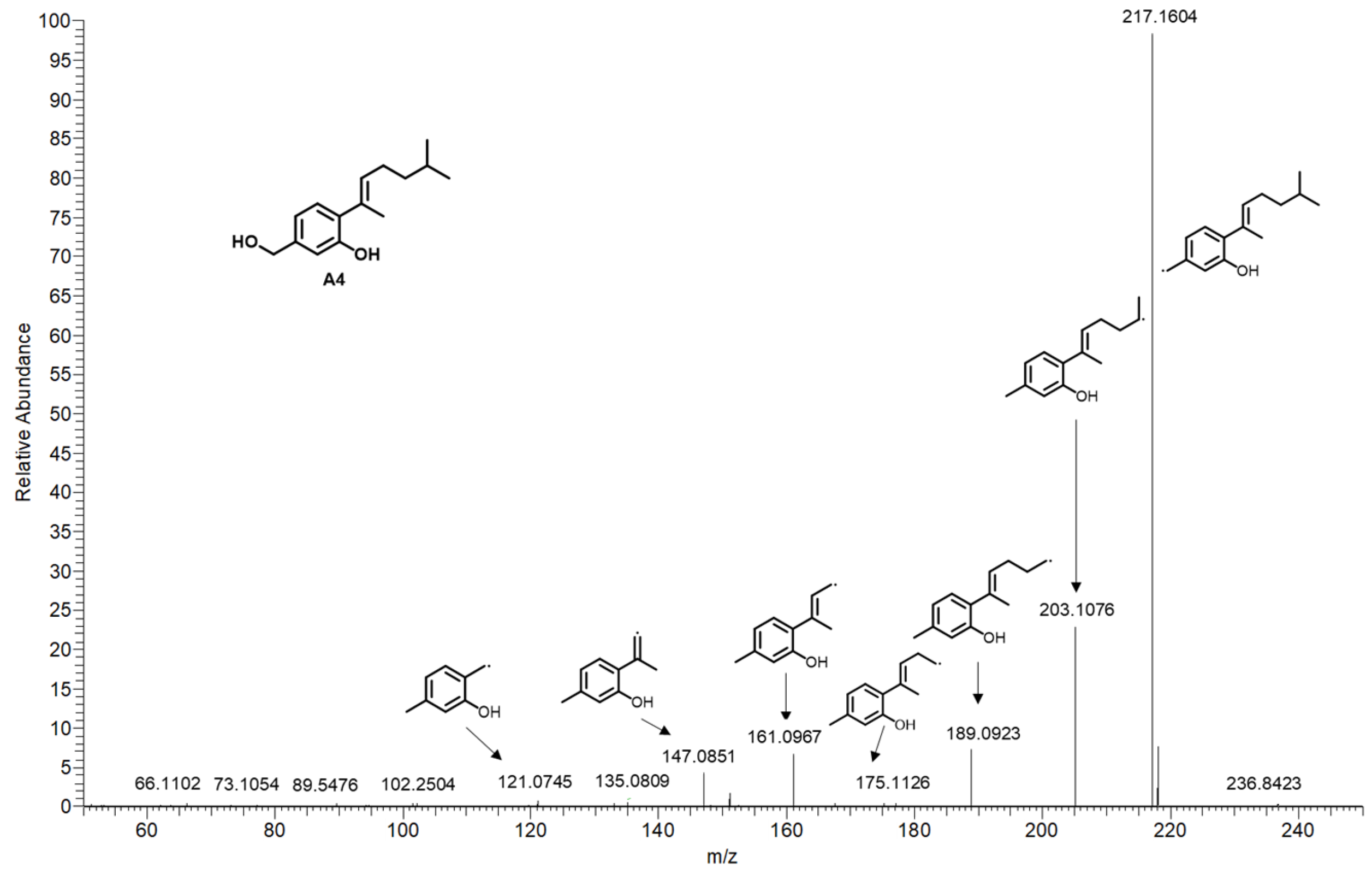
2.3. Identification of SAHA Biotransformation Products
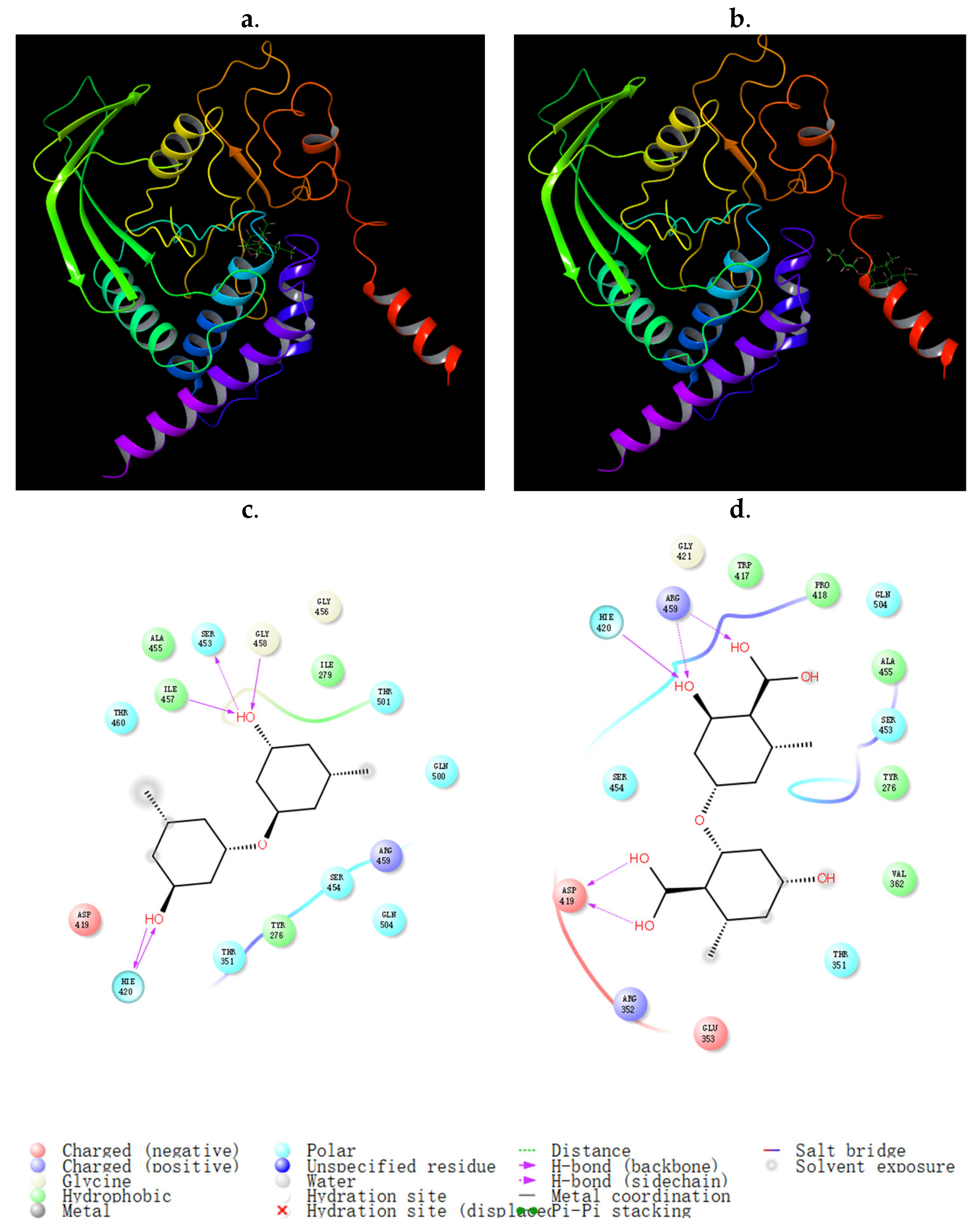
2.4. Biological Activity Assay
3. Discussion
4. Materials and Methods
4.1. Fungal Material
4.2. Extraction and Analysis by HPLC and LC-MS
4.3. Structural Annotation of Compounds
4.4. Isolation and Purification of the SMs
4.5. Computational Details
4.6. Bioactivity Evaluation
4.7. Molecular Docking
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Orfali, R.; Aboseada, M.A.; Abdel-Wahab, N.M.; Hassan, H.M.; Perveen, S.; Ameen, F.; Alturki, E.; Abdelmohsen, U.R. Recent updates on the bioactive compounds of the marine-derived genus Aspergillus. RSC Adv. 2021, 11, 17116–17150. [Google Scholar] [CrossRef]
- Alanzi, A.; Elhawary, E.A.; Ashour, M.L.; Moussa, A.Y. Aspergillus co-cultures: A recent insight into their secondary metabolites and microbial interactions. Arch. Pharmacal Res. 2023, 46, 273–298. [Google Scholar] [CrossRef]
- Ibrahim, S.; Mohamed, S.; Alsaadi, B.; Althubyani, M.; Awari, Z.; Hussein, H.; Aljohani, A.; Albasri, J.; Faraj, S.; Mohamed, G. Secondary metabolites, biological activities, and industrial and biotechnological importance of Aspergillus sydowii. Mar. Drugs 2023, 21, 441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, W.L.; Fang, Y.C.; Zhu, T.J.; Gu, Q.Q.; Zhu, W.M. Cytotoxic alkaloids and antibiotic nordammarane triterpenoids from the marine-derived fungus Aspergillus sydowi. J. Nat. Prod. 2008, 71, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Lin, X.P.; Qin, C.; Liao, S.R.; Wan, J.T.; Zhang, T.Y.; Liu, J.; Fredimoses, M.; Chen, H.; Yang, B.; et al. Antimicrobial and antiviral sesquiterpenoids from sponge-associated fungus, Aspergillus sydowii ZSDS1-F6. J. Antibiot. 2014, 67, 581–583. [Google Scholar] [CrossRef]
- Lanen, S.; Shen, B. Microbial genomics for the improvement of natural product discovery. Curr. Opin. Microbiol. 2006, 9, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Ronning, C.M.; Fedorova, N.D.; Bowyer, P.; Coulson, R.; Goldman, G.; Kim, H.S.; Turner, G.; Wortman, J.R.; Yu, J.; Anderson, M.J.; et al. Genomics of Aspergillus fumigatus. Rev. Iberoam. Micol. 2005, 22, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.F.; Somoza, A.D.; Keller, N.P.; Wang, C.C.C. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat. Prod. Rep. 2012, 29, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Fisch, K.M.; Gillaspy, A.F.; Gipson, M.; Henrikson, J.C.; Hoover, A.R.; Jackson, L.; Najar, F.Z.; Wagele, H.; Cichewicz, R.H. Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger. J. Ind. Microbiol. Biotechnol. 2009, 36, 1199–1213. [Google Scholar] [CrossRef]
- Zutz, C.; Gacek, A.; Sulyok, M.; Wagner, M.; Strauss, J.; Rychli, K. Small chemical chromatin effectors alter secondary metabolite production in Aspergillus clavatus. Toxins 2013, 5, 1723–1741. [Google Scholar] [CrossRef]
- Wang, L.; Li, M.; Tang, J.; Li, X. Eremophilane sesquiterpenes from a deep marine-derived fungus, Aspergillus sp. SCSIOW2, cultivated in the presence of epigenetic modifying agents. Molecules 2016, 21, 473. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xia, Z.; Tang, J.; Wu, J.; Tong, J.; Li, M.; Ju, J.; Chen, H.; Wang, L. Identification and biological evaluation of secondary metabolites from marine derived fungi-Aspergillus sp. SCSIOW3, cultivated in the presence of epigenetic modifying agents. Molecules 2017, 22, 1302. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sun, Y.; Liu, J.; Liu, W.; Xing, Y.; Xiu, Z.; Dong, Y. Metabolomic strategy to characterize the profile of secondary metabolites in Aspergillus aculeatus DL1011 regulated by chemical epigenetic agents. Molecules 2022, 28, 218. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, W.C.; Shi, X.; Zheng, H.Z.; Zheng, Z.H.; Lu, X.H.; Xing, Y.; Ji, K.; Liu, M.; Dong, Y.S. Inducing secondary metabolite production of Aspergillus sydowii through microbial co-culture with Bacillus subtilis. Microb. Cell Fact. 2021, 20, 42. [Google Scholar] [CrossRef] [PubMed]
- Julien, S.G.; Dubé, N.; Hardy, S.; Tremblay, M.L. Inside the human cancer tyrosine phosphatome. Nat. Rev. Cancer 2011, 11, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Zabolotny, J.M.; Haj, F.G.; Kim, Y.B.; Kim, H.J.; Shulman, G.I.; Kim, J.K.; Neel, B.G.; Kahn, B.B. Transgenic overexpression of protein-tyrosine phosphatase 1B in muscle causes insulin resistance, but overexpression with leukocyte antigen-related phosphatase does not additively impair insulin action. J. Biol. Chem. 2004, 279, 24844–24851. [Google Scholar] [CrossRef] [PubMed]
- Loh, K.; Fukushima, A.; Zhang, X.; Galic, S.; Briggs, D.; Enriori, P.J.; Simonds, S.; Wiede, F.; Reichenbach, A.; Hauser, C. Elevated hypothalamic TCPTP in obesity contributes to cellular leptin resistance. Cell Metab. 2012, 15, 925–926. [Google Scholar] [CrossRef]
- Dubois, M.J.; Bergeron, S.; Kim, H.J.; Dombrowski, L.; Perreault, M.; Fournès, B.; Faure, R.; Olivier, M.; Beauchemin, N.; Shulman, G.I.; et al. The SHP-1 protein tyrosine phosphatase negatively modulates glucose homeostasis. Nat. Med. 2006, 12, 549–556. [Google Scholar] [CrossRef]
- Reiko, U.; Susumu, K.; Takane, F. Stress compounds in the leaves of nicotiana undulata induced by tmv inoculation. Phytochemistry 1988, 27, 365–368. [Google Scholar]
- Liang, Z.; Zhang, T.; Zhang, X.; Zhang, J.; Zhao, C. An alkaloid and a steroid from the endophytic fungus Aspergillus fumigatus. Molecules 2015, 20, 1424–1433. [Google Scholar] [CrossRef]
- Huang, Z.; Nong, X.; Ren, Z.; Wang, J.; Zhang, X.; Qi, S. Anti-HSV-1, antioxidant and antifouling phenolic compounds from the deep-sea-derived fungus Aspergillus versicolor SCSIO 41502. Bioorg. Med. Chem. Lett. 2017, 27, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yang, J.; Shi, Y. Sesquiterpenoids and other constituents from the flower buds of Tussilago farfara. J. Asian Nat. Prod. Res. 2011, 13, 920–929. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Peng, S.; Yang, J.; Cong, Z.; Lin, X.; Liao, S.; Yang, B.; Zhou, X.; Zhou, X.; Liu, Y.; et al. Structurally diverse sesquiterpenoids and polyketides from a sponge-associated fungus Aspergillus sydowii SCSIO41301. Fitoterapia 2019, 135, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Zeng, X.; Feng, S.; Zhang, H.; Zhang, Y.; Luo, Z.; Xu, W.; Ma, X. Three new phomalone derivatives from a deep-sea-derived fungus Alternaria sp. MCCC 3A00467. Nat. Prod. Rep. 2020, 36, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Ahirrao, P.; Tambat, R.; Chandal, N.; Mahey, N.; Kamboj, A.; Jain, U.K.; Singh, I.P.; Jachak, S.M.; Nandanwar, H.S. MsrA efflux pump inhibitory activity of piper cubeba l.f. and its phytoconstituents against Staphylococcus aureus RN4220. Chem. Biodivers. 2020, 17, e2000144. [Google Scholar] [CrossRef] [PubMed]
- Wakana, D.; Kawahara, N.; Goda, Y. Two new pyrrolidine alkaloids, codonopsinol C and codonopiloside A, isolated from Codonopsis pilosula. Chem. Pharm. Bull. 2013, 61, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Rojas, I.S.; Lotina-Hennsen, B.; Mata, R. Effect of lichen metabolites on thylakoid electron transport and photophosphorylation in isolated spinach chloroplasts. J. Nat. Prod. 2000, 63, 1396–1399. [Google Scholar] [CrossRef]
- Ge, H.M.; Shen, Y.; Zhu, C.H.; Tan, S.H.; Ding, H.; Song, Y.C.; Tan, R.X. Penicidones A–C, three cytotoxic alkaloidal metabolites of an endophytic Penicillium sp. Phytochemistry 2008, 69, 571–576. [Google Scholar] [CrossRef]
- Hamao, U.; Norio, S.; Hiroshi, N.; Saburo, A.; Meiki, M.; Tomio, T. Isolation of lecanoric acid, an inhibitor of histidine decarboxylase from a fungus. J. Antibiot. 1974, 27, 587–596. [Google Scholar]
- Israilov, I.A.; Chelombit’ko, V.A.; Nazarova, L.E. Argemone alkaloids. Chem. Nat. Comp. 1986, 22, 742–743. [Google Scholar] [CrossRef]
- Wang, H.; Chen, K.; Xue, R.; Turghun, C.; Han, B. Identification of the chemical constituents in Cullen corylifolium ethanolic extract by LC-MS/MS and GC-MS. Nat. Prod. Rep. 2021, 37, 1392–1396. [Google Scholar] [CrossRef]
- George, R.P.; John, C.K.; Delbert, L.; Herald, D.L.; Davenport, R.; Pettit, R.K.; Tucker, B.E.; Schmidt, J.M. Isolation of Labradorins 1 and 2 from Pseudomonas syringae pv. coronafaciens. J. Nat. Prod. 2002, 65, 1793–1797. [Google Scholar]
- Ferrari, C.; Marion, L. Further alkaloids related to aspidoalbine and limaspermine. Can. J. Chem. 1964, 42, 2705–2709. [Google Scholar] [CrossRef]
- Yoshida, S.; Yoneyama, K.; Shiraishi, S.; Watanabe, A.; Takahashi, N. Chemical Structures of New Piericidins Produced by Streptomyces pactum. Agric. Biol. Chem. 1977, 41, 855–862. [Google Scholar] [CrossRef]
- Lee, M.; Song, J.Y.; Chin, Y.-W.; Sung, S.H. Anti-adipogenic diarylheptanoids from Alnus hirsuta f. sibirica on 3T3-L1 cells. Bioorg. Med. Chem. Lett. 2013, 23, 2069–2073. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Montanaro, S.; Ravelli, D.; Merli, D.; Fagnoni, M.; Albini, A. Decatungstate as photoredox catalyst: Benzylation of electron-poor olefins. Org. Lett. 2012, 14, 4218–4221. [Google Scholar] [CrossRef] [PubMed]
- Dudek, G.O.; Holm, R.H. Nuclear magnetic resonance studies of keto-enol equilibria. III. α,β-unsaturated-β-ketoamines. J. Am. Chem. Soc. 1962, 84, 2691–2696. [Google Scholar] [CrossRef]
- Alicea Velázquez, N.L.; Jakoncic, J.; Boggon, T.J. Structure-guided studies of the SHP-1/JAK1 interaction provide new insights into phosphatase catalytic domain substrate recognition. J. Struct. Biol. 2013, 181, 243–251. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Ren, H.; Liu, R.; Chen, L.; Zhu, T.; Zhu, W.M.; Gu, Q.Q. Two new hetero-spirocyclic γ-lactam derivatives from marine sediment-derived fungus Aspergillus sydowi D2–6. Arch. Pharmacal Res. 2010, 33, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Trisuwan, K.; Rukachaisirikul, V.; Kaewpet, M.; Phongpaichit, S.; Hutadilok-Towatana, N.; Preedanon, S.; Sakayaroj, J. Sesquiterpene and xanthone derivatives from the sea fan-derived fungus Aspergillus sydowii PSU-F154. J. Nat. Prod. 2011, 74, 1663–1667. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Raja, H.A.; Darveaux, B.A.; Chen, W.L.; Swanson, S.M.; Pearce, C.J.; Oberlies, N.H. New diketopiperazine dimer from a filamentous fungal isolate of Aspergillus sydowii. Magn. Reson. Chem. 2015, 53, 616–619. [Google Scholar] [CrossRef] [PubMed]
- Gesiane, d.S.L.; Aline, M.d.R.; Gabriel, F.d.S.; Alice, F.D.S.; Ivanildo, E.M.; Jacqueline, A.T. Metabolic response of Aspergillus sydowii to OSMAC modulation produces acetylcholinesterase inhibitors. Phytochem. Lett. 2018, 24, 39–45. [Google Scholar]
- Yang, S.Q.; Li, X.M.; Li, X.; Li, H.L.; Meng, L.H.; Wang, B.G. New citrinin analogues produced by coculture of the marine algal-derived endophytic fungal strains Aspergillus sydowii EN-534 and Penicillium citrinum EN-535. Phytochem. Lett. 2018, 25, 191–195. [Google Scholar] [CrossRef]
- Sun, Y.; Shi, X.; Xing, Y.; Ren, X.X.; Zhang, D.Y.; Li, X.; Xiu, Z.L.; Dong, Y.S. Co-culture of Aspergillus sydowii and Bacillus subtilis induces the production of antibacterial metabolites. Fungal Biol. 2022, 126, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.M.; Wei, C.K.; Chuang, D.W.; Mohamed, E.S.; Hsieh, C.T.; Asai, T.; Oshima, Y.; Hsieh, T.J.; Hwang, T.L.; Wu, Y.C.; et al. An epigenetic modifier enhances the production of anti-diabetic and anti-inflammatory sesquiterpenoids from Aspergillus sydowii. Bioorg. Med. Chem. 2013, 21, 3866–3872. [Google Scholar] [CrossRef] [PubMed]
- Pidroni, A.; Faber, B.; Brosch, G.; Bauer, I.; Graessle, S. A class 1 histone deacetylase as major regulator of secondary metabolite production in Aspergillus nidulans. Front. Microbiol. 2018, 9, 2212. [Google Scholar] [CrossRef]
- Shwab, E.K.; Bok, J.W.; Tribus, M.; Galehr, J.; Graessle, S.; Keller, N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef]
- Albright, J.C.; Henke, M.T.; Soukup, A.A.; McClure, R.A.; Thomson, R.J.; Keller, N.P.; Kelleher, N.L. Large-scale metabolomics reveals a complex response of Aspergillus nidulans to epigenetic perturbation. ACS Chem. Biol. 2015, 10, 1535–1541. [Google Scholar] [CrossRef]
- Henke, M.T.; Soukup, A.A.; Goering, A.W.; McClure, R.A.; Thomson, R.J.; Keller, N.P.; Kelleher, N.L. New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans. ACS Chem. Biol. 2016, 11, 2117–2123. [Google Scholar] [CrossRef]
- Allard, P.-M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Triastuti, A.; Vansteelandt, M.; Barakat, F.; Trinel, M.; Jargeat, P.; Fabre, N.; Amasifuen Guerra, C.A.; Mejia, K.; Valentin, A.; Haddad, M. How histone deacetylase inhibitors alter the secondary metabolites of botryosphaeria mamane, an endophytic fungus isolated from Bixa orellana. Chem. Biodivers. 2019, 16, e1800485. [Google Scholar]
- Anandan, S.K.; Ward, J.S.; Brokx, R.D.; Bray, M.R.; Patel, D.V.; Xiao, X.X. Mercaptoamide-based non-hydroxamic acid type histone deacetylase inhibitors. Bioorg. Med. Chem. Lett. 2005, 15, 1969–1972. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, M.V.; Konduktorov, K.A.; Shcherbakova, A.S.; Kochetkov, S.N. Synthesis of N′-propylhydrazide analogs of hydroxamic inhibitors of histone deacetylases (HDACs) and evaluation of their impact on activities of HDACs and replication of hepatitis C virus (HCV). Bioorg. Med. Chem. Lett. 2019, 29, 2369–2374. [Google Scholar] [CrossRef]
- Asai, T.; Chung, Y.M.; Sakurai, H.; Ozeki, T.; Chang, F.R.; Wu, Y.C.; Yamashita, K.; Oshima, Y. Highly oxidized ergosterols and isariotin analogs from an entomopathogenic fungus, Gibellula formosana, cultivated in the presence of epigenetic modifying agents. Tetrahedron 2012, 68, 5817–5823. [Google Scholar] [CrossRef]
- Tsugawa, H.; Nakabayashi, R.; Mori, T.; Yamada, Y.; Takahashi, M.; Rai, A.; Sugiyama, R.; Yamamoto, H.; Nakaya, T.; Yamazaki, M.; et al. A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 2019, 16, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Duhrkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Bocker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.C.; Yang, F.; Zhang, R.; Shi, X.; Lu, X.H.; Luan, Y.S.; Xiu, Z.L.; Dong, Y.S. Production of polyketides with anthelmintic activity by the fungus Talaromyces wortmannii using one strain-many compounds (OSMAC) method. Phytochem. Lett. 2016, 18, 157–161. [Google Scholar] [CrossRef]
- Sun, W.; Zhuang, C.; Li, X.; Zhang, B.; Lu, X.; Zheng, Z.; Dong, Y. Varic acid analogues from fungus as PTP1B inhibitors: Biological evaluation and structure-activity relationships. Bioorg. Med. Chem. Lett. 2017, 27, 3382–3385. [Google Scholar] [CrossRef]
- Dong, Y.S.; Yu, N.; Li, X.; Zhang, B.; Xing, Y.; Zhuang, C.; Xiu, Z.L. Dietary 5,6,7-trihydroxy-flavonoid aglycones and 1-deoxynojirimycin synergistically inhibit the recombinant maltase-glucoamylase subunit of alpha-glucosidase and lower postprandial blood glucose. J. Agric. Food Chem. 2020, 68, 8774–8787. [Google Scholar] [CrossRef]
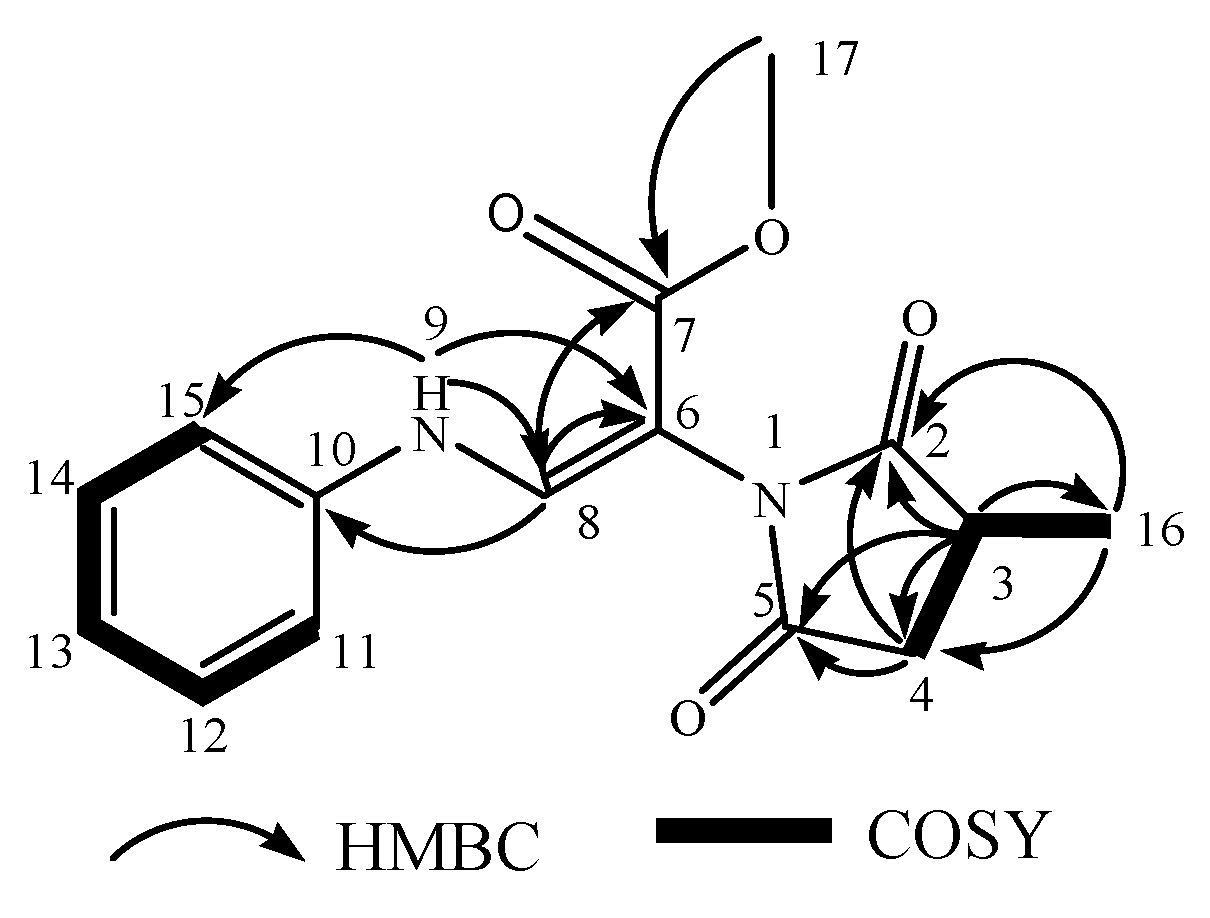
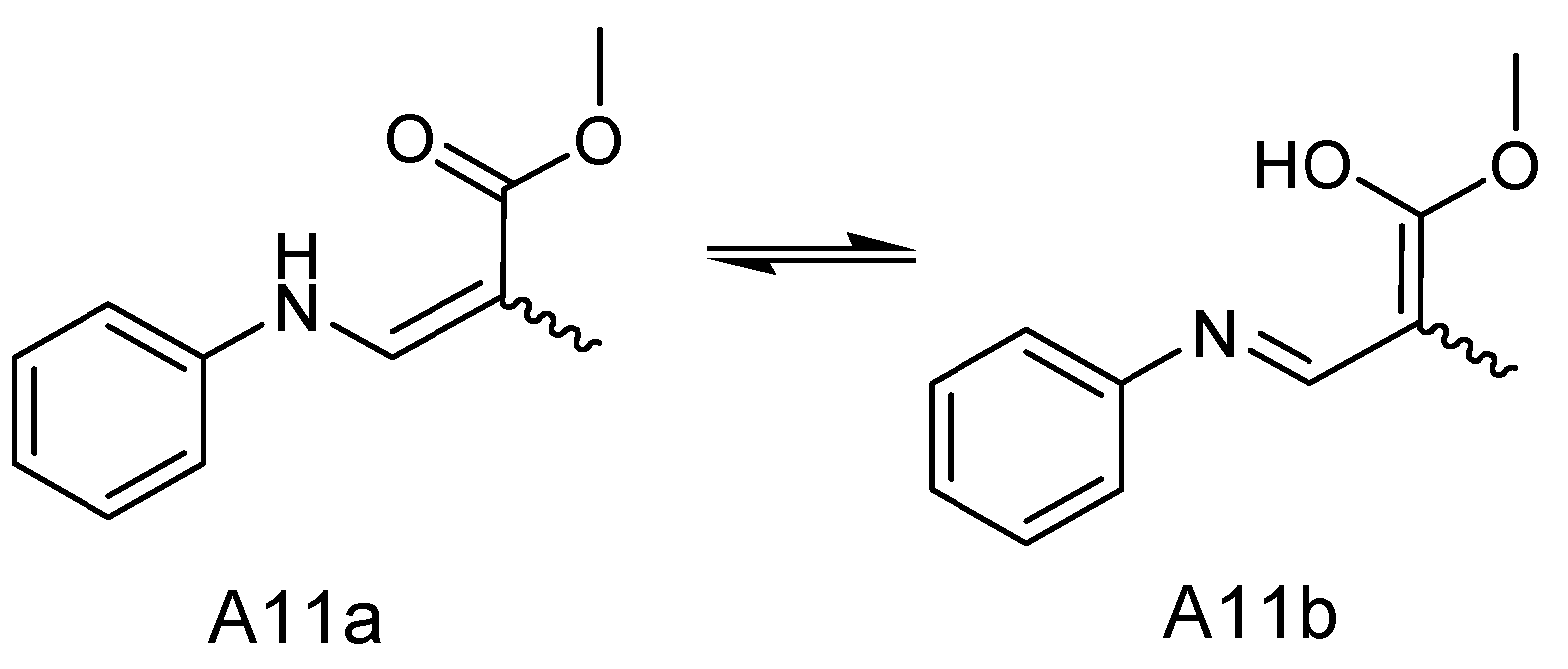

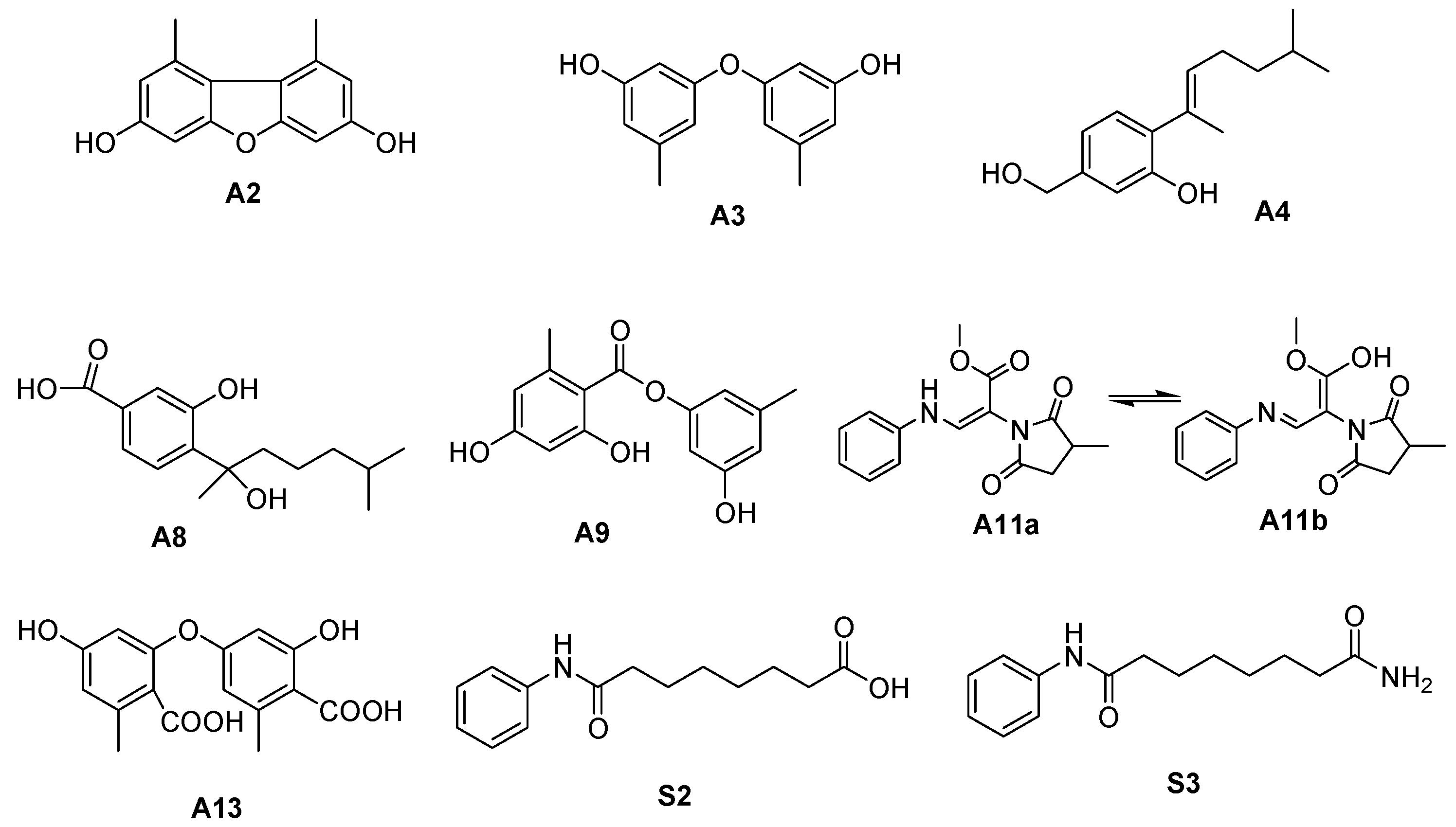
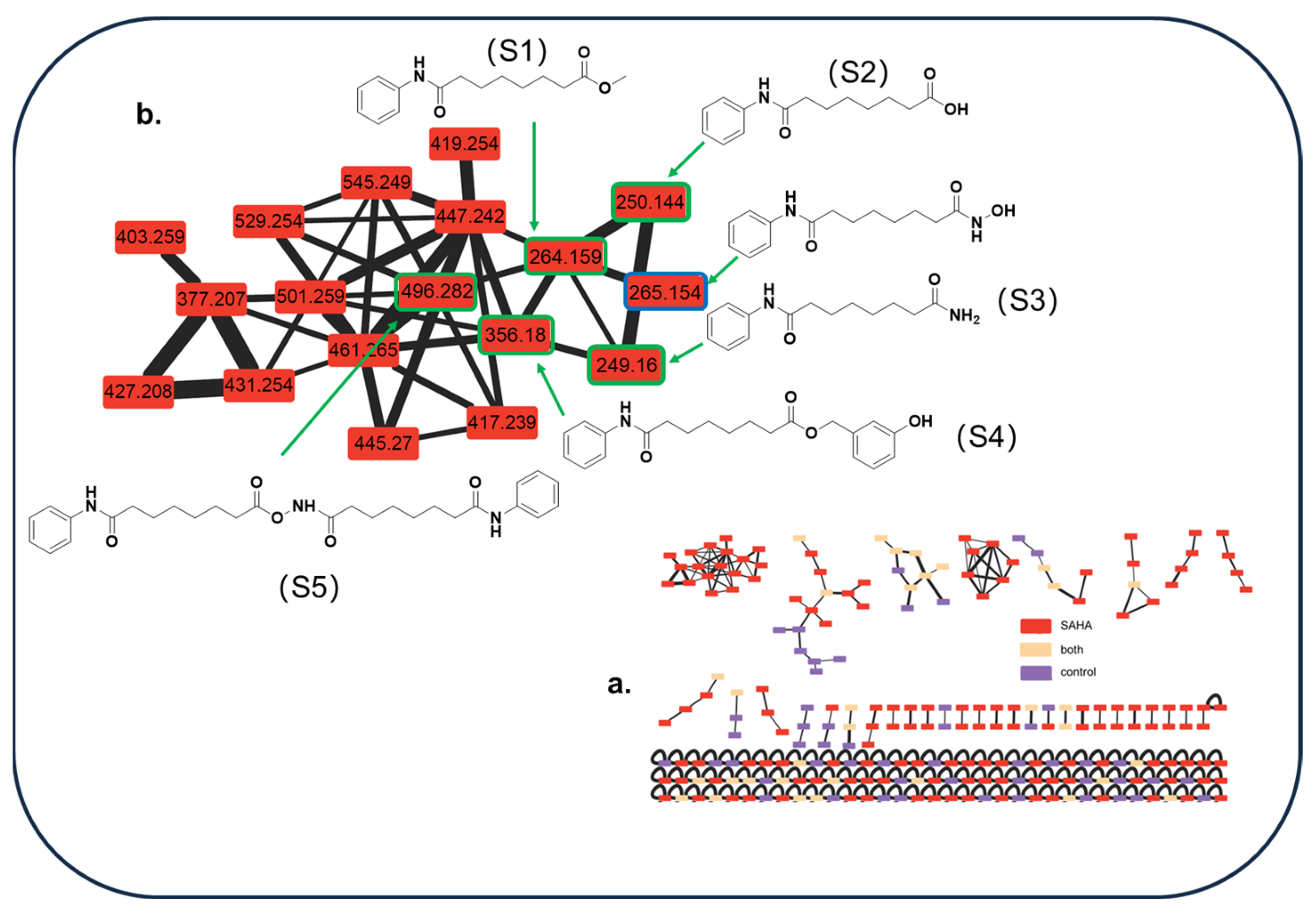
| No. | tR [min] | Observed [m/z] | Adduction | Molecular Formula | Error (ppm) | Identification | Originally Isolated from | First Report in A. sydowii | Identification Method (Level) |
|---|---|---|---|---|---|---|---|---|---|
| A1 | 24.472 | 217.1585 | [M+H]+ | C15H20O | −0.929 | (−)-1,2-dehydro-α-cyperone | Tussilago farfara [22] | Y | 2 |
| A2 | 26.640 | 229.0876 | [M+H]+ | C14H12O3 | −0.630 | 3,7-dihydroxy-1,9-dimethyldibenzofuran | A. sydowii [23] | N | 2, 4 |
| A3 | 22.818 | 231.1015 | [M+H]+ | C14H14O3 | −0.040 | diorcinol | A. sydowii [5] | N | 2, 4 |
| A4 | 24.341 | 235.1692 | [M+H]+ | C15H22O2 | −1.032 | aspergillusene A | A. sydowii [5] | N | 2, 4 |
| A5 | 20.616 | 239.1282 | [M+H]+ | C13H18O4 | 0.766 | 1-(3-ethyl-2,4-dihydroxy-6-methoxyphenyl) butan-1-one deoxyphomalone | Alternaria sp. [24] | Y | 1 |
| A6 | 21.301 | 253.1438 | [M+H]+ | C13H16O5 | −2.915 | methyl trimethoxycinnamate | Staphylococcus aureus [25] | Y | 2 |
| A7 | 7.868 | 256.1177 | [M+H]+ | C12H17NO5 | −2.958 | radicamine A | Codonopsis pilosula [26] | Y | 2 |
| A8 | 25.210 | 267.1586 | [M+H]+ | C15H22O4 | −3.758 | sydonic acid | A. sydowii [23] | N | 2 |
| A9 | 23.068 | 275.0912 | [M+H]+ | C15H14O5 | −2.866 | lecanorin | Parmotrema tinctorum [27] | Y | 2, 4 |
| A10 | 23.259 | 286.1441 | [M+Na]+ | C17H19NO3 | 0.872 | piperine | Staphylococcus aureus [25] | Y | 1 |
| A11 | 12.592 | 289.1183 | [M+H]+ | C15H16N2O4 | −1.874 | sydowimide A | A. sydowii | Y | 4 |
| A12 | 15.624 | 299.0903 | [M+H]+ | C17H14O5 | 0.329 | emodin-1,6-dimethyl ether | Penicillium sp. [28] | Y | 2 |
| A13 | 15.250 | 319.0812 | [M+H]+ | C16H14O7 | −1.779 | diorcinol acid | A. sydowii [21] | N | 2 |
| A14 | 28.042 | 319.0813 | [M+H]+ | C16H14O7 | −1.778 | lecanoric acid | Pyricularia [29] | Y | 2 |
| A15 | 28.391 | 328.1542 | [M+H]+ | C19H21NO4 | −2.173 | scoulerine | Argemone [30] | Y | 1 |
| A16 | 22.457 | 347.1251 | [M+Na]+ | C20H20O4 | −2.219 | bavachin | Cullen corylifolium [31] | Y | 1 |
| A17 | 22.909 | 393.2939 | [M+H]+ | C24H40O4 | −0.654 | deoxycholic acid | Pseudomonas syringae [32] | Y | 1 |
| A18 | 19.735 | 417.2390 | [M+H]+ | C23H32N2O5 | −0.402 | N-Acetyl-N-depropionyl-aspido-albinol | Aspidosperma album [33] | Y | 2 |
| A19 | 24.691 | 458.3265 | [M+H]+ | C28H43NO4 | 1.588 | 2-((2E,5E,7E,11E)-10-Hydroxy-3,5,7,9,11,13-hexamethyl-tetradeca-2,5,7,11-tetraenyl)-5,6-dimethoxy-3-methyl-pyridin-4-ol | Streptomyces pactum [34] | Y | 2 |
| A20 | 22.412 | 517.2373 | [M+Na]+ | C25H34O10 | −0.306 | rubranoside A | Alnus hirsuta f. sibirica [35] | Y | 1 |
| NO. | A11a | A11b | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | ||||
| 2 | 180.05 | 179.70 | ||
| 3 | 36.51 | 2.4 (1H, m) | 36.13 | 2.4 (1H, m) |
| 4 | 34.58 | 3.01 (2H, m) | 34.67 | 3.01 (2H, m) |
| 5 | 175.98 | 175.86 | ||
| 6 | 95.62 | 95.68 | ||
| 7 | 164.19 | 164.25 | ||
| 8 | 140.41 | 8.08 (1H, d, 10) | 140.41 | 8.08 (1H, d, 10) |
| NH | 9.22 (d, J = 13.5 Hz, 1H) | 9.17 (d, J = 13.6 Hz, 1H) | ||
| 10 | 139.59 | 139.67 | ||
| 11 | 116.01 | 7.16 (1H, m) | 116.27 | 7.16 (1H, m) |
| 12 | 129.61 | 7.33 (1H, m) | 129.61 | 7.33 (1H, m) |
| 13 | 122.80 | 7.03 (1H, m) | 122.90 | 7.03 (1H, m) |
| 14 | 129.61 | 7.33 (1H, m) | 129.61 | 7.33 (1H, m) |
| 15 | 116.01 | 7.16 (1H, m) | 116.27 | 7.16 (1H, m) |
| 16 | 16.64 | 1.25 (d, J = 6.8 Hz, 3H) | 15.25 | 1.33 (d, J = 6.7 Hz, 3H) |
| 17 | 51.24 | 3.62 (3H, s) | 51.24 | 3.62 (3H, s) |
| IC50 (μM) | ||||
|---|---|---|---|---|
| SHP1 | TCPTP | PTP1B | CD45 | |
| A3 | 0.96 ± 0.02 | >20 | >20 | 4.03 ± 0.13 |
| A4 | 8.69 ± 0.26 | >20 | 9.65 ± 0.73 | >20 |
| A9 | 6.01 ± 0.18 | >20 | 7.60 ± 0.21 | 7.40 ± 0.08 |
| A11 | 1.50 ± 0.09 | 2.40 ± 0.65 | >20 | 18.83 ± 0.04 |
| A13 | 2.16 ± 0.23 | >20 | >20 | 4.46 ± 0.16 |
| Site | Site Score | Docking Glide Score (kcal/mol) | ||
|---|---|---|---|---|
| A3 | A13 | |||
| SHP1 | 2 | 0.927 | −5.622 | −4.588 |
| 1 | 0.903 | −4.356 | −4.135 | |
| 3 | 0.706 | −3.002 | −4.832 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, X.; Li, X.; He, X.; Zhang, D.; Quan, C.; Xiu, Z.; Dong, Y. Chemical Epigenetic Regulation Secondary Metabolites Derived from Aspergillus sydowii DL1045 with Inhibitory Activities for Protein Tyrosine Phosphatases. Molecules 2024, 29, 670. https://doi.org/10.3390/molecules29030670
Shi X, Li X, He X, Zhang D, Quan C, Xiu Z, Dong Y. Chemical Epigenetic Regulation Secondary Metabolites Derived from Aspergillus sydowii DL1045 with Inhibitory Activities for Protein Tyrosine Phosphatases. Molecules. 2024; 29(3):670. https://doi.org/10.3390/molecules29030670
Chicago/Turabian StyleShi, Xuan, Xia Li, Xiaoshi He, Danyang Zhang, Chunshan Quan, Zhilong Xiu, and Yuesheng Dong. 2024. "Chemical Epigenetic Regulation Secondary Metabolites Derived from Aspergillus sydowii DL1045 with Inhibitory Activities for Protein Tyrosine Phosphatases" Molecules 29, no. 3: 670. https://doi.org/10.3390/molecules29030670
APA StyleShi, X., Li, X., He, X., Zhang, D., Quan, C., Xiu, Z., & Dong, Y. (2024). Chemical Epigenetic Regulation Secondary Metabolites Derived from Aspergillus sydowii DL1045 with Inhibitory Activities for Protein Tyrosine Phosphatases. Molecules, 29(3), 670. https://doi.org/10.3390/molecules29030670







