Bioactive α-Pyrone Analogs from the Endophytic Fungus Diaporthe sp. CB10100: α-Glucosidase Inhibitory Activity, Molecular Docking, and Molecular Dynamics Studies
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structural Elucidation
2.2. Analysis of Secondary Metabolite Biosynthetic Potential
2.3. α-Glucosidase Inhibition Activity
2.4. Antibacterial Assay
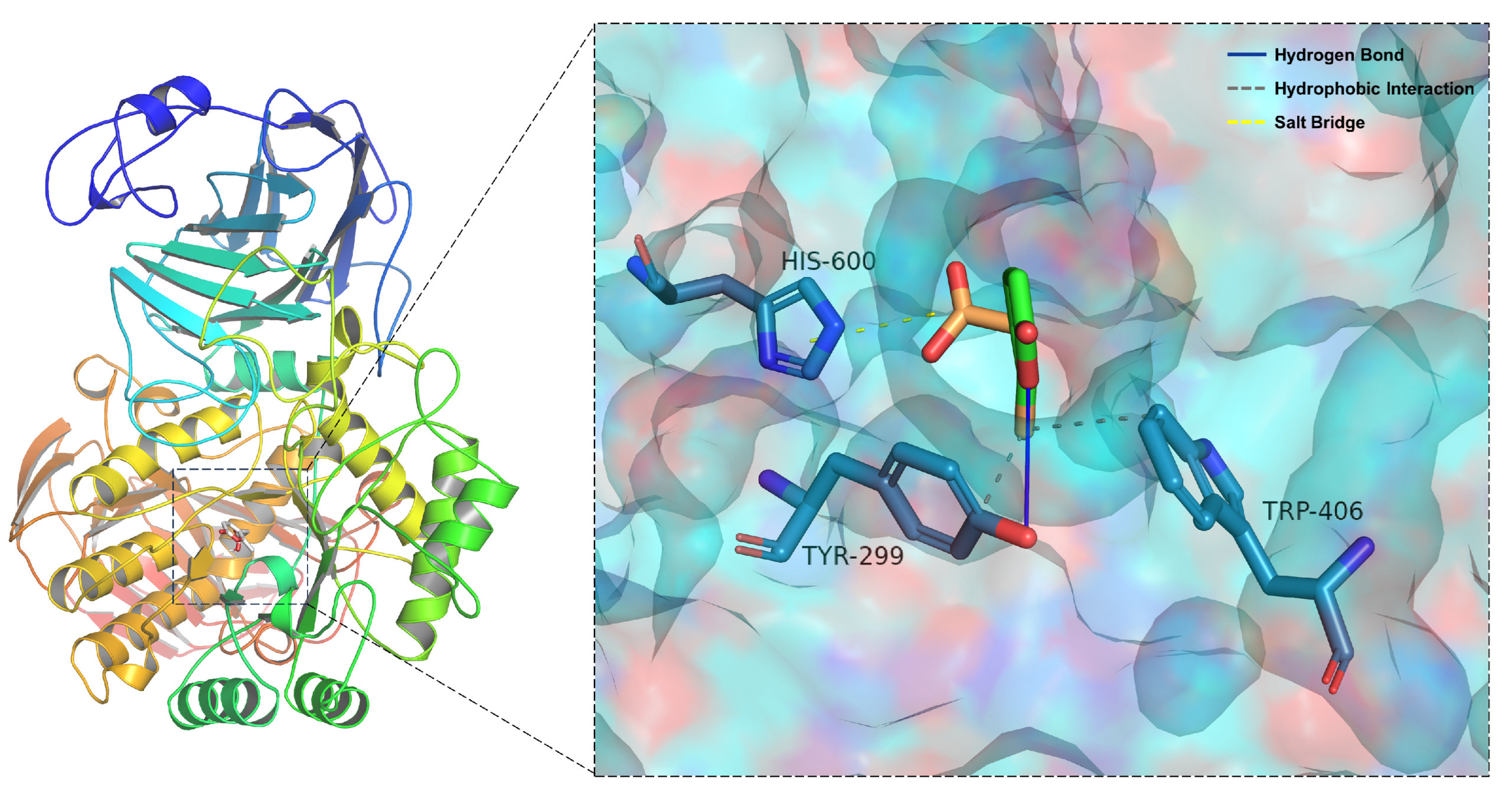
2.5. Molecular Docking
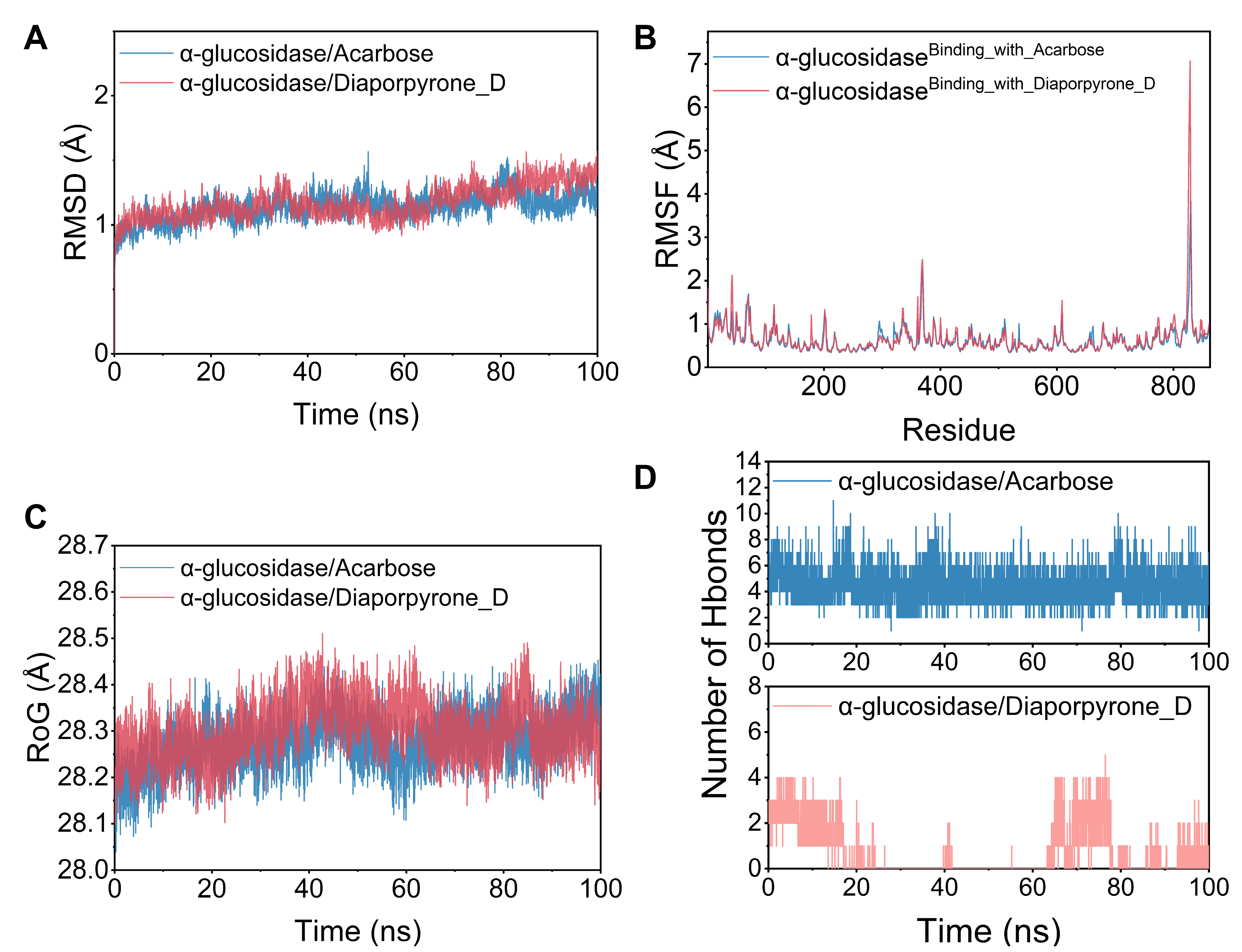
2.6. Molecular Dynamics Simulations
3. Materials and Methods
3.1. General Methods
3.2. Fungal Strain
3.3. Fermentation of Diaporthe sp. CB10100
3.4. Isolation of Compounds 3–4
Diaporpyrone F (3)
3.5. α-Glucosidase Inhibition Assay
3.6. Antibacterial Assay
3.7. Molecular Docking Analysis
3.8. Molecular Dynamic Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Su, J.; Tang, L.; Luo, Y.; Xu, J.; Ouyang, S. Research Progress on Drugs for Diabetes Based on Insulin Receptor/Insulin Receptor Substrate. Biochem. Pharmacol. 2023, 217, 115830. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and Regional Diabetes Prevalence Estimates for 2019 and Projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th Edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef]
- Majety, P.; Lozada Orquera, F.A.; Edem, D.; Hamdy, O. Pharmacological Approaches to the Prevention of Type 2 Diabetes Mellitus. Front. Endocrinol. 2023, 14, 1118848. [Google Scholar] [CrossRef] [PubMed]
- Blahova, J.; Martiniakova, M.; Babikova, M.; Kovacova, V.; Mondockova, V.; Omelka, R. Pharmaceutical Drugs and Natural Therapeutic Products for the Treatment of Type 2 Diabetes Mellitus. Pharmaceuticals 2021, 14, 806. [Google Scholar] [CrossRef]
- Sohretoglu, D.; Renda, G.; Arroo, R.; Xiao, J.; Sari, S. Advances in the Natural α-Glucosidase Inhibitors. eFood 2023, 4, e112. [Google Scholar] [CrossRef]
- Agrawal, S.; Bhatt, A. Microbial Endophytes: Emerging Trends and Biotechnological Applications. Curr. Microbiol. 2023, 80, 249. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, L.; Pan, Y.; Zheng, X.; Liang, X.; Sheng, L.; Zhang, D.; Sun, Q.; Wang, Q. Research Advances on Endophytic Fungi and Their Bioactive Metabolites. Bioprocess. Biosyst. Eng. 2023, 46, 165–170. [Google Scholar] [CrossRef]
- Kandasamy, G.D.; Kathirvel, P. Insights into Bacterial Endophytic Diversity and Isolation with a Focus on Their Potential Applications -A Review. Microbiol. Res. 2023, 266, 127256. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.Y.; Liu, X.; Cui, J.L.; Wang, J.H.; Wang, M.L.; Zhang, G. Endophytic Fungi and Their Bioactive Secondary Metabolites in Medicinal Leguminosae Plants: Nearly Untapped Medical Resources. FEMS Microbiol. Lett. 2022, 369, fnac052. [Google Scholar] [CrossRef]
- Jiang, L.; Ma, Q.; Li, A.; Sun, R.; Tang, G.; Huang, X.; Pu, H. Bioactive Secondary Metabolites Produced by Fungi of the Genus Diaporthe (Phomopsis): Structures, Biological Activities, and Biosynthesis. Arab. J. Chem. 2023, 16, 105062. [Google Scholar] [CrossRef]
- Chang, J.L.; Pei, J.; Zhou, Y.H.; Ouyang, Q.X.; Qin, C.L.; Hu, J.Y.; Meng, X.G.; Ruan, H.L. Diaporaustalides A–L, Austalide Meroterpenoids from a Plant Endophytic Diaporthe sp. J. Nat. Prod. 2024, 87, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Khan, B.; Li, Y.; Wei, W.; Liu, G.; Xiao, C.; He, B.; Zhang, C.; Rajput, N.A.; Ye, Y.; Yan, W. Chemical Investigation of Endophytic Diaporthe Unshiuensis YSP3 Reveals New Antibacterial and Cytotoxic Agents. J. Fungi 2023, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Aldino, I.Y.; Rivera-Chávez, J.; Morales-Jiménez, J. Integrating Taxonomic and Chemical Diversity of Mangrove-Associated Ascomycetes to Discover or Repurpose Bioactive Natural Products. J. Nat. Prod. 2023, 86, 2423–2434. [Google Scholar] [CrossRef] [PubMed]
- McGlacken, G.P.; Fairlamb, I.J.S. 2-Pyrone Natural Products and Mimetics: Isolation, Characterisation and Biological Activity. Nat. Prod. Rep. 2005, 22, 369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, H.; Chen, Y.; Wei, S.; Zhang, W.; Tan, H. Cytospones E-J from the Endophytic Fungus Cytospora Rhizophorae. Fitoterapia 2022, 163, 105324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, B.; Cai, L.; Liu, L. New Dibenzo-α-Pyrone Derivatives with α-Glucosidase Inhibitory Activities from the Marine-Derived Fungus Alternaria Alternata. Mar. Drugs 2022, 20, 778. [Google Scholar] [CrossRef] [PubMed]
- Pu, H.; Liu, J.; Wang, Y.; Peng, Y.; Zheng, W.; Tang, Y.; Hui, B.; Nie, C.; Huang, X.; Duan, Y.; et al. Bioactive α-Pyrone Derivatives from the Endophytic Fungus Diaporthe sp. CB10100 as Inducible Nitric Oxide Synthase Inhibitors. Front. Chem. 2021, 9, 679592. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Zhang, R.; Dong, L.; Chi, J.; Huang, F.; Dong, L.; Zhang, M.; Jia, X. α-Glucosidase Inhibitors from Brown Rice Bound Phenolics Extracts (BRBPE): Identification and Mechanism. Food Chem. 2022, 372, 131306. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.T.; Deng, X.Y.; Chen, J.; Liang, Q.M.; Zhang, K.; Li, D.L.; Wu, P.P.; Zheng, X.; Zhou, R.P.; Jiang, Z.Y.; et al. Synthesis and Biological Evaluation of Coumarin Derivatives as α-Glucosidase Inhibitors. Eur. J. Med. Chem. 2020, 189, 112013. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Sanner, M.F. Python: A Programming Language for Software Integration and Development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An Overview of the Amber Biomolecular Simulation Package. WIREs Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Sagui, C.; Darden, T.A. Molecular Dynamics Simulations of Biomolecules: Long-Range Electrostatic Effects. Annu. Rev. Biophys. Biomolec. Struct. 1999, 28, 155–179. [Google Scholar] [CrossRef]
- Kräutler, V.; Van Gunsteren, W.F.; Hünenberger, P.H. A Fast SHAKE Algorithm to Solve Distance Constraint Equations for Small Molecules in Molecular Dynamics Simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Larini, L.; Mannella, R.; Leporini, D. Langevin Stabilization of Molecular-Dynamics Simulations of Polymers by Means of Quasisymplectic Algorithms. J. Chem. Phys. 2007, 126, 104101. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the Performance of the MM/PBSA and MM/GBSA Methods. 1. The Accuracy of Binding Free Energy Calculations Based on Molecular Dynamics Simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zheng, Y.; Fong, P.; Mao, S.; Wang, Q. The Application of the MM/GBSA Method in the Binding Pose Prediction of FGFR Inhibitors. Phys. Chem. Chem. Phys. 2020, 22, 9656–9663. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Rastelli, G.; Del Rio, A.; Degliesposti, G.; Sgobba, M. Fast and Accurate Predictions of Binding Free Energies Using MM-PBSA and MM-GBSA. J. Comput. Chem. 2010, 31, 797–810. [Google Scholar] [CrossRef]
- Nguyen, H.; Roe, D.R.; Simmerling, C. Improved Generalized Born Solvent Model Parameters for Protein Simulations. J. Chem. Theory Comput. 2013, 9, 2020–2034. [Google Scholar] [CrossRef]
- Weiser, J.; Shenkin, P.S.; Still, W.C. Approximate Atomic Surfaces from Linear Combinations of Pairwise Overlaps (LCPO). J. Comput. Chem. 1999, 20, 217–230. [Google Scholar] [CrossRef]
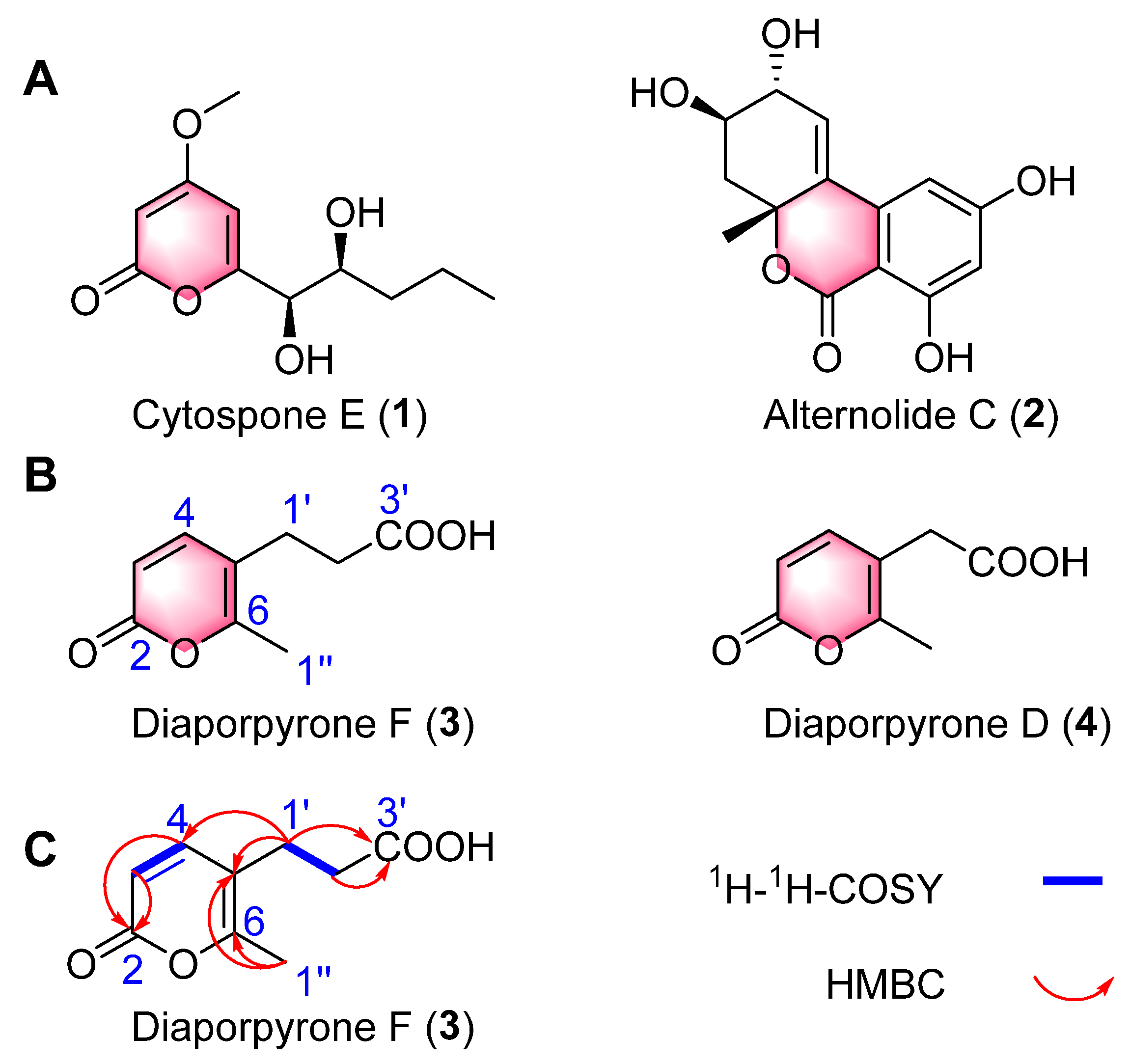
| Position | Diaporpyrone F (3) | |
|---|---|---|
| δC, Type | δH (J in Hz) | |
| 2 | 161.8, C | |
| 3 | 112.4, CH | 6.12 (d, J = 9.3 Hz) |
| 4 | 148.1, CH | 7.46 (d, J = 9.4 Hz) |
| 5 | 114.7, C | |
| 6 | 158.9, C | |
| 1′ | 24.7, CH2 | 2.49 (m) |
| 2′ | 34.8, CH2 | 2.33 (t, J = 7.4 Hz) |
| 3′ | 174.6, C | |
| 1″ | 17.1, CH3 | 2.20, s |
| α-Pyrone BGC | α-Pyrone BGC | α-Pyrone BGC | |||
|---|---|---|---|---|---|
| Diaporthe ampelina | ·· | Diaporthe citri | · | Diaporthe longicolla | ·· |
| Diaporthe amygdali | ·· | Diaporthe citriasiana | · | Diaporthe nobilis | ·· |
| Diaporthe aspalathi | ·· | Diaporthe citrichinensis | · | Diaporthe sp. DP-2020a | ·· |
| Diaporthe batatas | ·· | Diaporthe destruens | · | Diaporthe sp. HANT25 | ···· |
| Diaporthe capsici | ·· | Diaporthe eres | ·· | Diaporthe sp. NJD1 | ·· |
| Diaporthe caulivora | · | Diaporthe ilicicola | ·· | Diaporthe vexans | ·· |
| Activity | α-Glucosidase/Bacteria | Acarbose/Levofloxacin | 3 | 4 |
|---|---|---|---|---|
| % inhibition (800 μM) | α-glucosidase | 57.33 | - | 46.4 |
| antibacterial activities (μg/mL) | MRSA | 0.5 | >64 | >64 |
| M. smegmatis | 1 | >64 | >64 | |
| K. pneumoniae | 2 | >64 | >64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Ma, Q.; Wu, G.; Zhong, Y.; Feng, B.; Huang, P.; Li, A.; Tang, G.; Huang, X.; Pu, H. Bioactive α-Pyrone Analogs from the Endophytic Fungus Diaporthe sp. CB10100: α-Glucosidase Inhibitory Activity, Molecular Docking, and Molecular Dynamics Studies. Molecules 2024, 29, 1768. https://doi.org/10.3390/molecules29081768
Wang Z, Ma Q, Wu G, Zhong Y, Feng B, Huang P, Li A, Tang G, Huang X, Pu H. Bioactive α-Pyrone Analogs from the Endophytic Fungus Diaporthe sp. CB10100: α-Glucosidase Inhibitory Activity, Molecular Docking, and Molecular Dynamics Studies. Molecules. 2024; 29(8):1768. https://doi.org/10.3390/molecules29081768
Chicago/Turabian StyleWang, Zhong, Qingxian Ma, Guangling Wu, Yani Zhong, Bin Feng, Pingzhi Huang, Aijie Li, Genyun Tang, Xueshuang Huang, and Hong Pu. 2024. "Bioactive α-Pyrone Analogs from the Endophytic Fungus Diaporthe sp. CB10100: α-Glucosidase Inhibitory Activity, Molecular Docking, and Molecular Dynamics Studies" Molecules 29, no. 8: 1768. https://doi.org/10.3390/molecules29081768




