Aptamer Technologies in Neuroscience, Neuro-Diagnostics and Neuro-Medicine Development
Abstract
:1. Introduction
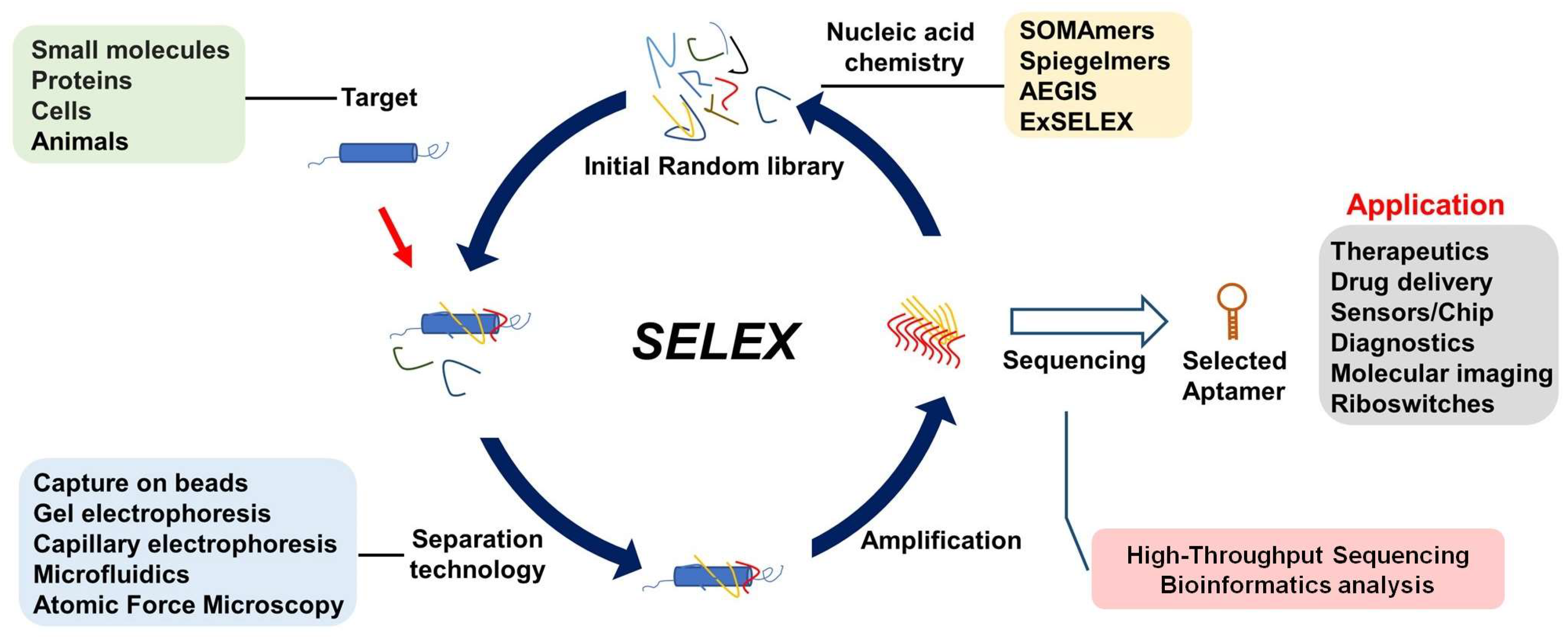
2. Discovery of Target-Specific Aptamers Using SELEX
- Pacification or synthesis of targets suitable for aptamer selection, or efficient identification of the specific target when a mixture system is used in selection, as in Cell-Selex.
- Highly efficient separation of high binding affinity and specific aptamer from library pool; the ultimate goal is single-round selection.
- Chemically modified nucleic acid to improve limited chemical diversity, relatively low binding affinity, and resistance to nuclease. Development of the corresponding polymerase or methods for the amplification of modified nucleic acid.
- High-throughput selection of many targets at one time and high-throughput sequencing for aptamer evolution.
- AI-driven machine learning for aptamer discovery to avoid tedious manual operation and lower cost.
- Discovery of useful aptamers with more function not just binding to target; functionally directed selection offers a promising avenue to develop functional aptamers.
3. Comparing the Advantages and Disadvantages of Aptamers, Antibodies, and Small Molecules as Neuro-Therapeutics
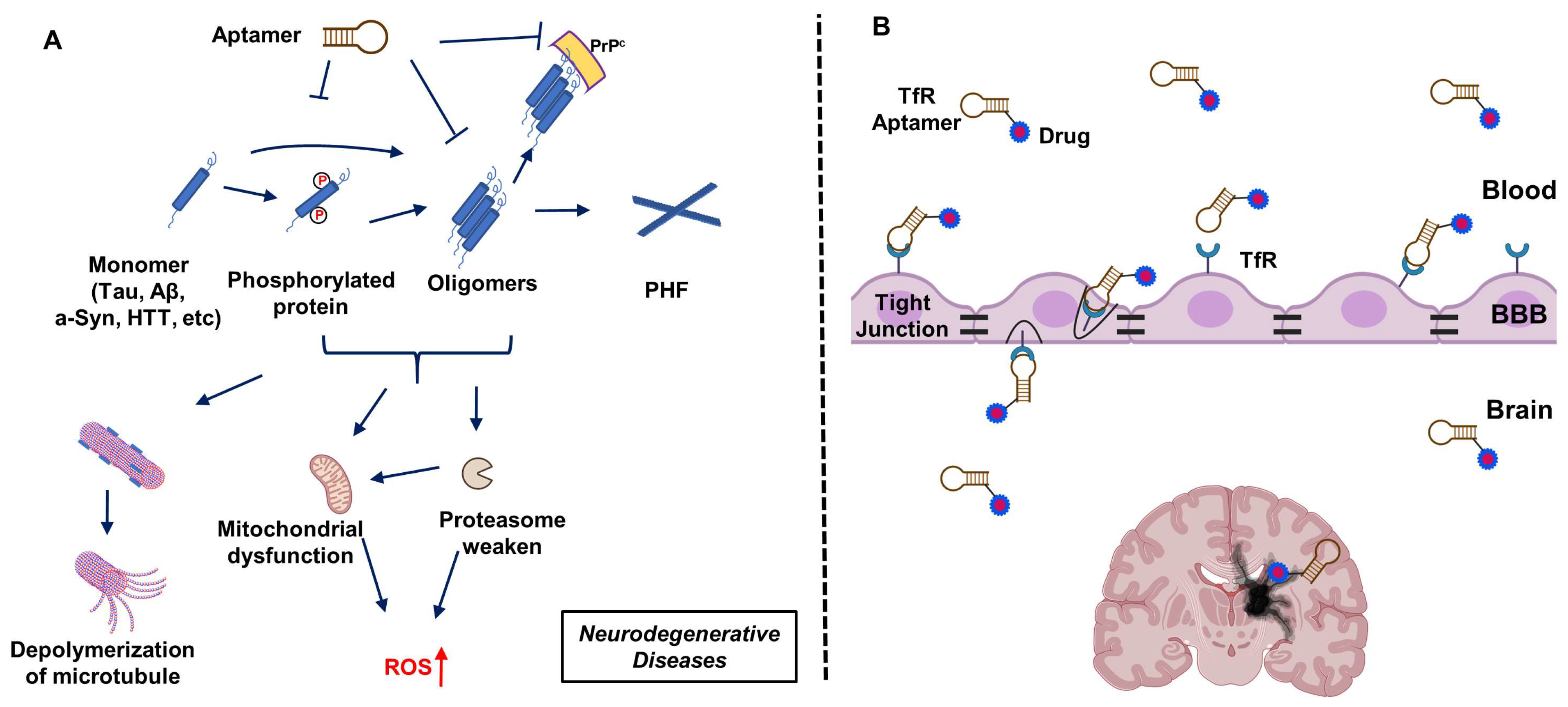
4. Aptamers Targeting Neuro-Medically Relevant Targets
5. In Vitro Neuro-Diagnostics Using Aptamers
6. In Vivo Imaging Using Neuro-Aptamers
7. Applying Experience from Non-CNS Therapeutic Aptamers towards Neuro-Aptamer Development
8. Aptamers Targeting Transferrin Receptor 1 to Facilitate Drug Transport across the BBB
9. Other Challenges in Aptamer-Based Therapeutic Development
- Nuclease sensitivity
- Renal excretion
- Binding affinity limited to nanomolar level
- Increase the chemical complexity of aptamers [5].
- Transfer across the BBB
- Multivalent aptamer hybrid with TfR aptamer [110] (see above).
- Therapeutic ability
- Functionally guided SELEX (our group’s unpublished work).
- Toxicity
- More sophisticated mechanistic research is needed [175].
10. Critical Limitations and Potential Solutions
10.1. Selection Process
10.2. Nuclease Resistance and Renal Filtration
10.3. Improvement in Binding Affinities
10.4. Binding Does Not Always Equate to Therapeutic Functions
11. Summary/Prospective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef]
- Gawande, B.N.; Rohloff, J.C.; Carter, J.D.; von Carlowitz, I.; Zhang, C.; Schneider, D.J.; Janjic, N. Selection of DNA aptamers with two modified bases. Proc. Natl. Acad. Sci. USA 2017, 114, 2898–2903. [Google Scholar] [CrossRef]
- Kimoto, M.; Yamashige, R.; Matsunaga, K.-i.; Yokoyama, S.; Hirao, I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013, 31, 453–457. [Google Scholar] [CrossRef]
- Sefah, K.; Yang, Z.; Bradley, K.M.; Hoshika, S.; Jiménez, E.; Zhang, L.; Zhu, G.; Shanker, S.; Yu, F.; Turek, D. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA 2014, 111, 1449–1454. [Google Scholar] [CrossRef]
- Chen, Z.; Lichtor, P.A.; Berliner, A.P.; Chen, J.C.; Liu, D.R. Evolution of sequence-defined highly functionalized nucleic acid polymers. Nat. Chem. 2018, 10, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Berezhnoy, A.; Stewart, C.A.; Mcnamara II, J.O.; Thiel, W.; Giangrande, P.; Trinchieri, G.; Gilboa, E. Isolation and optimization of murine IL-10 receptor blocking oligonucleotide aptamers using high-throughput sequencing. Mol. Ther. 2012, 20, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- Kacherovsky, N.; Cardle, I.I.; Cheng, E.L.; Yu, J.L.; Baldwin, M.L.; Salipante, S.J.; Jensen, M.C.; Pun, S.H. Traceless aptamer-mediated isolation of CD8+ T cells for chimeric antigen receptor T-cell therapy. Nat. Biomed. Eng. 2019, 3, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M.; Blind, M.; Mayer, G. Intramers as promising new tools in functional proteomics. Chem. Biol. 2001, 8, 931–939. [Google Scholar] [CrossRef]
- Kobeissy, F.H.; Guingab-Cagmat, J.D.; Zhang, Z.; Moghieb, A.; Glushakova, O.Y.; Mondello, S.; Boutté, A.M.; Anagli, J.; Rubenstein, R.; Bahmad, H. Neuroproteomics and systems biology approach to identify temporal biomarker changes post experimental traumatic brain injury in rats. Front. Neurol. 2016, 7, 198. [Google Scholar] [CrossRef]
- Jaber, Z.; Aouad, P.; Al Medawar, M.; Bahmad, H.; Abou-Abbass, H.; Ghandour, H.; Mondello, S.; Kobeissy, F. Role of systems biology in brain injury biomarker discovery: Neuroproteomics application. Methods Mol. Biol. 2016, 1462, 157–174. [Google Scholar]
- Razafsha, M.; Khaku, A.; Azari, H.; Alawieh, A.; Behforuzi, H.; Fadlallah, B.; Kobeissy, F.H.; Wang, K.K.; Gold, M.S. Biomarker identification in psychiatric disorders: From neuroscience to clinical practice. J. Psychiatr. Pract. 2015, 21, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Kobeissy, F.H.; Guingab-Cagmat, J.D.; Razafsha, M.; O’Steen, L.; Zhang, Z.; Hayes, R.L.; Chiu, W.-T.; Wang, K.K. Leveraging biomarker platforms and systems biology for rehabilomics and biologics effectiveness research. PMR 2011, 3, S139–S147. [Google Scholar] [CrossRef] [PubMed]
- Feigin, V.L.; Nichols, E.; Alam, T.; Bannick, M.S.; Beghi, E.; Blake, N.; Culpepper, W.J.; Dorsey, E.R.; Elbaz, A.; Ellenbogen, R.G. Global, regional, and national burden of neurological disorders, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 459–480. [Google Scholar] [CrossRef] [PubMed]
- Martier, R.; Konstantinova, P. Gene therapy for neurodegenerative diseases: Slowing down the ticking clock. Front. Neurosci. 2020, 14, 580179. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Preeti, K.; Fernandes, V.; Khatri, D.K.; Singh, S.B. Role of MicroRNAs, Aptamers in Neuroinflammation and Neurodegenerative Disorders. Cell. Mol. Neurobiol. 2022, 42, 2075–2095. [Google Scholar] [CrossRef]
- Hauser, S.L.; Cree, B.A. Treatment of multiple sclerosis: A review. Am. J. Med. 2020, 133, 1380–1390.e2. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Alzheimer disease and aducanumab: Adjusting our approach. Nat. Rev. Neurol. 2019, 15, 365–366. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, J.; Antonelli, A.C.; Afridi, A.; Vatsia, S.; Joshi, G.; Romanov, V.; Murray, I.V.; Khan, S.A. Protein misfolding and aggregation in neurodegenerative diseases: A review of pathogeneses, novel detection strategies, and potential therapeutics. Rev. Neurosci. 2019, 30, 339–358. [Google Scholar] [CrossRef]
- Zhou, G.; Wilson, G.; Hebbard, L.; Duan, W.; Liddle, C.; George, J.; Qiao, L. Aptamers: A promising chemical antibody for cancer therapy. Oncotarget 2016, 7, 13446. [Google Scholar] [CrossRef]
- Iwagawa, T.; Ohuchi, S.P.; Watanabe, S.; Nakamura, Y. Selection of RNA aptamers against mouse embryonic stem cells. Biochimie 2012, 94, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, A.; Takamura, Y.; Nishigaki, K.; Biyani, M. Competitive non-SELEX for the selective and rapid enrichment of DNA aptamers and its use in electrochemical aptasensor. Sci. Rep. 2019, 9, 6642. [Google Scholar] [CrossRef]
- Vorobyeva, M.A.; Davydova, A.S.; Vorobjev, P.E.; Pyshnyi, D.V.; Venyaminova, A.G. Key aspects of nucleic acid library design for in vitro selection. Int. J. Mol. Sci. 2018, 19, 470. [Google Scholar] [CrossRef]
- Randrianjatovo-Gbalou, I.; Rosario, S.; Sismeiro, O.; Varet, H.; Legendre, R.; Coppée, J.-Y.; Huteau, V.; Pochet, S.; Delarue, M. Enzymatic synthesis of random sequences of RNA and RNA analogues by DNA polymerase theta mutants for the generation of aptamer libraries. Nucleic Acids Res. 2018, 46, 6271–6284. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, A.; DeRosa, M.C. Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Lan, T.; Shi, H.; Lu, Y. Portable detection of melamine in milk using a personal glucose meter based on an in vitro selected structure-switching aptamer. Anal. Chem. 2015, 87, 7676–7682. [Google Scholar] [CrossRef]
- Bayat, P.; Nosrati, R.; Alibolandi, M.; Rafatpanah, H.; Abnous, K.; Khedri, M.; Ramezani, M. SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie 2018, 154, 132–155. [Google Scholar] [CrossRef]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’donoghue, M.B.; Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef]
- Krüger, A.; de Jesus Santos, A.P.; de Sá, V.; Ulrich, H.; Wrenger, C. Aptamer applications in emerging viral diseases. Pharmaceuticals 2021, 14, 622. [Google Scholar] [CrossRef]
- Cheng, C.; Chen, Y.H.; Lennox, K.A.; Behlke, M.A.; Davidson, B.L. In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol. Ther. Nucleic Acids 2013, 2, e67. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Bowser, M.T. Capillary Electrophoresis–SELEX Selection of Catalytic DNA Aptamers for a Small-Molecule Porphyrin Target. Anal. Chem. 2013, 85, 1525–1530. [Google Scholar] [CrossRef] [PubMed]
- Ngubane, N.A.; Gresh, L.; Pym, A.; Rubin, E.J.; Khati, M. Selection of RNA aptamers against the M. tuberculosis EsxG protein using surface plasmon resonance-based SELEX. Biochem. Biophys. Res. Commun. 2014, 449, 114–119. [Google Scholar] [CrossRef]
- Challa, S.; Tzipori, S.; Sheoran, A. Selective Evolution of Ligands by Exponential Enrichment to Identify RNA Aptamers against Shiga Toxins. J. Nucleic Acids 2014, 2014, 214929. [Google Scholar] [CrossRef]
- Tsai, R.Y.; Reed, R.R. Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol. Cell. Biol. 1998, 18, 6447–6456. [Google Scholar] [CrossRef]
- Jing, M.; Bowser, M.T. Isolation of DNA aptamers using micro free flow electrophoresis. Lab. A Chip 2011, 11, 3703–3709. [Google Scholar] [CrossRef]
- Takenaka, M.; Okumura, Y.; Amino, T.; Miyachi, Y.; Ogino, C.; Kondo, A. DNA-duplex linker for AFM-SELEX of DNA aptamer against human serum albumin. Bioorganic Med. Chem. Lett. 2017, 27, 954–957. [Google Scholar] [CrossRef]
- Miyachi, Y.; Shimizu, N.; Ogino, C.; Kondo, A. Selection of DNA aptamers using atomic force microscopy. Nucleic Acids Res. 2010, 38, e21. [Google Scholar] [CrossRef]
- Wang, J.; Gong, Q.; Maheshwari, N.; Eisenstein, M.; Arcila, M.L.; Kosik, K.S.; Soh, H.T. Particle display: A quantitative screening method for generating high-affinity aptamers. Angew. Chem. 2014, 126, 4896–4901. [Google Scholar] [CrossRef]
- Platt, M.; Rowe, W.; Wedge, D.C.; Kell, D.B.; Knowles, J.; Day, P.J. Aptamer evolution for array-based diagnostics. Anal. Biochem. 2009, 390, 203–205. [Google Scholar] [CrossRef] [PubMed]
- Paniel, N.; Istamboulié, G.; Triki, A.; Lozano, C.; Barthelmebs, L.; Noguer, T. Selection of DNA aptamers against penicillin G using Capture-SELEX for the development of an impedimetric sensor. Talanta 2017, 162, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.V.; Musheev, M.U.; Drabovich, A.P.; Jitkova, J.V.; Krylov, S.N. Non-SELEX: Selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat. Protoc. 2006, 1, 1359–1369. [Google Scholar] [CrossRef]
- Ouellet, E.; Foley, J.H.; Conway, E.M.; Haynes, C. Hi-Fi SELEX: A high-fidelity digital-PCR based therapeutic aptamer discovery platform. Biotechnol. Bioeng. 2015, 112, 1506–1522. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Qian, J.; Xiao, Y.; Viel, L.; Gerdon, A.E.; Lagally, E.T.; Atzberger, P.; Tarasow, T.M.; Heeger, A.J.; Soh, H.T. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. USA 2009, 106, 2989–2994. [Google Scholar] [CrossRef] [PubMed]
- Mercier, M.-C.; Dontenwill, M.; Choulier, L. Selection of nucleic acid aptamers targeting tumor cell-surface protein biomarkers. Cancers 2017, 9, 69. [Google Scholar] [CrossRef]
- Wang, B.; Pan, X.; Teng, I.-T.; Li, X.; Kobeissy, F.; Wu, Z.Y.; Zhu, J.; Cai, G.; Yan, H.; Yan, X.; et al. Functional Selection of Tau Oligomerization-Inhibiting Aptamers. Angew. Chem. 2024, e202402007. [Google Scholar]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjic, N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165): Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, T.; Romesberg, F.E. Evolved polymerases facilitate selection of fully 2′-OMe-modified aptamers. Chem. Sci. 2017, 8, 8179–8182. [Google Scholar] [CrossRef]
- Freund, N.; Taylor, A.I.; Arangundy-Franklin, S.; Subramanian, N.; Peak-Chew, S.-Y.; Whitaker, A.M.; Freudenthal, B.D.; Abramov, M.; Herdewijn, P.; Holliger, P. A two-residue nascent-strand steric gate controls synthesis of 2′-O-methyl-and 2′-O-(2-methoxyethyl)-RNA. Nat. Chem. 2022, 15, 1–10. [Google Scholar] [CrossRef]
- Yan, X.; Gao, X.; Zhang, Z. Isolation and characterization of 2′-amino-modified RNA aptamers for human TNFα. Genom. Proteom. Bioinform. 2004, 2, 32–42. [Google Scholar] [CrossRef]
- Rose, K.M.; Alves Ferreira-Bravo, I.; Li, M.; Craigie, R.; Ditzler, M.A.; Holliger, P.; DeStefano, J.J. Selection of 2′-deoxy-2′-fluoroarabino nucleic acid (FANA) aptamers that bind HIV-1 integrase with picomolar affinity. ACS Chem. Biol. 2019, 14, 2166–2175. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, S.; Chaput, J.C. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 2012, 4, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Wlotzka, B.; Leva, S.; Eschgfäller, B.; Burmeister, J.; Kleinjung, F.; Kaduk, C.; Muhn, P.; Hess-Stumpp, H.; Klussmann, S. In vivo properties of an anti-GnRH Spiegelmer: An example of an oligonucleotide-based therapeutic substance class. Proc. Natl. Acad. Sci. USA 2002, 99, 8898–8902. [Google Scholar] [CrossRef]
- Abeydeera, N.D.; Egli, M.; Cox, N.; Mercier, K.; Conde, J.N.; Pallan, P.S.; Mizurini, D.M.; Sierant, M.; Hibti, F.-E.; Hassell, T. Evoking picomolar binding in RNA by a single phosphorodithioate linkage. Nucleic Acids Res. 2016, 44, 8052–8064. [Google Scholar] [CrossRef]
- Schmidt, K.S.; Borkowski, S.; Kurreck, J.; Stephens, A.W.; Bald, R.; Hecht, M.; Friebe, M.; Dinkelborg, L.; Erdmann, V.A. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004, 32, 5757–5765. [Google Scholar] [CrossRef]
- Eremeeva, E.; Fikatas, A.; Margamuljana, L.; Abramov, M.; Schols, D.; Groaz, E.; Herdewijn, P. Highly stable hexitol based XNA aptamers targeting the vascular endothelial growth factor. Nucleic Acids Res. 2019, 47, 4927–4939. [Google Scholar] [CrossRef]
- Kimoto, M.; Matsunaga, K.-I.; Hirao, I. DNA aptamer generation by genetic alphabet expansion SELEX (ExSELEX) using an unnatural base pair system. Nucleic Acid Aptamers Sel. Charact. Appl. 2016, 1380, 47–60. [Google Scholar]
- Zhang, L.; Yang, Z.; Sefah, K.; Bradley, K.M.; Hoshika, S.; Kim, M.-J.; Kim, H.-J.; Zhu, G.; Jiménez, E.; Cansiz, S.; et al. Evolution of Functional Six-Nucleotide DNA. J. Am. Chem. Soc. 2015, 137, 6734–6737. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Tolle, F.; Rosenthal, M.; Brändle, G.M.; Ewers, J.; Mayer, G. Identification and characterization of nucleobase-modified aptamers by click-SELEX. Nat. Protoc. 2018, 13, 1153–1180. [Google Scholar] [CrossRef]
- Hoinka, J.; Berezhnoy, A.; Dao, P.; Sauna, Z.E.; Gilboa, E.; Przytycka, T.M. Large scale analysis of the mutational landscape in HT-SELEX improves aptamer discovery. Nucleic Acids Res. 2015, 43, 5699–5707. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, M.; Zhu, T.F. Directed evolution and selection of biostable l-DNA aptamers with a mirror-image DNA polymerase. Nat. Biotechnol. 2022, 40, 1601–1609. [Google Scholar] [CrossRef] [PubMed]
- Bashir, A.; Yang, Q.; Wang, J.; Hoyer, S.; Chou, W.; McLean, C.; Davis, G.; Gong, Q.; Armstrong, Z.; Jang, J. Machine learning guided aptamer refinement and discovery. Nat. Commun. 2021, 12, 2366. [Google Scholar] [CrossRef]
- Singh, S.; Tank, N.K.; Dwiwedi, P.; Charan, J.; Kaur, R.; Sidhu, P.; Chugh, V.K. Monoclonal antibodies: A review. Curr. Clin. Pharmacol. 2018, 13, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, B.; Pleitt, K.; Shave, E.; Baker, K.; Lua, L.H. Progression of continuous downstream processing of monoclonal antibodies: Current trends and challenges. Biotechnol. Bioeng. 2018, 115, 2893–2907. [Google Scholar] [CrossRef] [PubMed]
- Aljohani, M.M.; Cialla-May, D.; Popp, J.; Chinnappan, R.; Al-Kattan, K.; Zourob, M. Aptamers: Potential diagnostic and therapeutic agents for blood diseases. Molecules 2022, 27, 383. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Yu, S.; Zheng, Y.; Zheng, Y.; Yang, H.; Zhang, J. Aptamer and its applications in neurodegenerative diseases. Cell. Mol. Life Sci. 2017, 74, 683–695. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Agrawal, S.; Joshi, M.; Christoforidis, J.B. Vitreous inflammation associated with intravitreal anti-VEGF pharmacotherapy. Mediat. Inflamm. 2013, 2013, 943409. [Google Scholar] [CrossRef]
- Fu, Z.; Xiang, J. Aptamers, the nucleic acid antibodies, in cancer therapy. Int. J. Mol. Sci. 2020, 21, 2793. [Google Scholar] [CrossRef]
- Swayze, E.E.; Siwkowski, A.M.; Wancewicz, E.V.; Migawa, M.T.; Wyrzykiewicz, T.K.; Hung, G.; Monia, B.P.; Bennett, C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007, 35, 687–700. [Google Scholar] [CrossRef]
- Ganson, N.J.; Povsic, T.J.; Sullenger, B.A.; Alexander, J.H.; Zelenkofske, S.L.; Sailstad, J.M.; Rusconi, C.P.; Hershfield, M.S. Pre-existing anti-polyethylene glycol antibody linked to first-exposure allergic reactions to pegnivacogin, a PEGylated RNA aptamer. J. Allergy Clin. Immunol. 2016, 137, 1610–1613.e7. [Google Scholar] [CrossRef] [PubMed]
- Roesler, T.W.; Marvian, A.T.; Brendel, M.; Nykaenen, N.-P.; Hoellerhage, M.; Schwarz, S.C.; Hopfner, F.; Koeglsperger, T.; Respondek, G.; Schweyer, K. Four-repeat tauopathies. Prog. Neurobiol. 2019, 180, 101644. [Google Scholar] [CrossRef]
- Ma, J.; Gao, J.; Wang, J.; Xie, A. Prion-like mechanisms in Parkinson’s disease. Front. Neurosci. 2019, 13, 552. [Google Scholar] [CrossRef]
- Hernández-Jiménez, M.; Abad-Santos, F.; Cotgreave, I.; Gallego, J.; Jilma, B.; Flores, A.; Jovin, T.G.; Vivancos, J.; Molina, C.A.; Montaner, J. APRIL: A double-blind, placebo-controlled, randomized, Phase Ib/IIa clinical study of ApTOLL for the treatment of acute ischemic stroke. Front. Neurol. 2023, 14, 1127585. [Google Scholar] [CrossRef]
- Yun, Y.; Yang, X.; Tan, S.; Wang, P.; Ji, Y.; Sun, X. Targeting upregulated RNA binding protein RCAN1. 1: A promising strategy for neuroprotection in acute ischemic stroke. CNS Neurosci. Ther. 2022, 28, 1814–1828. [Google Scholar] [CrossRef] [PubMed]
- Zacco, E.; Kantelberg, O.; Milanetti, E.; Armaos, A.; Panei, F.P.; Gregory, J.; Jeacock, K.; Clarke, D.J.; Chandran, S.; Ruocco, G. Probing TDP-43 condensation using an in silico designed aptamer. Nat. Commun. 2022, 13, 3306. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, Q.-A.; Chen, Q.; Lee, T.H.; Huang, Y.; Wetsel, W.C.; Michelotti, G.A.; Sullenger, B.A.; Zhang, X. Targeting inhibition of GluR1 Ser845 phosphorylation with an RNA aptamer that blocks AMPA receptor trafficking. J. Neurochem. 2009, 108, 147–157. [Google Scholar] [CrossRef]
- Lennarz, S.; Alich, T.C.; Kelly, T.; Blind, M.; Beck, H.; Mayer, G. Selective Aptamer-Based Control of Intraneuronal Signaling. Angew. Chem. 2015, 127, 5459–5463. [Google Scholar] [CrossRef]
- Kahsai, A.W.; Wisler, J.W.; Lee, J.; Ahn, S.; Cahill III, T.J.; Dennison, S.M.; Staus, D.P.; Thomsen, A.R.; Anasti, K.M.; Pani, B. Conformationally selective RNA aptamers allosterically modulate the β2-adrenoceptor. Nat. Chem. Biol. 2016, 12, 709–716. [Google Scholar] [CrossRef]
- Teng, I.-T.; Li, X.; Yadikar, H.A.; Yang, Z.; Li, L.; Lyu, Y.; Pan, X.; Wang, K.K.; Tan, W. Identification and characterization of DNA aptamers specific for phosphorylation epitopes of tau protein. J. Am. Chem. Soc. 2018, 140, 14314–14323. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, E.; Choi, W.H.; Lee, J.; Lee, J.H.; Lee, H.; Kim, D.-E.; Suh, Y.H.; Lee, M.J. Inhibitory RNA aptamers of tau oligomerization and their neuroprotective roles against proteotoxic stress. Mol. Pharm. 2016, 13, 2039–2048. [Google Scholar] [CrossRef]
- Lisi, S.; Fiore, E.; Scarano, S.; Pascale, E.; Boehman, Y.; Ducongé, F.; Chierici, S.; Minunni, M.; Peyrin, E.; Ravelet, C. Non-SELEX isolation of DNA aptamers for the homogeneous-phase fluorescence anisotropy sensing of tau Proteins. Anal. Chim. Acta 2018, 1038, 173–181. [Google Scholar] [CrossRef]
- Ylera, F.; Lurz, R.; Erdmann, V.A.; Fürste, J.P. Selection of RNA aptamers to the Alzheimer’s disease amyloid peptide. Biochem. Biophys. Res. Commun. 2002, 290, 1583–1588. [Google Scholar] [CrossRef]
- Takahashi, T.; Tada, K.; Mihara, H. RNA aptamers selected against amyloid β-peptide (Aβ) inhibit the aggregation of Aβ. Mol. Biosyst. 2009, 5, 986–991. [Google Scholar] [CrossRef]
- Song, G.; Shui, R.; Wang, D.; Fang, R.; Yuan, T.; Li, L.; Feng, J.; Gao, F.; Shen, Q.; Gong, J. Aptamer-conjugated graphene oxide-based surface assisted laser desorption ionization mass spectrometry for selective extraction and detection of Aβ1–42 in an Alzheimer’s disease SH-SY5 cell model. Front. Aging Neurosci. 2022, 14, 993281. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, P.; Li, S.; Geng, X.; Zou, L.; Jin, M.; Zou, Q.; Wang, Q.; Yang, X.; Wang, K. Development of DNA aptamer as a β-amyloid aggregation inhibitor. ACS Appl. Bio Mater. 2020, 3, 8611–8618. [Google Scholar] [CrossRef]
- Murakami, K.; Obata, Y.; Sekikawa, A.; Ueda, H.; Izuo, N.; Awano, T.; Takabe, K.; Shimizu, T.; Irie, K. An RNA aptamer with potent affinity for a toxic dimer of amyloid β42 has potential utility for histochemical studies of Alzheimer’s disease. J. Biol. Chem. 2020, 295, 4870–4880. [Google Scholar] [CrossRef]
- Rahimi, F.; Murakami, K.; Summers, J.L.; Chen, C.-H.B.; Bitan, G. RNA aptamers generated against oligomeric Aβ40 recognize common amyloid aptatopes with low specificity but high sensitivity. PLoS ONE 2009, 4, e7694. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Qu, J.; Xue, F.; Zheng, Y.; Yang, B.; Chang, Y.; Yang, H.; Zhang, J. Novel DNA aptamers for Parkinson’s disease treatment inhibit α-synuclein aggregation and facilitate its degradation. Mol. Ther. Nucleic Acids 2018, 11, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Tsukakoshi, K.; Abe, K.; Sode, K.; Ikebukuro, K. Selection of DNA aptamers that recognize α-synuclein oligomers using a competitive screening method. Anal. Chem. 2012, 84, 5542–5547. [Google Scholar] [CrossRef] [PubMed]
- Hmila, I.; Sudhakaran, I.P.; Ghanem, S.S.; Vaikath, N.N.; Poggiolini, I.; Abdesselem, H.; El-Agnaf, O.M. Inhibition of α-Synuclein Seeding-Dependent Aggregation by ssDNA Aptamers Specific to C-Terminally Truncated α-Synuclein Fibrils. ACS Chem. Neurosci. 2022, 13, 3330–3341. [Google Scholar] [CrossRef]
- Rock, M.; Zouganelis, G.D.; de Andrade, A.F.B.; Drake, S.J.; Alexiou, A.; Albrakati, A.; Batiha, G.E.-S.; Illingworth, T.A. Development and validation of anti-human Alpha synuclein DNA aptamer using computer modelling techniques—An in silico study. J. Integr. Neurosci. 2022, 21, 5. [Google Scholar] [CrossRef]
- Tsukakoshi, K.; Harada, R.; Sode, K.; Ikebukuro, K. Screening of DNA aptamer which binds to α-synuclein. Biotechnol. Lett. 2010, 32, 643–648. [Google Scholar] [CrossRef]
- Lobanova, E.; Whiten, D.; Ruggeri, F.S.; Taylor, C.G.; Kouli, A.; Xia, Z.; Emin, D.; Zhang, Y.P.; Lam, J.Y.; Williams-Gray, C.H. Imaging protein aggregates in the serum and cerebrospinal fluid in Parkinson’s disease. Brain 2022, 145, 632–643. [Google Scholar] [CrossRef]
- Mannironi, C.; Di Nardo, A.; Fruscoloni, P.; Tocchini-Valentini, G. In vitro selection of dopamine RNA ligands. Biochemistry 1997, 36, 9726–9734. [Google Scholar] [CrossRef]
- Fernández, G.; Moraga, A.; Cuartero, M.I.; García-Culebras, A.; Peña-Martínez, C.; Pradillo, J.M.; Hernández-Jiménez, M.; Sacristán, S.; Ayuso, M.I.; Gonzalo-Gobernado, R. TLR4-binding DNA aptamers show a protective effect against acute stroke in animal models. Mol. Ther. 2018, 26, 2047–2059. [Google Scholar] [CrossRef] [PubMed]
- Camorani, S.; Esposito, C.L.; Rienzo, A.; Catuogno, S.; Iaboni, M.; Condorelli, G.; De Franciscis, V.; Cerchia, L. Inhibition of receptor signaling and of glioblastoma-derived tumor growth by a novel PDGFRβ aptamer. Mol. Ther. 2014, 22, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Monaco, I.; Camorani, S.; Colecchia, D.; Locatelli, E.; Calandro, P.; Oudin, A.; Niclou, S.; Arra, C.; Chiariello, M.; Cerchia, L. Aptamer functionalization of nanosystems for glioblastoma targeting through the blood–brain barrier. J. Med. Chem. 2017, 60, 4510–4516. [Google Scholar] [CrossRef] [PubMed]
- Rozenblum, G.T.; Kaufman, T.; Vitullo, A.D. Myelin basic protein and a multiple sclerosis-related MBP-peptide bind to oligonucleotides. Mol. Ther. Nucleic Acids 2014, 3, e192. [Google Scholar] [CrossRef] [PubMed]
- Vorobjeva, M.A.; Krasitskaya, V.V.; Fokina, A.A.; Timoshenko, V.V.; Nevinsky, G.A.; Venyaminova, A.G.; Frank, L.A. RNA aptamer against autoantibodies associated with multiple sclerosis and bioluminescent detection probe on its basis. Anal. Chem. 2014, 86, 2590–2594. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Bing, T.; Liu, Y.; Zhang, N.; Shen, L.; Liu, X.; Wang, J.; Shangguan, D. Imaging of neurite network with an anti-L1CAM aptamer generated by neurite-SELEX. J. Am. Chem. Soc. 2018, 140, 18066–18073. [Google Scholar] [CrossRef]
- Iida, M.; Mashima, T.; Yamaoki, Y.; So, M.; Nagata, T.; Katahira, M. The anti-prion RNA aptamer R12 disrupts the Alzheimer’s disease-related complex between prion and amyloid β. FEBS J. 2019, 286, 2355–2365. [Google Scholar] [CrossRef]
- Ogasawara, D.; Hasegawa, H.; Kaneko, K.; Sode, K.; Ikebukuro, K. Screening of DNA aptamer against mouse prion protein by competitive selection. Prion 2007, 1, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Proske, D.; Gilch, S.; Wopfner, F.; Schätzl, H.M.; Winnacker, E.L.; Famulok, M. Prion-protein-specific aptamer reduces PrPSc formation. Chembiochem 2002, 3, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Matos, C.O.; Passos, Y.M.; do Amaral, M.J.; Macedo, B.; Tempone, M.H.; Bezerra, O.C.; Moraes, M.O.; Almeida, M.S.; Weber, G.; Missailidis, S. Liquid-liquid phase separation and fibrillation of the prion protein modulated by a high-affinity DNA aptamer. FASEB J. 2020, 34, 365–385. [Google Scholar] [CrossRef]
- Mashima, T.; Lee, J.-H.; Kamatari, Y.O.; Hayashi, T.; Nagata, T.; Nishikawa, F.; Nishikawa, S.; Kinoshita, M.; Kuwata, K.; Katahira, M. Development and structural determination of an anti-PrPC aptamer that blocks pathological conformational conversion of prion protein. Sci. Rep. 2020, 10, 4934. [Google Scholar] [CrossRef] [PubMed]
- Georges, J.; Qi, X.; Liu, X.; Zhou, Y.; Woolf, E.C.; Valeri, A.; Al-Atrache, Z.; Belykh, E.; Feuerstein, B.G.; Preul, M. Provision of rapid and specific ex vivo diagnosis of central nervous system lymphoma from rodent xenograft biopsies by a fluorescent aptamer. J. Neurosurg. 2020, 134, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Huang, N.; Zhang, X.; Zhou, T.; Tan, Y.; Pi, J.; Pi, L.; Cheng, S.; Zheng, H.; Cheng, Y. Aptamer-conjugated PEGylated quantum dots targeting epidermal growth factor receptor variant III for fluorescence imaging of glioma. Int. J. Nanomed. 2017, 12, 3899. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.; Zhang, Y.; Zhang, C.; Huang, L.; Tan, S.; Wang, P.; Vilariño-Gúell, C.; Song, W.; Sun, X. Regulator of calcineurin 1 is a novel RNA-binding protein to regulate neuronal apoptosis. Mol. Psychiatry 2021, 26, 1361–1375. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Zhao, H.; Zhu, T.; Yang, Z.; Xu, H.; Fu, Y.; Lin, F.; Pan, X.; Li, L. Enhanced in vivo blood–brain barrier penetration by circular Tau–transferrin receptor bifunctional aptamer for tauopathy therapy. J. Am. Chem. Soc. 2020, 142, 3862–3872. [Google Scholar] [CrossRef]
- McCann, H.; Stevens, C.H.; Cartwright, H.; Halliday, G.M. α-Synucleinopathy phenotypes. Park. Relat. Disord. 2014, 20, S62–S67. [Google Scholar] [CrossRef]
- Ren, X.; Zhao, Y.; Xue, F.; Zheng, Y.; Huang, H.; Wang, W.; Chang, Y.; Yang, H.; Zhang, J. Exosomal DNA aptamer targeting α-synuclein aggregates reduced neuropathological deficits in a mouse Parkinson’s disease model. Mol. Ther. Nucleic Acids 2019, 17, 726–740. [Google Scholar] [CrossRef] [PubMed]
- Shamili, F.H.; Alibolandi, M.; Rafatpanah, H.; Abnous, K.; Mahmoudi, M.; Kalantari, M.; Taghdisi, S.M.; Ramezani, M. Immunomodulatory properties of MSC-derived exosomes armed with high affinity aptamer toward mylein as a platform for reducing multiple sclerosis clinical score. J. Control Release 2019, 299, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, M.M. Multiple sclerosis review. Pharm. Ther. 2012, 37, 175. [Google Scholar]
- Qu, W.; Yuan, B.; Liu, J.; Liu, Q.; Zhang, X.; Cui, R.; Yang, W.; Li, B. Emerging role of AMPA receptor subunit GluA1 in synaptic plasticity: Implications for Alzheimer’s disease. Cell Prolif. 2021, 54, e12959. [Google Scholar] [CrossRef] [PubMed]
- Kiefel, H.; Bondong, S.; Hazin, J.; Ridinger, J.; Schirmer, U.; Riedle, S.; Altevogt, P. L1CAM: A major driver for tumor cell invasion and motility. Cell Adh Migr. 2012, 6, 374–384. [Google Scholar] [CrossRef]
- Shi, S.; Fu, W.; Lin, S.; Tian, T.; Li, S.; Shao, X.; Zhang, Y.; Zhang, T.; Tang, Z.; Zhou, Y. Targeted and effective glioblastoma therapy via aptamer-modified tetrahedral framework nucleic acid-paclitaxel nanoconjugates that can pass the blood brain barrier. Nanomed. Nanotechnol. Biol. Med. 2019, 21, 102061. [Google Scholar] [CrossRef]
- Wang, F.; Zhou, Y.; Cheng, S.; Lou, J.; Zhang, X.; He, Q.; Huang, N.; Cheng, Y. Gint4.T-modified DNA tetrahedrons loaded with doxorubicin inhibits glioma cell proliferation by targeting PDGFRβ. Nanoscale Res. Lett. 2020, 15, 150. [Google Scholar] [CrossRef] [PubMed]
- Hong, P.; Li, W.; Li, J. Applications of aptasensors in clinical diagnostics. Sensors 2012, 12, 1181–1193. [Google Scholar] [CrossRef]
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.; Tang, T.-H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015, 64, 392–403. [Google Scholar] [CrossRef]
- Wu, Z.-S.; Guo, M.-M.; Zhang, S.-B.; Chen, C.-R.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Reusable electrochemical sensing platform for highly sensitive detection of small molecules based on structure-switching signaling aptamers. Anal. Chem. 2007, 79, 2933–2939. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.Y.; Plaxco, K.W.; Heeger, A.J. Aptamer-based electrochemical detection of picomolar platelet-derived growth factor directly in blood serum. Anal. Chem. 2007, 79, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Liss, M.; Petersen, B.; Wolf, H.; Prohaska, E. An aptamer-based quartz crystal protein biosensor. Anal. Chem. 2002, 74, 4488–4495. [Google Scholar] [CrossRef] [PubMed]
- Barbour, C.; Kosa, P.; Komori, M.; Tanigawa, M.; Masvekar, R.; Wu, T.; Johnson, K.; Douvaras, P.; Fossati, V.; Herbst, R. Molecular-based diagnosis of multiple sclerosis and its progressive stage. Ann. Neurol. 2017, 82, 795–812. [Google Scholar] [CrossRef] [PubMed]
- Farrar, C.T.; William, C.M.; Hudry, E.; Hashimoto, T.; Hyman, B.T. RNA aptamer probes as optical imaging agents for the detection of amyloid plaques. PLoS ONE 2014, 9, e89901. [Google Scholar] [CrossRef] [PubMed]
- Di Cera, E. Thrombin. Mol. Asp. Med. 2008, 29, 203–254. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, L.; Zhang, F.; Zhang, Z.; Tang, J.; Xie, J. High-sensitive determination of human α-thrombin by its 29-mer aptamer in affinity probe capillary electrophoresis. Electrophoresis 2008, 29, 2570–2577. [Google Scholar] [CrossRef]
- Padmanabhan, K.; Padmanabhan, K.; Ferrara, J.; Sadler, J.E.; Tulinsky, A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993, 268, 17651–17654. [Google Scholar] [CrossRef]
- Gambhir, S.S. Molecular imaging of cancer with positron emission tomography. Nat. Rev. Cancer 2002, 2, 683–693. [Google Scholar] [CrossRef]
- Charlton, J.; Sennello, J.; Smith, D. In vivo imaging of inflammation using an aptamer inhibitor of human neutrophil elastase. Chem. Biol. 1997, 4, 809–816. [Google Scholar] [CrossRef]
- Grogna, M.; Cloots, R.; Luxen, A.; Jérôme, C.; Desreux, J.-F.; Detrembleur, C. Design and synthesis of novel DOTA (Gd 3+)–polymer conjugates as potential MRI contrast agents. J. Mater. Chem. 2011, 21, 12917–12926. [Google Scholar] [CrossRef]
- Tan, M.; Wu, X.; Jeong, E.-K.; Chen, Q.; Lu, Z.-R. Peptide-targeted nanoglobular Gd-DOTA monoamide conjugates for magnetic resonance cancer molecular imaging. Biomacromolecules 2010, 11, 754–761. [Google Scholar] [CrossRef]
- Sullenger, B.A.; Gallardo, H.F.; Ungers, G.E.; Gilboa, E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell 1990, 63, 601–608. [Google Scholar] [CrossRef]
- Ng, E.W.M.; Shima, D.T.; Calias, P.; Cunningham, E.T.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Hughes, Q.W.; Le, B.T.; Gilmore, G.; Baker, R.I.; Veedu, R.N. Construction of a Bivalent Thrombin Binding Aptamer and Its Antidote with Improved Properties. Molecules 2017, 22, 1770. [Google Scholar] [CrossRef]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 2020, 155, 1420–1431. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, S.-i.; Taguchi, K.; Fukami, K. DNA-aptamers raised against AGEs as a blocker of various aging-related disorders. Glycoconj. J. 2016, 33, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Helmling, S.; Maasch, C.; Eulberg, D.; Buchner, K.; Schröder, W.; Lange, C.; Vonhoff, S.; Wlotzka, B.; Tschöp, M.H.; Rosewicz, S. Inhibition of ghrelin action in vitro and in vivo by an RNA-Spiegelmer. Proc. Natl. Acad. Sci. USA 2004, 101, 13174–13179. [Google Scholar] [CrossRef] [PubMed]
- Vater, A.; Sell, S.; Kaczmarek, P.; Maasch, C.; Buchner, K.; Pruszynska-Oszmalek, E.; Kolodziejski, P.; Purschke, W.G.; Nowak, K.W.; Strowski, M.Z. A mixed mirror-image DNA/RNA aptamer inhibits glucagon and acutely improves glucose tolerance in models of type 1 and type 2 diabetes. J. Biol. Chem. 2013, 288, 21136–21147. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yang, Y.; Hong, H.; Zhang, Y.; Cai, W.; Fang, D. Aptamers as therapeutics in cardiovascular diseases. Curr. Med. Chem. 2011, 18, 4169–4174. [Google Scholar] [CrossRef] [PubMed]
- Maier, K.E.; Jangra, R.K.; Shieh, K.R.; Cureton, D.K.; Xiao, H.; Snapp, E.L.; Whelan, S.P.; Chandran, K.; Levy, M. A new transferrin receptor aptamer inhibits new world hemorrhagic fever mammarenavirus entry. Mol. Ther. Nucleic Acids 2016, 5, e321. [Google Scholar] [CrossRef]
- Li, W.; Feng, X.; Yan, X.; Liu, K.; Deng, L. A DNA Aptamer Against Influenza A Virus: An Effective Inhibitor to the Hemagglutinin–Glycan Interactions. Nucleic Acid Ther. 2016, 26, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Soule, E.E.; Bompiani, K.M.; Woodruff, R.S.; Sullenger, B.A. Targeting two coagulation cascade proteases with a bivalent aptamer yields a potent and antidote-controllable anticoagulant. Nucleic Acid Ther. 2016, 26, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Nonaka, Y.; Miyakawa, S.; Fujiwara, M.; Nakamura, Y. Dual therapeutic action of a neutralizing anti-FGF2 aptamer in bone disease and bone cancer pain. Mol. Ther. 2016, 24, 1974–1986. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lan, X. Aptamer oligonucleotides: Novel potential therapeutic agents in autoimmune disease. Nucleic Acid Ther. 2015, 25, 173–179. [Google Scholar] [CrossRef]
- Zhu, G.; Ye, M.; Donovan, M.J.; Song, E.; Zhao, Z.; Tan, W. Nucleic acid aptamers: An emerging frontier in cancer therapy. Chem. Commun. 2012, 48, 10472–10480. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Hwang, K.; Li, J.; Torabi, S.-F.; Lu, Y. DNA aptamer technology for personalized medicine. Curr. Opin. Chem. Eng. 2014, 4, 79–87. [Google Scholar] [CrossRef]
- Ballabh, P.; Braun, A.; Nedergaard, M. The blood–brain barrier: An overview: Structure, regulation, and clinical implications. Neurobiol. Dis. 2004, 16, 1–13. [Google Scholar] [CrossRef]
- Abbott, N.J.; Patabendige, A.A.; Dolman, D.E.; Yusof, S.R.; Begley, D.J. Structure and function of the blood–brain barrier. Neurobiol. Dis. 2010, 37, 13–25. [Google Scholar] [CrossRef]
- Gabathuler, R. Approaches to transport therapeutic drugs across the blood–brain barrier to treat brain diseases. Neurobiol. Dis. 2010, 37, 48–57. [Google Scholar] [CrossRef]
- Wang, R.E.; Wu, H.; Niu, Y.; Cai, J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011, 18, 4126–4138. [Google Scholar] [CrossRef] [PubMed]
- Kariolis, M.S.; Wells, R.C.; Getz, J.A.; Kwan, W.; Mahon, C.S.; Tong, R.; Kim, D.J.; Srivastava, A.; Bedard, C.; Henne, K.R. Brain delivery of therapeutic proteins using an Fc fragment blood-brain barrier transport vehicle in mice and monkeys. Sci. Transl. Med. 2020, 12, eaay1359. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.J.; Zhang, Y.; Kenrick, M.; Hoyte, K.; Luk, W.; Lu, Y.; Atwal, J.; Elliott, J.M.; Prabhu, S.; Watts, R.J. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci. Transl. Med. 2011, 3, ra44–ra84. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Burkhart, A.; Thomsen, L.B.; Andresen, T.L.; Moos, T. Targeting the transferrin receptor for brain drug delivery. Prog. Neurobiol. 2019, 181, 101665. [Google Scholar] [CrossRef]
- Chen, C.-h.B.; Dellamaggiore, K.R.; Ouellette, C.P.; Sedano, C.D.; Lizadjohry, M.; Chernis, G.A.; Gonzales, M.; Baltasar, F.E.; Fan, A.L.; Myerowitz, R. Aptamer-based endocytosis of a lysosomal enzyme. Proc. Natl. Acad. Sci. USA 2008, 105, 15908–15913. [Google Scholar] [CrossRef]
- Macdonald, J.; Houghton, P.; Xiang, D.; Duan, W.; Shigdar, S. Truncation and mutation of a transferrin receptor aptamer enhances binding affinity. Nucleic Acid. Ther. 2016, 26, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, J.; Denoyer, D.; Henri, J.; Jamieson, A.; Burvenich, I.J.; Pouliot, N.; Shigdar, S. Bifunctional aptamer–doxorubicin conjugate crosses the blood–brain barrier and selectively delivers its payload to EpCAM-positive tumor cells. Nucleic Acid. Ther. 2020, 30, 117–128. [Google Scholar] [CrossRef]
- Wilner, S.E.; Wengerter, B.; Maier, K.; Magalhães, M.d.L.B.; Del Amo, D.S.; Pai, S.; Opazo, F.; Rizzoli, S.O.; Yan, A.; Levy, M. An RNA alternative to human transferrin: A new tool for targeting human cells. Mol. Ther. Nucleic Acids 2012, 1, e21. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, Z.; Bai, H.; Fu, T.; Yang, C.; Hu, X.; Liu, Q.; Champanhac, C.; Teng, I.-T.; Ye, M. DNA aptamer selected against pancreatic ductal adenocarcinoma for in vivo imaging and clinical tissue recognition. Theranostics 2015, 5, 985. [Google Scholar] [CrossRef]
- Wu, X.; Liu, H.; Han, D.; Peng, B.; Zhang, H.; Zhang, L.; Li, J.; Liu, J.; Cui, C.; Fang, S. Elucidation and structural modeling of CD71 as a molecular target for cell-specific aptamer binding. J. Am. Chem. Soc. 2019, 141, 10760–10769. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, J.; Bing, T.; Liu, X.; Shangguan, D. Transferrin receptor-mediated internalization and intracellular fate of conjugates of a DNA aptamer. Mol. Ther. Nucleic Acids 2022, 27, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, M.; Nilsen-Hamilton, M.; Ilgu, M. Aptamer applications in neuroscience. Pharmaceuticals 2021, 14, 1260. [Google Scholar] [CrossRef] [PubMed]
- Söylemez, F.; Türkseven, Ç.H. Aptamers and possible effects on neurodegeneration. In Neuroprotection-New Approaches and Prospects; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar]
- Burmeister, P.E.; Wang, C.; Killough, J.R.; Lewis, S.D.; Horwitz, L.R.; Ferguson, A.; Thompson, K.M.; Pendergrast, P.S.; McCauley, T.G.; Kurz, M. 2-Deoxy purine, 2-O-methyl pyrimidine (dRmY) aptamers as candidate therapeutics. Oligonucleotides 2006, 16, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef] [PubMed]
- Healy, J.M.; Lewis, S.D.; Kurz, M.; Boomer, R.M.; Thompson, K.M.; Wilson, C.; McCauley, T.G. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res. 2004, 21, 2234–2246. [Google Scholar] [CrossRef]
- Burmeister, P.E.; Lewis, S.D.; Silva, R.F.; Preiss, J.R.; Horwitz, L.R.; Pendergrast, P.S.; McCauley, T.G.; Kurz, J.C.; Epstein, D.M.; Wilson, C. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 2005, 12, 25–33. [Google Scholar] [CrossRef]
- Lee, C.H.; Lee, S.-H.; Kim, J.H.; Noh, Y.-H.; Noh, G.-J.; Lee, S.-W. Pharmacokinetics of a cholesterol-conjugated aptamer against the hepatitis C virus (HCV) NS5B protein. Mol. Ther. Nucleic Acids 2015, 4, e254. [Google Scholar] [CrossRef]
- Heo, K.; Min, S.-W.; Sung, H.J.; Kim, H.G.; Kim, H.J.; Kim, Y.H.; Choi, B.K.; Han, S.; Chung, S.; Lee, E.S. An aptamer-antibody complex (oligobody) as a novel delivery platform for targeted cancer therapies. J. Control Release 2016, 229, 1–9. [Google Scholar] [CrossRef]
- Willis, M.C.; Collins, B.; Zhang, T.; Green, L.S.; Sebesta, D.P.; Bell, C.; Kellogg, E.; Gill, S.C.; Magallanez, A.; Knauer, S. Liposome-anchored vascular endothelial growth factor aptamers. Bioconj. Chem. 1998, 9, 573–582. [Google Scholar] [CrossRef]
- Zhou, J.; Soontornworajit, B.; Martin, J.; Sullenger, B.A.; Gilboa, E.; Wang, Y. A hybrid DNA aptamer–dendrimer nanomaterial for targeted cell labeling. Macromol. Biosci. 2009, 9, 831–835. [Google Scholar] [CrossRef]
- Zheng, D.; Seferos, D.S.; Giljohann, D.A.; Patel, P.C.; Mirkin, C.A. Aptamer Nano-flares for Molecular Detection in Living Cells. Nano Lett. 2009, 9, 3258–3261. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Feng, K.; Li, L.; Yang, L.; Pan, X.; Yazd, H.S.; Cui, C.; Li, J.; Moroz, L.; Sun, Y. Lipid–oligonucleotide conjugates for bioapplications. Natl. Sci. Rev. 2020, 7, 1933–1953. [Google Scholar] [CrossRef] [PubMed]
- Borbas, K.E.; Ferreira, C.S.; Perkins, A.; Bruce, J.I.; Missailidis, S. Design and synthesis of mono-and multimeric targeted radiopharmaceuticals based on novel cyclen ligands coupled to anti-MUC1 aptamers for the diagnostic imaging and targeted radiotherapy of cancer. Bioconj. Chem. 2007, 18, 1205–1212. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Kumar Kulabhusan, P.; Hussain, B.; Yüce, M. Current perspectives on aptamers as diagnostic tools and therapeutic agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef]
- Ni, S.; Zhuo, Z.; Pan, Y.; Yu, Y.; Li, F.; Liu, J.; Wang, L.; Wu, X.; Li, D.; Wan, Y. Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl. Mater. Interfaces 2020, 13, 9500–9519. [Google Scholar] [CrossRef]
- Eilers, A.; Witt, S.; Walter, J. Aptamer-modified nanoparticles in medical applications. Aptamers Biotechnol. 2020, 174, 161–193. [Google Scholar]
- Bohrmann, L.; Burghardt, T.; Haynes, C.; Saatchi, K.; Häfeli, U.O. Aptamers used for molecular imaging and theranostics-recent developments. Theranostics 2022, 12, 4010. [Google Scholar] [CrossRef]
- Kohlberger, M.; Gadermaier, G. SELEX: Critical factors and optimization strategies for successful aptamer selection. Biotechnol. Appl. Biochem. 2022, 69, 1771–1792. [Google Scholar] [CrossRef]


| Consideration | Small Molecule | Aptamer | Antibody |
|---|---|---|---|
| Structure |  |  | 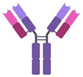 |
| Size | <1 kDa | 10~25 kDa | ~150 kDa |
| Stability | Stable at R.T. | Stable at R.T. | Sensitive to temperature and pH changes |
| Cost | Low | Low | High |
| Immunogenicity | Low | Low | High |
| Ligand specificity | Medium (Kd nM to uM) | High (Kd pM to nM) | High (Kd pM to nM) |
| Toxicity | Mid-high toxicity | Not observed | Immune reaction |
| Administration | Oral, i.v. | i.v., s.c. | i.v., s.c. |
| Tissue penetration | Variable | Slow | Slow |
| Target | Aptamers (Name) | Dissociation Constant | Disease | Utility, Key Results | Ref. |
|---|---|---|---|---|---|
| GluR1 Ser845 | A1, A2, A3 (RNA) | 28–57 nM | Protein phosphorylation-related diseases | A2 binds GluR1 that inhibits GluR1/GluR1 containing AMPA receptor trafficking to the cell surface and abrogates forskolin-stimulated phosphorylation at GluR1 Ser845 | [78] |
| (MAPK) Erk1/2 | C5 (RNA) | 10 nM | CNS disorders, Alzheimer’s disease, Stroke, Epilepsy | C5 selective to inhibit the mitogen-activated kinase pathway in neurons | [79] |
| β2-adrenoceptor (β2-AR) (GPCR) | A1, A2, A13, and A16 (RNA) | 30.4–258.5 nM | - | RNA aptamers as allosteric GPCR modulators | [80] |
| Tau protein | IT(1–6), IT2a (DNA) | 5.5–68 nM | Traumatic brain injury, Alzheimer’s disease | Aptamers inhibit tau phosphorylation and oligomerization | [81] |
| Tau-1 (RNA) | ~200 nM | [82] | |||
| Aptamer 314 (DNA) | 13 ± 3 nM | Aptamer binds Tau protein | [83] | ||
| β-Amyloid protein | β aptamers, e.g., β55 (RNA) | 29–48 nM | Alzheimer’s disease | β55 aptamer binds amyloid plaques in AD brain tissue | [84] |
| E1, E2, N1, N2, G2, etc. (RNA) | 10.9–21.6 μM | N2 aptamer is used to build a luminescent aptamer-ruthenium complex system for the detection of Aβ | [85] | ||
| Apt-GO (RNA) | 0.1–10 μM | Apt-GO selectively detects Aβ1–42 in an AD SH-SY5 cell model | [86] | ||
| Aβ42 | Aβ7-92-1H1 | 63.4 nM | Inhibits Aβ42 aggregation | [87] | |
| Aβ42 dimer | E22P–AbD43 (RNA) | 20 ± 6.0 nM | Aptamer inhibits the nucleation phase of the dimer and its associated neurotoxicity in SH-SY5Y human neuroblastoma cells. | [88] | |
| Aβ40 oligomers | KM (RNA) | - | Aptamers bind with Aβ40 fibrils that may serve as amyloid recognition tools | [89] | |
| α-synuclein protein | F5R1 (DNA) | 2.40 nM | Parkinson’s disease | Blocks the aberrant cellular effects of the overexpressed α-synuclein in cells | [90] |
| T-SO508 (DNA) | 68 nM | T-SO508 can bind to soluble α-synuclein oligomers | [91] | ||
| Apt11(DNA) | - | Parkinson’s disease and dementia with Lewy bodies | Apt11 aptamer binds to α-syn fibrils and inhibits α-syn aggregation in the in vitro model of PD and DLB. | [92] | |
| TMG-79 (DNA) | - | TMG-79 aptamer detects α-syn in Lewy body and PD-associated dementia. | [93] | ||
| M5-15 (DNA) | 14.3 nM | M5-15 aptamer detects α-syn in Lewy body and PD-associated dementia. | [94] | ||
| α-synuclein & amyloid-β (Aβ) | AD-PAINT (DNA) | 500 nM–2 μM | Parkinson’s disease | AD-PAINT aptamer binds to fibrillar aggregates of α-syn and Aβ aggregates detected in both serum and CSF in PD | [95] |
| Dopamine | dopa2 (129 nucleotides); dopa2/c.1 | 2.8 µM 1.6 µM | Parkinson’s disease | Dopa2 and dopa2/c.1 are characterized to bind a dopamine affinity column; the dopamine binding site is obtained by secondary selection | [96] |
| Toll-like receptor 4 (TLR4) | ApTLR#1R, ApTLR#4F (DNA) | - | Stroke disease | Aptamers have a TLR4 antagonistic effect | [74] |
| ApTOLL (DNA) | 20 nM | Acute ischemic stroke | ApTOLL aptamer binds and antagonizes TLR4 and improves functional outcomes in AIS patients | [75] | |
| Platelet-derived growth factor receptor β (PDGFRβ) | Gint4.T aptamer (RNA) | 9.6 nM | Glioma | Aptamer binds to human DGFRβ ectodomain, causing a strong inhibition of ligand-dependent receptor activation | [97,98] |
| Myelin basic protein | MBPcl3 MBPcl9 (DNA) | - | Multiple Sclerosis | MBPc13 detects myelin-rich regions in paraffin-embedded mouse brain tissue; aptamer was found more sensitive than a commercial antibody. MBPcl3 blocks the binding of the antibody to MBP | [99] |
| Myelin basic protein (MBP) autoantibody | Apt2-9c (RNA) | 1.2 ± 0.1 nM | Multiple Sclerosis | Apt2-9c provides proof-of-principle for the detection of MS-specific autoantibodies | [100] |
| L1-CAM (Neurites) | yly12 (DNA) | 3.51 nM | Neurite-surface targeting and inhibitory effect on neurite outgrowth between cells | [101] | |
| Prion protein (PrP) | R12 (RNA) | ~10 nM | Creutzfeldt-Jakob disease; prion diseases | R12 binding to PrP results in the dissociation of PrP with Aβ. | [102] |
| clone 4–9 (DNA) | 113 nM | binds to PrP | [103] | ||
| DP7 (RNA) | 0.1–1.7 µM | Prion-protein-specific aptamer reduces PrPSc formation | [104] | ||
| A1 (DNA) | 232 nM | Aptamers modulate phase separation and promote PrP fibrillation | [105] | ||
| R24 (RNA) | 18 nM | R24 exhibited the lowest recorded IC50 and the highest anti-prion activity | [106] | ||
| Crossing BBB (target unknown) | A15 (RNA) | - | Neurological disorders or diseases. | In vivo SELEX (brain-penetrating aptamers) | [31] |
| CD20+B cells | TD05 (DNA) | 256 nM | Glioma | TD05-488 can diagnose CNS lymphoma within 11 min of biopsy from xenograft brain tumor models | [107] |
| U87 glioma cell line/EGFRvIII | QD-A32 (DNA) | - | Glioma | QD-apt can penetrate the BBB and then selectively accumulate in the tumors through binding to EGFRvIII | [108] |
| The Regulator of calcineurin 1 (RCAN1) | R1SR13 (RNA) | 0.3 µM | Down syndrome and Alzheimer’s disease | Inhibits the regulatory function of RCAN1 in NFAT and NF-kB signaling pathways | [109] |
| 0.23–30 nM | Acute ischemic stroke | R1SR13 aptamer alleviates the RCAN1.1 L-induced neuronal apoptosis in the human SHSY-5Y neuroblastoma cells and in the mouse model of AIS | [76] | ||
| TAR-DNA-Binding Protein 43 (TDP-43) | Apt-1 (DNA) | 1 μM | Amyotrophic Lateral Sclerosis | Apt-1 aptamer binds to TDP-43 in the ALS model. | [77] |
| Transferrin Aptamer Name, Nucleotide Sequence | 2-D Structure | Ref. |
|---|---|---|
| Mouse transferrin receptor-specific GS24, reduced to 50 nucleotides. Sequence (5′-3′): GCGTGTGCACACGGTCACAGTTAGTATCGCTACGTTCTTTGGTAGTCCGTTCGG |  | [155] |
| Target Mouse TfR Aptamer name: TfRA1 Truncated GS24; 14 nucleotides Sequence (5′-3′): GCGTGTGCACACGC | 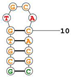 | [156] |
| Human transferrin receptor specific C2. targets the apical domain of the human transferrin receptor (hTfR) Sequence (5′-3′): CAUCUCACAGAUCAAUCCAAGGCACCUCGUUAAAGGACGACUCCCUUACAUGCGAGAUG | 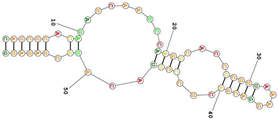 | [158] |
| Aptamer name: Min2/Waz (RNA) non-competitive for Transferrin. Targets the apical domain of the human transferrin receptor (hTfR) Sequence (5′-3′): GGGUUCUACGAUAAACGGUUAAUGACCAGCUUAUGGCUGGCAGUUCCC | 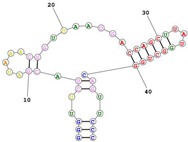 | [141] |
| Human TfR (cell-SELEX) (XQ-2d Shares a Similar Binding Site on CD71 with Transferrin) Aptamer name: XQ-2d (DNA) Sequence (5′-3′): ACTCATAGGGTTAGGGGCTGCTGGCCAGATACTCAGATGGTAGGGTTACTATGAGC |  | [159,160] |
| Human TfR (cell-SELEX) Aptamer name: HG1-9 (DNA) HG1-9 aptamer binds human TfR with affinity (Kd < 20 nM) and completes a same bind site of human TfR with transferrin. Sequence (5′-3′): GGATAGGGATTCTGTTGGTCGGCTGGTTGGTATCC | 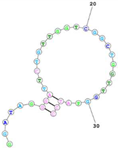 | [161] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Kobeissy, F.; Golpich, M.; Cai, G.; Li, X.; Abedi, R.; Haskins, W.; Tan, W.; Benner, S.A.; Wang, K.K.W. Aptamer Technologies in Neuroscience, Neuro-Diagnostics and Neuro-Medicine Development. Molecules 2024, 29, 1124. https://doi.org/10.3390/molecules29051124
Wang B, Kobeissy F, Golpich M, Cai G, Li X, Abedi R, Haskins W, Tan W, Benner SA, Wang KKW. Aptamer Technologies in Neuroscience, Neuro-Diagnostics and Neuro-Medicine Development. Molecules. 2024; 29(5):1124. https://doi.org/10.3390/molecules29051124
Chicago/Turabian StyleWang, Bang, Firas Kobeissy, Mojtaba Golpich, Guangzheng Cai, Xiaowei Li, Reem Abedi, William Haskins, Weihong Tan, Steven A. Benner, and Kevin K. W. Wang. 2024. "Aptamer Technologies in Neuroscience, Neuro-Diagnostics and Neuro-Medicine Development" Molecules 29, no. 5: 1124. https://doi.org/10.3390/molecules29051124
APA StyleWang, B., Kobeissy, F., Golpich, M., Cai, G., Li, X., Abedi, R., Haskins, W., Tan, W., Benner, S. A., & Wang, K. K. W. (2024). Aptamer Technologies in Neuroscience, Neuro-Diagnostics and Neuro-Medicine Development. Molecules, 29(5), 1124. https://doi.org/10.3390/molecules29051124









