Camel-Derived Nanobodies as Potent Inhibitors of New Delhi Metallo-β-Lactamase-1 Enzyme
Abstract
:1. Introduction
2. Results and Discussion
2.1. Library Construction
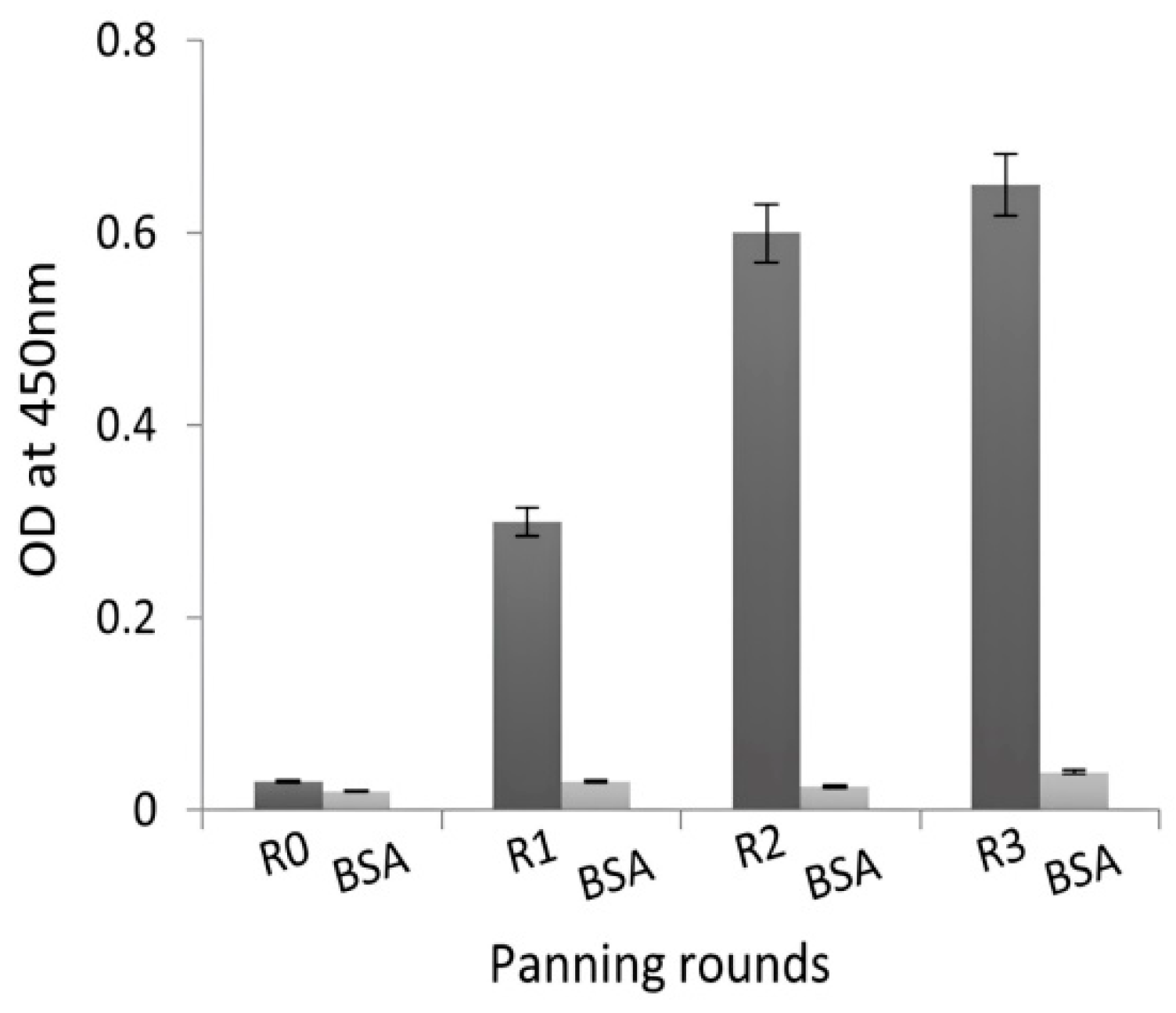
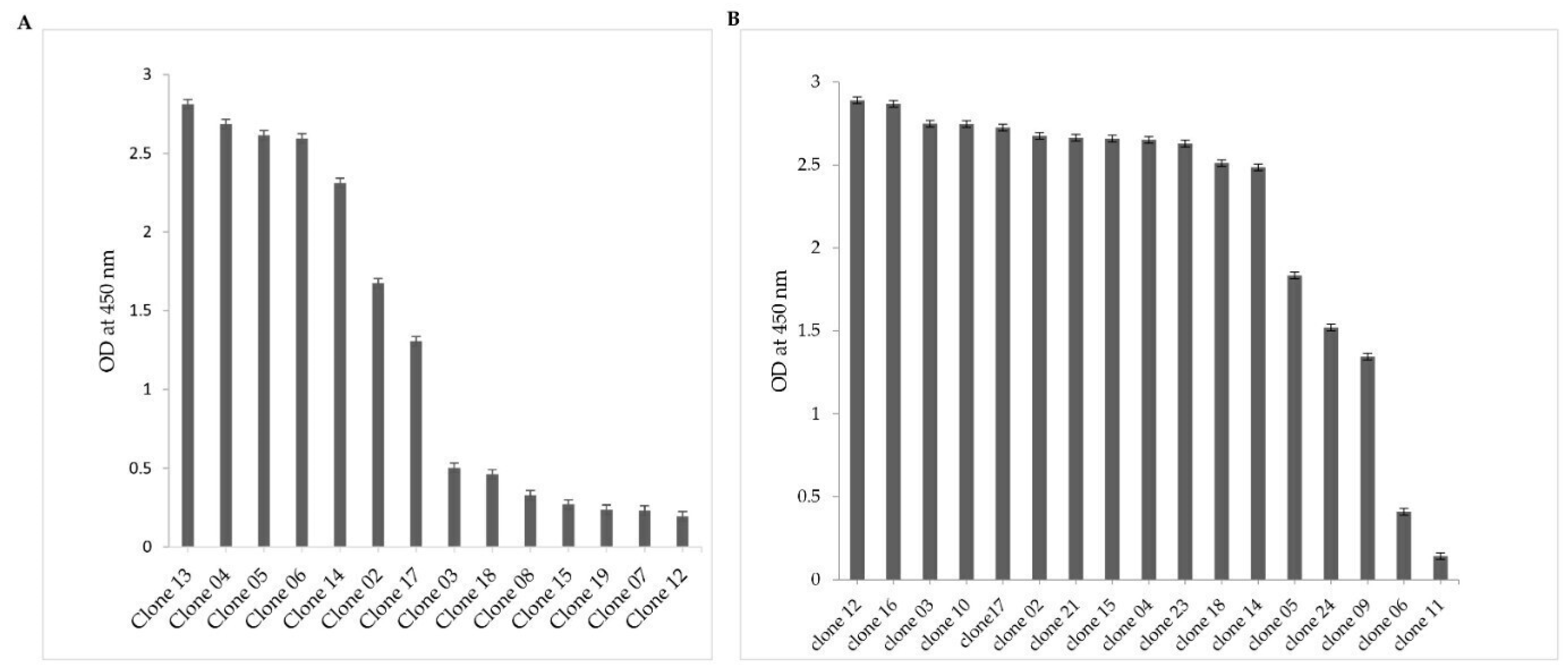
2.2. Selection of NDM-1-Specific VHH Fragments after Phage Display and Biopanning
2.3. VHH Sequence Analysis
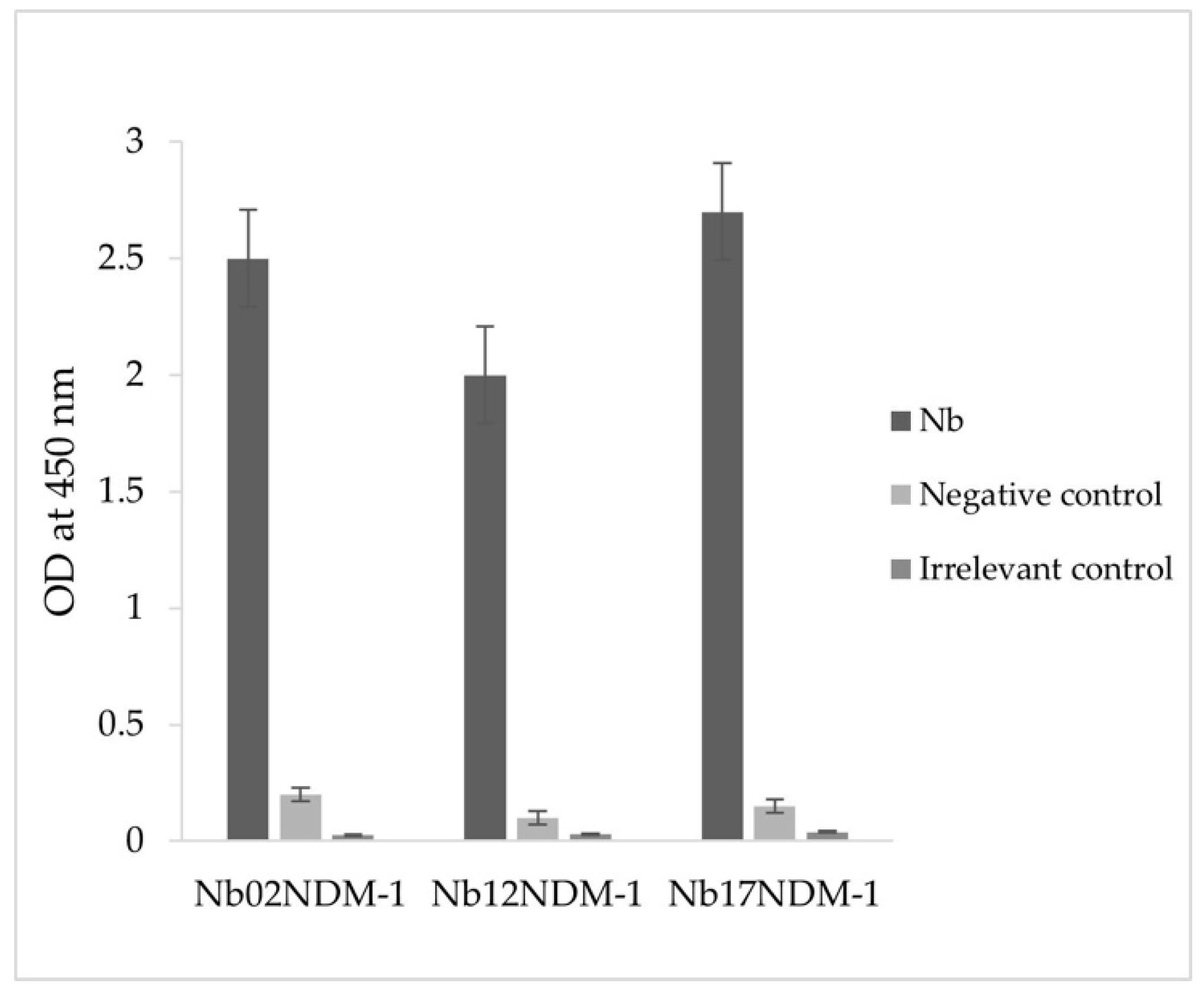
2.4. Production and Purification of Nbs
2.5. Inhibition Kinetic Assays
3. Materials and Methods
3.1. Enzyme, Vector, and Strains
3.2. Library Construction
3.3. Selection of NDM-1-Specific VHH Fragments Using Biopanning Phage Display
3.4. Screening for NDM-1-Specific Nbs
3.5. VHH Sequence Analysis
3.6. Nb Production and Purification
3.7. Inhibition Kinetic Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef]
- Solomon, S.L.; Oliver, K.B. Antibiotic resistance threats in the United States: Stepping back from the brink. Am. Fam. Physician 2014, 89, 938–941. [Google Scholar] [PubMed]
- Lupo, A.; Coyne, S.; Berendonk, T.U. Origin and evolution of antibiotic resistance: The common mechanisms of emergence and spread in water bodies. Front. Microbiol. 2012, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Jernigan, J.A.; Hatfield, K.M.; Wolford, H.; Nelson, R.E.; Olubajo, B.; Reddy, S.C.; McCarthy, N.; Paul, P.; McDonald, L.C.; Kallen, A.; et al. Multidrug-resistant bacterial infections in US hospitalized patients, 2012–2017. N. Eng. J. Med. 2020, 382, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-Lactamase DataBase (BLDB)—Structure and Function. Enzyme Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar]
- Bahr, G.; González, L.J.; Vila, A.J. Metallo-β-lactamases in the Age of Multidrug Resistance: From Structure and Mechanism to Evolution, Dissemination, and Inhibitor Design. Chem. Rev. 2021, 121, 7957–8094. [Google Scholar] [CrossRef]
- Garau, G.; García-Sáez, I.; Bebrone, C.; Anne, C.; Mercuri, P.; Galleni, M.; Frère, J.M.; Dideberg, O. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 2004, 48, 2347–2349. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; MaNally, A.; Zong, Z. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef] [PubMed]
- Krahn, T.; Wibberg, D.; Maus, I.; Winkler, A.; Bontron, S.; Sczyrba, A.; Nordmann, P.; Pulher, A.; Poirel, L.; Schluter, A. Intraspecies transfer of the chromosomal Acinetobacter baumannii blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 2016, 60, 3032–3040. [Google Scholar] [CrossRef]
- Tanner, W.D.; Atkinson, R.M.; Goel, R.K.; Toleman, M.A.; Benson, L.S.; Porucznik, C.A.; Van Derslice, J.A. Horizontal transfer of the blaNDM-1 gene to Pseudomonas aeruginosa and Acinetobacter baumannii in biofilms. FEMS Microbiol. Lett. 2017, 1, 364. [Google Scholar] [CrossRef]
- Politi, L.; Gartzonika, K.; Spanakis, N.; Zarkotou, O.; Poulou, A.; Skoura, L.; Vrioni, G.; Tsakris, A. Emergence of NDM-1-producing Klebsiella pneumoniae in Greece: Evidence of a widespread clonal outbreak. J. Antimicrob. Chemother. 2019, 74, 2197–2202. [Google Scholar] [CrossRef]
- Wailan, A.M.; Paterson, D.L. The spread and acquisition of NDM-1: A multifactorial problem. Expert Rev. Anti. Infect. Ther. 2014, 12, 91–115. [Google Scholar] [CrossRef]
- Mojica, M.F.; Rossi, M.A.; Vila, A.J.; Bonomo, R.A. The urgent need for metallo-β-lactamase inhibitors: An unattended global threat. Lancet Infect. Dis. 2022, 22, e28–e34. [Google Scholar] [CrossRef]
- Wang, T.; Xu, K.; Zhao, L.; Tong, R.; Xiong, L.; Shi, J. Recent research and development of NDM-1 inhibitors. Eur. J. Med. Chem. 2021, 223, 113667. [Google Scholar] [CrossRef]
- Liu, B.; Trout, R.E.L.; Chu, G.H.; McGarry, D.; Jackson, R.W.; Hamrick, J.C.; Daigle, D.M.; Cusick, S.M.; Pozzi, C.; De Luca, F.; et al. Discovery of Taniborbactam (VNRX-5133): A Broad-Spectrum Serine- and Metallo-β-lactamase Inhibitor for Carbapenem-Resistant Bacterial Infections. Med. Chem. 2020, 63, 2789–2801. [Google Scholar] [CrossRef]
- Piccirilli, A.; Segatore, B.; Brisdelli, F.; Amicosante, G.; Perilli, M. Potent inhibitory activity of taniborbactam towards NDM-1 and NDM-1Q119X mutants, and in vitro activity of cefepime/taniborbactam against MBLs producing Enterobacterales. Int. J. Antimicrob. Agents. 2021, 57, 106228. [Google Scholar] [CrossRef] [PubMed]
- Tsivkovski, R.; Totrov, M.; Lomovskaya, O. Biochemical characterization of QPX7728, a new ultrabroad-spectrum beta-lactamase inhibitor of serine and metallo-β-lactamases. Antimicrob. Agents Chemother. 2020, 64, e00130-20. [Google Scholar] [CrossRef] [PubMed]
- Kaplon, H.; Chenoweth, A.; Crescioli, S.; Reichert, J.M. Antibodies to watch in 2022. MAbs 2022, 14, 2014296. [Google Scholar] [CrossRef] [PubMed]
- Transue, T.R.; De Genst, E.; Ghahroudi, M.A.; Wyns, L.; Muyldermans, S. Camel single-domain antibody inhibits enzyme by mimicking carbohydrate substrate. Proteins Struct. Funct. Bioinform. 1998, 32, 515–522. [Google Scholar] [CrossRef]
- Sohier, J.S.; Laurent, C.; Chevigné, A.; Pardon, E.; Srinivasan, V.; Wernery, U.; Lassaux, P.; Steyaert, J.; Galleni, M. Allosteric inhibition of VIM metallo-β-lactamases by a camelid nanobody. Biochem. J. 2013, 450, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Ben Abderrazek, R.; Chammam, S.; Ksouri, A.; Perilli, M.; Dhaouadi, S.; Mdini, I.; Benlasfar, Z.; Amicosante, G.; Bouhaouala-Zahar, B.; Piccirilli, A. Inhibitory Potential of Polyclonal Camel Antibodies against New Delhi Metallo-β-lactamase-1 (NDM-1). Molecules 2020, 25, 4453. [Google Scholar] [CrossRef] [PubMed]
- Dhaouadi, S.; Ben Abderrazek, R.; Loustau, T.; Abou-Faycal, C.; Ksouri, A.; Erne, W.; Murdamoothoo, D.; Mörgelin, M.; Kungl, A.; Jung, A.; et al. Novel human tenascin-C function-blocking camel single domain nanobodies. Front. Immunol. 2021, 12, 635166. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Pillay, T.S.; Muyldermans, S. Application of Single-Domain Antibodies (“Nanobodies”) to Laboratory Diagnosis. Ann. Lab. Med. 2021, 41, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Anand, T.; Virmani, N.; Bera, B.C.; Vaid, R.K.; Vashisth, M.; Bardajatya, P.; Kumar, A.; Tripathi, B.N. Phage display technique as a tool for diagnosis and antibody selection for coronaviruses. Curr. Microbiol. 2021, 78, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, M.P.; Pommié, C.; Ruiz, M.; Giudicelli, V.; Foulquier, E.; Truong, L.; Thouvenin-Contet, V.; Lefranc, G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003, 27, 55–77. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, L.S.; Colwell, L.J. Comparative analysis of nanobody sequence and structure data. Proteins 2018, 86, 697–706. [Google Scholar] [CrossRef]
- Zavrtanik, U.; Lukan, J.; Loris, R.; Lah, J.; Hadži, S. Structural Basis of Epitope Recognition by Heavy-Chain Camelid Antibodies. Mol. Biol. 2018, 430, 4369–4386. [Google Scholar] [CrossRef]
- Cortez-Retamozo, V.; Backmann, N.; Senter, P.D.; Wernery, U.; De Baetselier, P.; Muyldermans, S.; Revets, H. Efficient cancer therapy with a nanobody-based conjugate. Cancer Res. 2004, 64, 2853–2857. [Google Scholar] [CrossRef]
- De Genst, E.; Silence, K.; Ghahroudi, M.A.; Decanniere, K.; Loris, R.; Kinne, J.; Wyns, L.; Muyldermans, S. Strong in vivo maturation compensates for structurally restricted H3 loops in antibody repertoires. Biol. Chem. 2005, 280, 14114–14121. [Google Scholar] [CrossRef]
- Truong, T.T.T.; Huynh, V.Q.; Vo, N.T.; Nguyen, H.D. Studying the characteristics of nanobody CDR regions based on sequence analysis in combination with 3D structures. J. Genet. Eng. Biotechnol. 2022, 20, 157. [Google Scholar] [CrossRef]
- Vincke, C.; Muyldermans, S. Introduction to heavy chain antibodies and derived Nanobodies. In Single Domain Antibodies: Methods Protocols; Springer: Berlin/Heidelberg, Germany, 2012; pp. 15–26. [Google Scholar]
- Yang, M.; Zhu, G.; Korza, G.; Sun, X.; Setlow, P.; Li, J. Engineering Bacillus subtilis as a versatile and stable platform for production of nanobodies. Appl. Environ. Microbiol. 2020, 86, e02938-19. [Google Scholar] [CrossRef]
- Noël, F.; Malpertuy, A.; de Brevern, A.G. Global analysis of VHHs framework regions with a structural alphabet. Biochimie 2016, 131, 11–19. [Google Scholar] [CrossRef]
- De Genst, E.; Silence, K.; Decanniere, K.; Conrath, K.; Loris, R.; Kinne, J.; Muyldermans, S.; Wyns, L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. USA 2006, 103, 4586–4591. [Google Scholar] [CrossRef]
- Marquardt, A.; Muyldermans, S.; Przybylski, M. A synthetic camel anti-lysozyme peptide antibody (peptibody) with flexible loop structure identified by high-resolution affinity mass spectrometry. Chem. Eur. J. 2006, 12, 1915–1923. [Google Scholar] [CrossRef]
- Piccirilli, A.; Criscuolo, E.; Brisdelli, F.; Mercuri, P.S.; Cherubini, S.; De Sciscio, M.L.; Maccarrone, M.; Galleni, M.; Amicosante, G.; Perilli, M. Exploring the role of L10 loop in New Delhi metallo-β-lactamase (NDM-1): Kinetic and dynamic studies. Molecules 2021, 26, 5489. [Google Scholar] [CrossRef]
- Vincke, C.; Gutiérrez, C.; Wernery, U.; Devoogdt, N.; Hassanzadeh-Ghassabeh, G.; Muyldermans, S. Generation of single domain antibody fragments derived from camelids and generation of manifold constructs. Methods Mol. Biol. 2012, 907, 145–176. [Google Scholar] [PubMed]
- Yan, J.; Wang, P.; Zhu, M.; Li, G.; Romão, E.; Xiong, S.; Wan, Y. Characterization and applications of Nanobodies against human procalcitonin selected from a novel naive Nanobody phage display library. Nanobiotechnology 2015, 13, 33. [Google Scholar] [CrossRef] [PubMed]
- Hmila, I.; Saerens, D.; Benlasfar, Z.; Conrath, K.; El Ayeb, M.; Muyldermans, S.; Bouhaouala-Zahar, B. VHH, bivalent domains and chimeric heavy chain-only antibodies with high neutralizing efficacy for scorpion toxin AahI’. Mol. Immunol. 2008, 45, 3847–3856. [Google Scholar] [CrossRef] [PubMed]
- Ben Abderrazek, R.; Ksouri, A.; Idoudi, F.; Dhaouadi, S.; Hamdi, E.; Vincke, C.; Farah, A.; Benlasfar, Z.; Majdoub, H.; El Ayeb, M.; et al. Neutralizing dromedary-derived nanobodies against BotI-like toxin from the most hazardous scorpion venom in the Middle East and North Africa Region. Front. Immunol. 2022, 13, 863012. [Google Scholar] [CrossRef] [PubMed]
- De Meester, F.; Joris, B.; Reckinger, G. Automated analysis of enzyme inactivation phenomena: Application to β-lactamases and DD-peptidases. Biochem. Pharmacol. 1987, 36, 2393–2403. [Google Scholar] [CrossRef] [PubMed]
- Bottoni, C.; Perilli, M.; Marcoccia, F.; Piccirilli, A.; Pellegrini, C.; Colapietro, M.; Sabatini, A.; Celenza, G.; Kerff, F.; Amicosante, G.; et al. Kinetic studies on CphA Mutants Reveal the role of the P158-P172 Loop in activity versus carbapenems. Antimicrob. Agents Chemother. 2016, 60, 3123–3126. [Google Scholar] [CrossRef] [PubMed]

| MBLs + Nbs | Km (μM) | kcat (s−1) |
|---|---|---|
| NDM-1 | 80 ± 2 | 75 ± 1 |
| NDM-1 + Nb02 (9 nM) | 71 ± 3 | 65 ± 2 |
| NDM-1 + Nb02 (30 nM) | 40 ± 1 | 37 ± 0.5 |
| NDM-1 + Nb12 (400 nM) | 65 ± 4 | 58 ± 2 |
| NDM-1 + Nb12 (700 nM) | 34 ± 2 | 31 ± 1 |
| NDM-1 + Nb17 (5 nM) | 53 ± 3 | 36 ± 0.5 |
| NDM-1 + Nb17 (10 nM) | 35 ± 1 | 21 ± 0.8 |
| NDM-1 | Ki (nM) | IC50 (nM) |
|---|---|---|
| Nb02NDM-1 | 4.7 ± 0.3 | 26.0 ± 1 |
| Nb12NDM-1 | 117 ± 8 | 485 ± 15 |
| Nb17NDM-1 | 3.7 ± 0.5 | 7.6 ± 1.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Abderrazek, R.; Hamdi, E.; Piccirilli, A.; Dhaouadi, S.; Muyldermans, S.; Perilli, M.; Bouhaouala-Zahar, B. Camel-Derived Nanobodies as Potent Inhibitors of New Delhi Metallo-β-Lactamase-1 Enzyme. Molecules 2024, 29, 1431. https://doi.org/10.3390/molecules29071431
Ben Abderrazek R, Hamdi E, Piccirilli A, Dhaouadi S, Muyldermans S, Perilli M, Bouhaouala-Zahar B. Camel-Derived Nanobodies as Potent Inhibitors of New Delhi Metallo-β-Lactamase-1 Enzyme. Molecules. 2024; 29(7):1431. https://doi.org/10.3390/molecules29071431
Chicago/Turabian StyleBen Abderrazek, Rahma, Emna Hamdi, Alessandra Piccirilli, Sayda Dhaouadi, Serge Muyldermans, Mariagrazia Perilli, and Balkiss Bouhaouala-Zahar. 2024. "Camel-Derived Nanobodies as Potent Inhibitors of New Delhi Metallo-β-Lactamase-1 Enzyme" Molecules 29, no. 7: 1431. https://doi.org/10.3390/molecules29071431
APA StyleBen Abderrazek, R., Hamdi, E., Piccirilli, A., Dhaouadi, S., Muyldermans, S., Perilli, M., & Bouhaouala-Zahar, B. (2024). Camel-Derived Nanobodies as Potent Inhibitors of New Delhi Metallo-β-Lactamase-1 Enzyme. Molecules, 29(7), 1431. https://doi.org/10.3390/molecules29071431








