Machine Learning-Based Spectral Analyses for Camellia japonica Cultivar Identification
Abstract
:1. Introduction
- Test two distinct machine learning-based approaches for discriminating C. japonica cultivars using NIR and FTIR spectroscopy, respectively, as follows:
- Approach 1: Select spectral ranges based on known absorption bands of biological molecules and apply a partial least squares discriminant analysis (PLSDA) for classification.
- Approach 2: Use forward feature selection taking advantage of five statistical selectors (ANOVA, chi-squared, FDR, mutual information, and FER), followed by principal component analysis (PCA) and testing multiple classifiers for prediction.
- Compare and discuss the performance of the two infrared-based techniques together with both data analyses applied approaches by analyzing their capability to accurately identify each one of the studied Camellia japonica cultivars.
2. Materials and Methods
2.1. Camellia japonica Samples
2.2. Infrared Spectra Acquisition
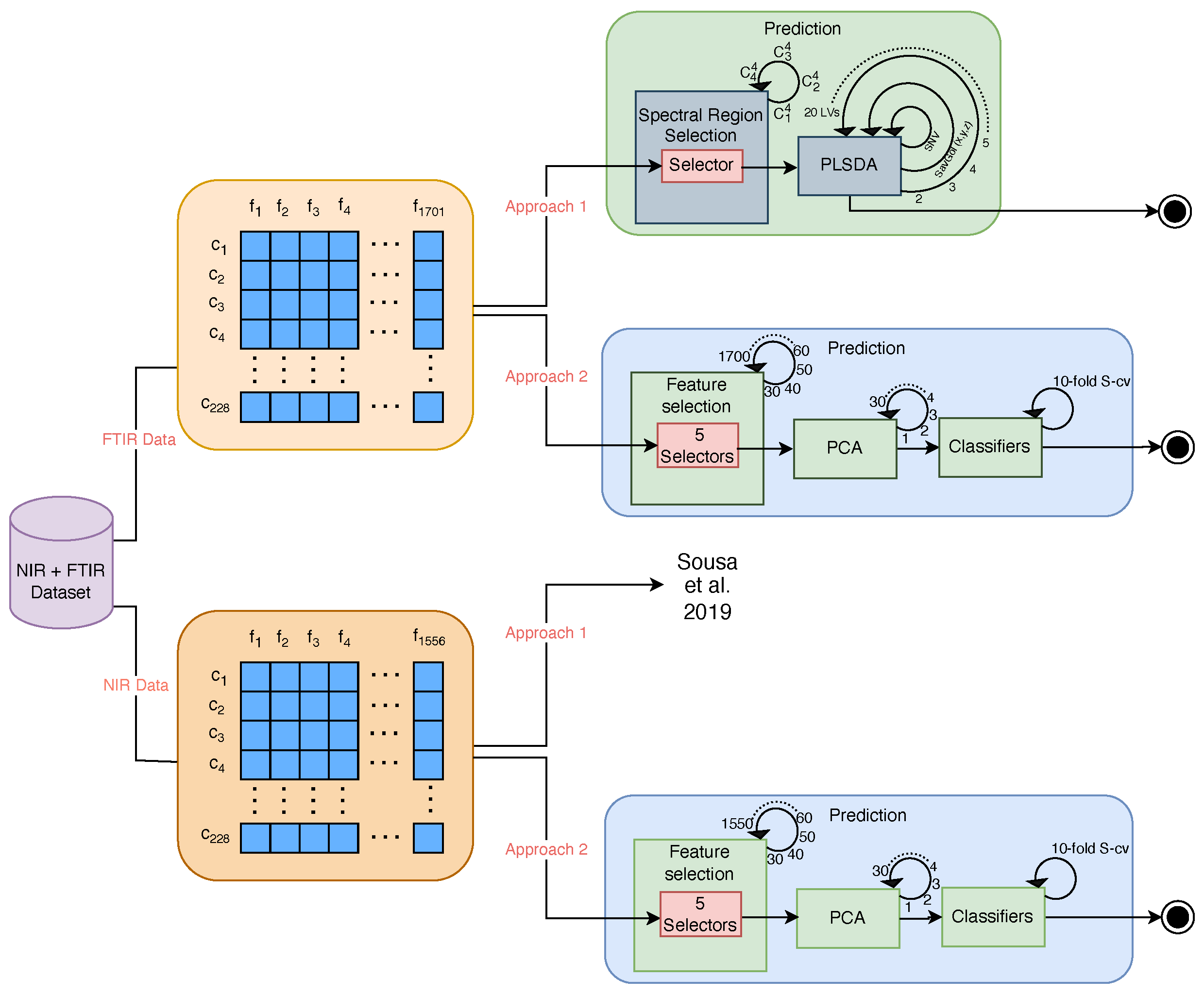
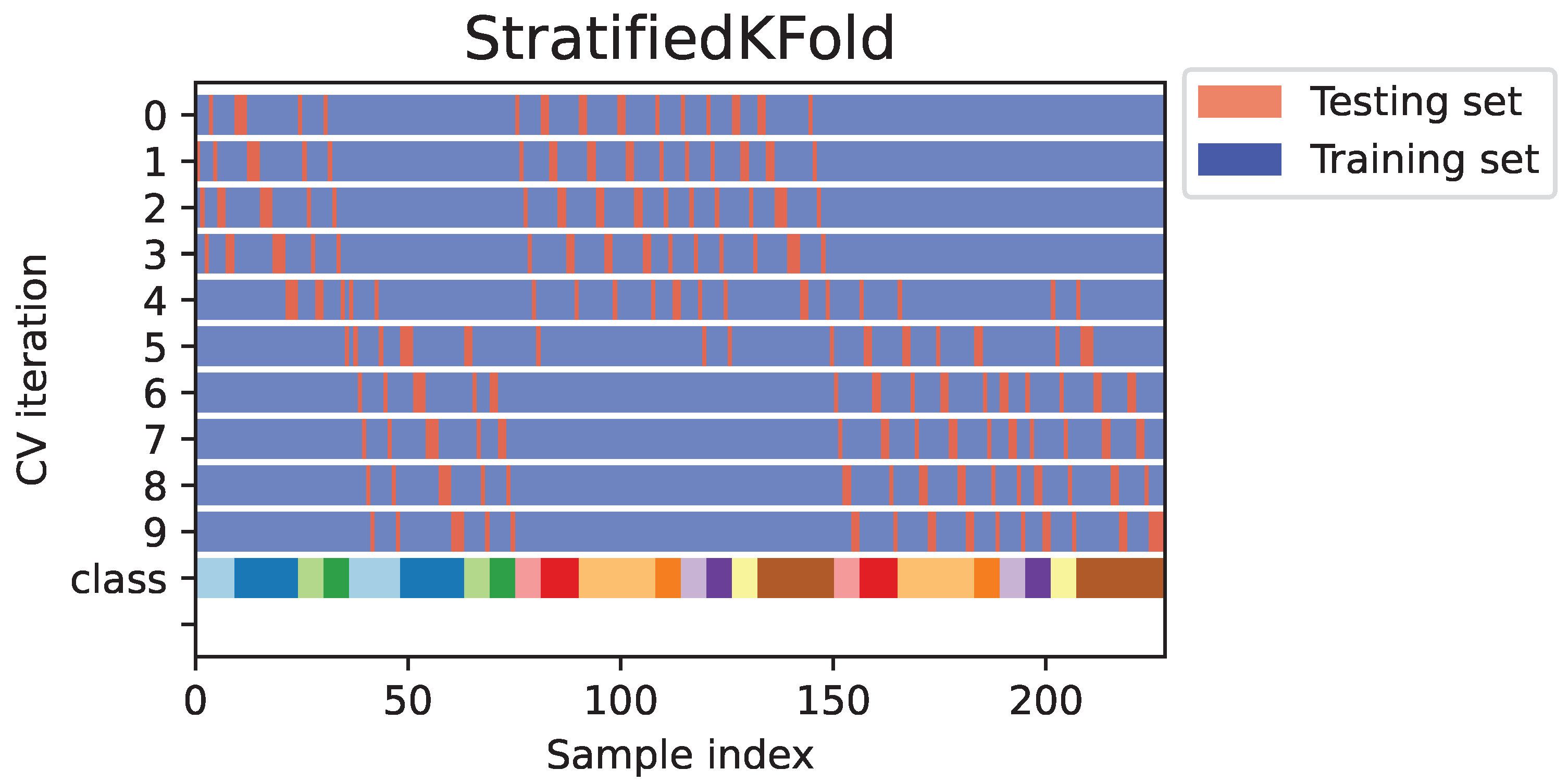
2.3. Data Analysis and Prediction
2.3.1. Approach 1—Unblinding Spectral Region Selection Combined with PLSDA
2.3.2. Approach 2—Blinding Forward Spectral Frequencies’ Selection Combined with Scikit-Learn ML Models
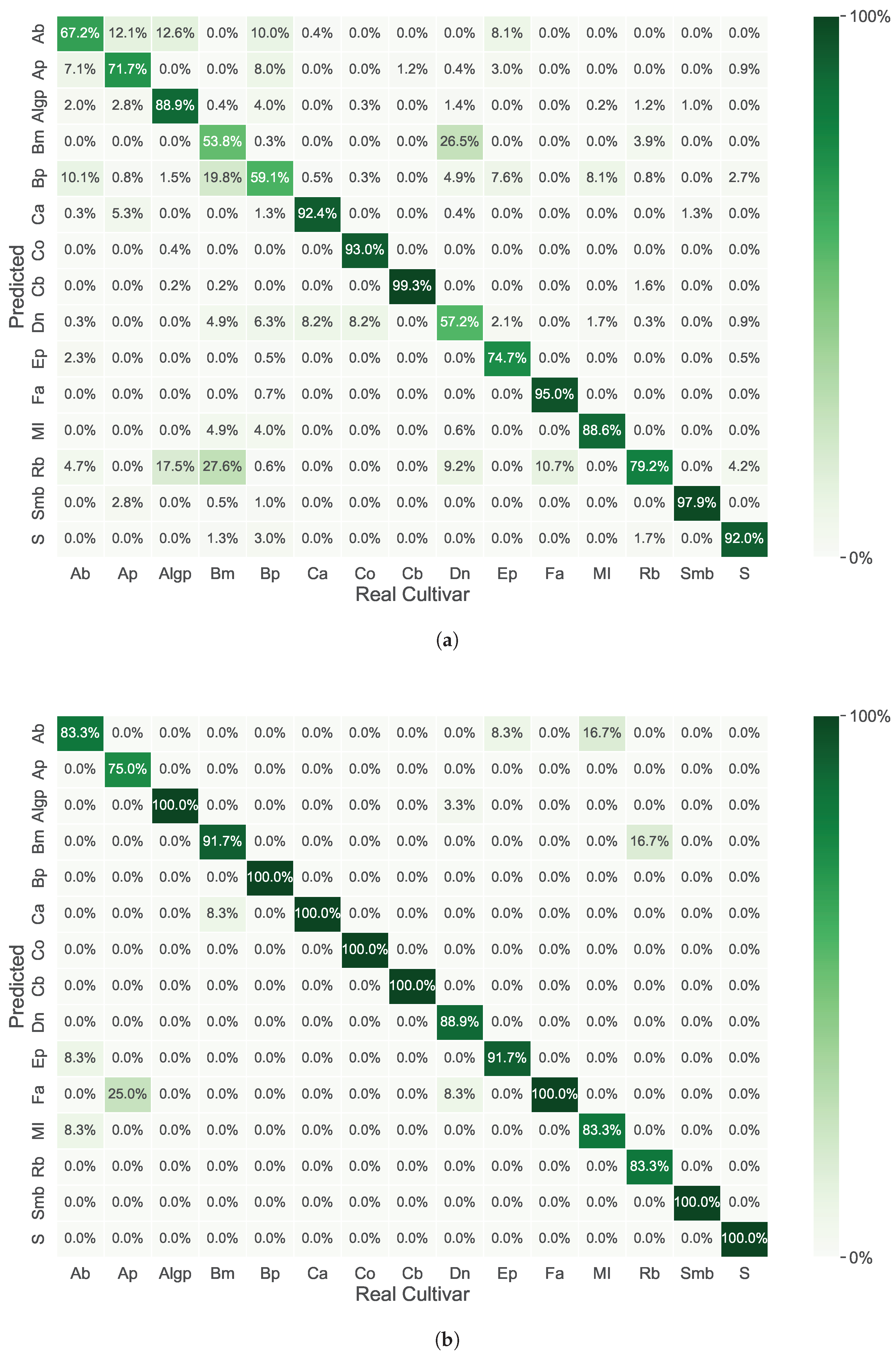
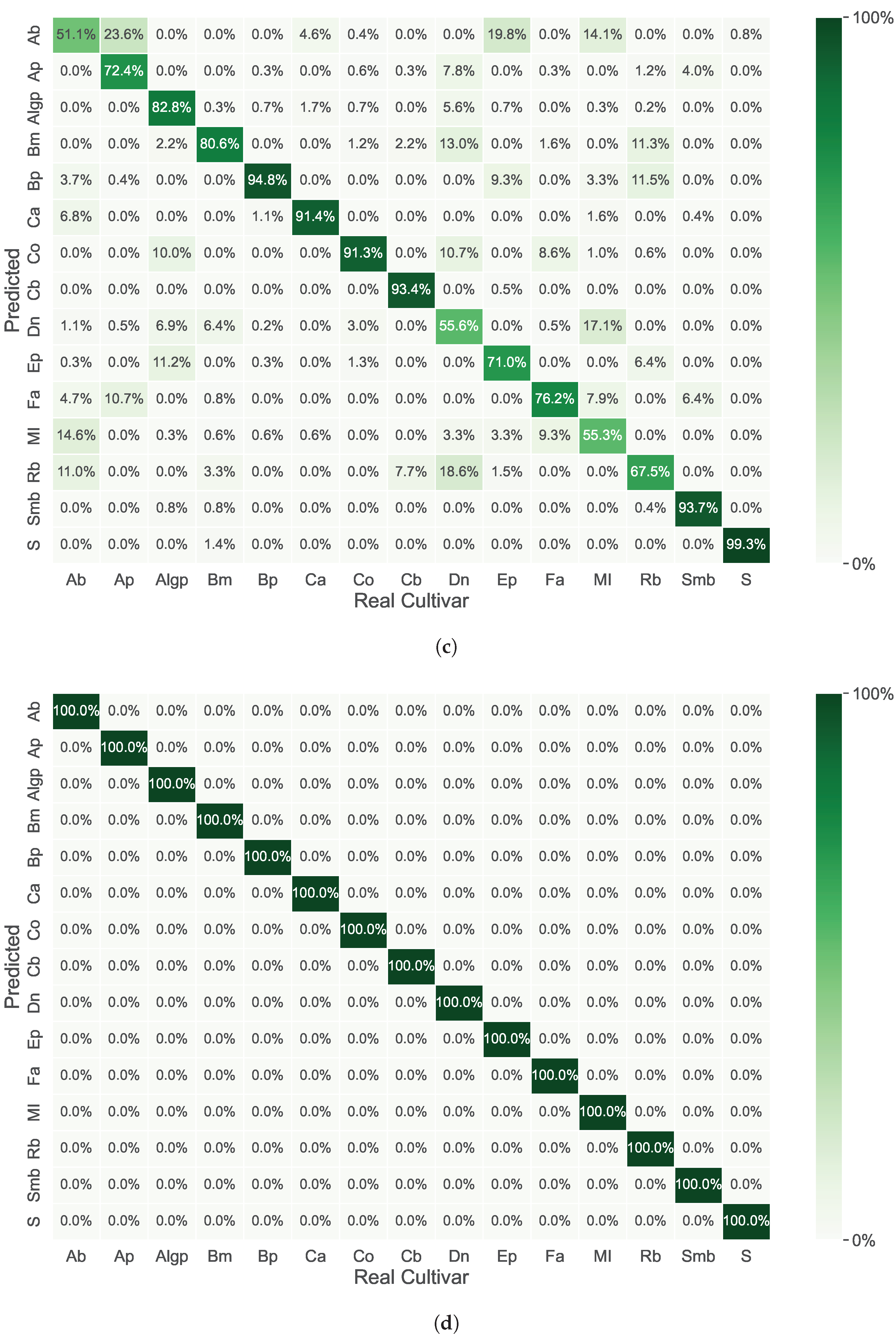
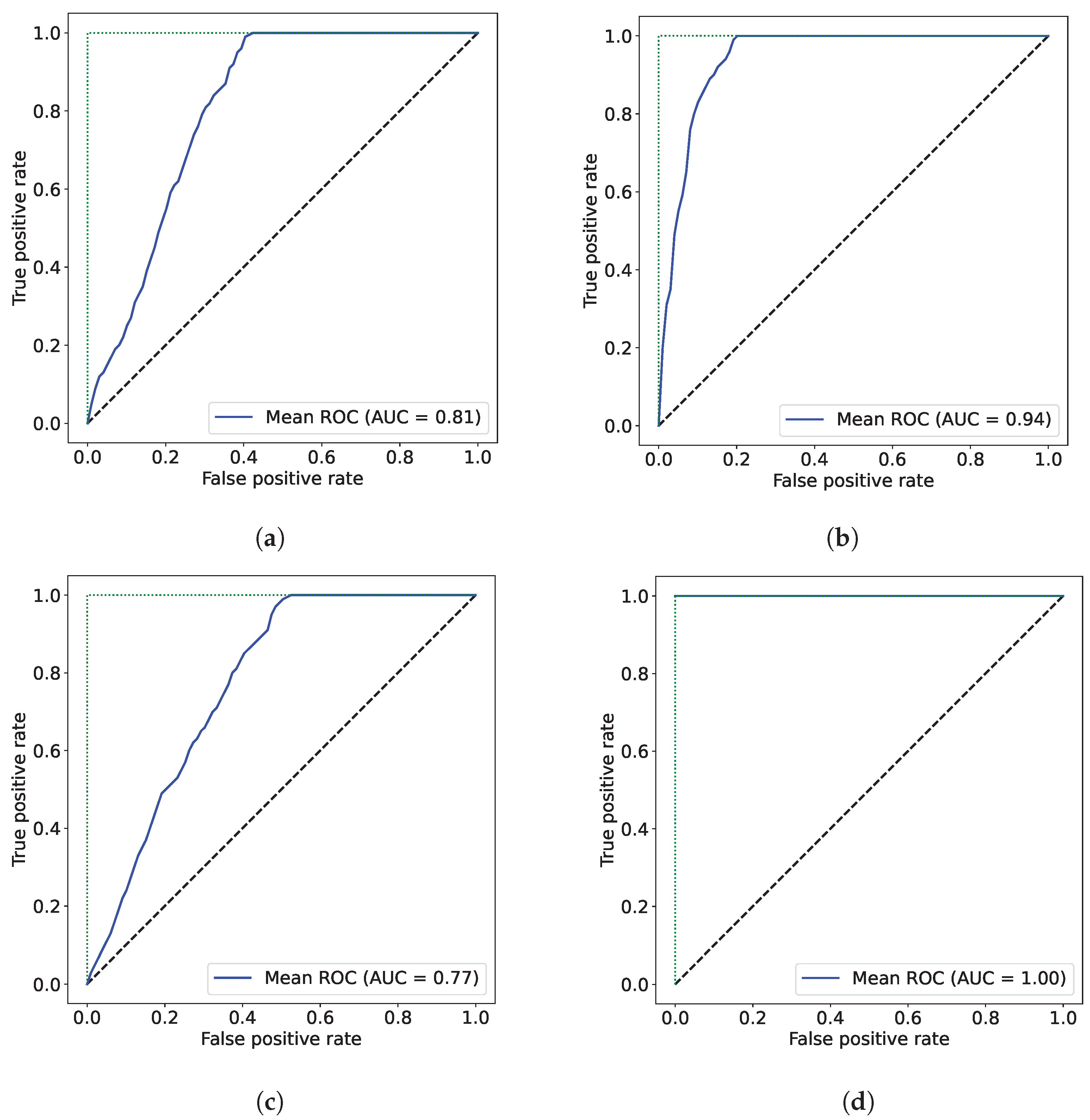
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Páscoa, R.N.M.J.; Teixeira, A.M.; Sousa, C. Antioxidant capacity of Camellia japonica cultivars assessed by near- and mid-infrared spectroscopy. Planta 2018, 249, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhuang, H.; Shen, Y.; Wang, Y.; Wang, Z. The Dataset of Camellia Cultivars Names in the World. Biodivers. Data J. 2021, 9, e61646. [Google Scholar] [CrossRef] [PubMed]
- Mugnai, S.; Pandolfi, C.; Azzarello, E.; Masi, E.; Mancuso, S. Camellia japonica L. genotypes identified by an artificial neural network based on phyllometric and fractal parameters. Plant Syst. Evol. 2007, 270, 95–108. [Google Scholar] [CrossRef]
- Heitkam, T.; Petrasch, S.; Zakrzewski, F.; Kögler, A.; Wenke, T.; Wanke, S.; Schmidt, T. Next-generation sequencing reveals differentially amplified tandem repeats as a major genome component of Northern Europe’s oldest Camellia japonica. Chromosome Res. 2015, 23, 791–806. [Google Scholar] [CrossRef]
- Sousa, C.; Quintelas, C.; Augusto, C.; Ferreira, E.C.; Páscoa, R.N. Discrimination of Camellia japonica cultivars and chemometric models: An interlaboratory study. Comput. Electron. Agric. 2019, 159, 28–33. [Google Scholar] [CrossRef]
- Han, B.; Yan, H.; Chen, C.; Yao, H.; Dai, J.; Chen, N. A rapid identification of four medicinal chrysanthemum varieties with near infrared spectroscopy. Pharmacogn. Mag. 2014, 10, 353. [Google Scholar] [CrossRef]
- Sousa, C.; Silva, L.; Grosso, F.; Nemec, A.; Lopes, J.; Peixe, L. Discrimination of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex species by Fourier transform infrared spectroscopy. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1345–1353. [Google Scholar] [CrossRef]
- Vaz, M.; Meirinhos-Soares, L.; Sousa, C.; Ramirez, M.; Melo-Cristino, J.; Lopes, J. Serotype discrimination of encapsulated Streptococcus pneumoniae strains by Fourier-transform infrared spectroscopy and chemometrics. J. Microbiol. Methods 2013, 93, 102–107. [Google Scholar] [CrossRef]
- Antamis, T.; Drosou, A.; Vafeiadis, T.; Nizamis, A.; Ioannidis, D.; Tzovaras, D. Interpretability of deep neural networks: A review of methods, classification and hardware. Neurocomputing 2024, 601, 128204. [Google Scholar] [CrossRef]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Alsberg, B.; Kell, D.; Goodacre, R. Variable Selection in Discriminant Partial Least-Squares Analysis. Anal. Chem. 1998, 70, 4126–4133. [Google Scholar] [CrossRef] [PubMed]
- Næs, T.; Isaksson, T.; Fearn, T.; Davies, T. A User-Friendly Guide to Multivariate Calibration and Classification; IM Publications Open: Chichester, UK, 2017. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Páscoa, R.; Lopo, M.; Teixeira dos Santos, C.; Graça, A.; Lopes, J. Exploratory study on vineyards soil mapping by visible/near-infrared spectroscopy of grapevine leaves. Comput. Electron. Agric. 2016, 127, 15–25. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Widodo, S.; Brawijaya, H.; Samudi, S. Stratified K-fold cross validation optimization on machine learning for prediction. Sinkron 2022, 7, 2407–2414. [Google Scholar] [CrossRef]
- Silva, S.; Tobaldini-Valerio, F.; Costa-de Oliveira, S.; Henriques, M.; Azeredo, J.; Ferreira, E.C.; Lopes, J.A.; Sousa, C. Discrimination of clinically relevant Candida species by Fourier-transform infrared spectroscopy with attenuated total reflectance (FTIR-ATR). RSC Adv. 2016, 6, 92065–92072. [Google Scholar] [CrossRef]
- Páscoa, R.N.; Moreira, S.; Lopes, J.A.; Sousa, C. Citrus species and hybrids depicted by near- and mid-infrared spectroscopy. J. Sci. Food Agric. 2018, 98, 3953–3961. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Tardaguila, J.; Fernández-Novales, J.; Diago, M.P. Data Mining and NIR Spectroscopy in Viticulture: Applications for Plant Phenotyping under Field Conditions. Sensors 2016, 16, 236. [Google Scholar] [CrossRef]
- Machado, J.C.; Faria, M.A.; Ferreira, I.M.; Páscoa, R.N.; Lopes, J.A. Varietal discrimination of hop pellets by near and mid infrared spectroscopy. Talanta 2018, 180, 69–75. [Google Scholar] [CrossRef]
- Buitrago, M.F.; Skidmore, A.K.; Groen, T.A.; Hecker, C.A. Connecting infrared spectra with plant traits to identify species. ISPRS J. Photogramm. Remote Sens. 2018, 139, 183–200. [Google Scholar] [CrossRef]
- Li, Y.; Via, B.K.; Han, F.; Li, Y.; Pei, Z. Comparison of various chemometric methods on visible and near-infrared spectral analysis for wood density prediction among different tree species and geographical origins. Front. Plant Sci. 2023, 14, 1121287. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tang, X.; Shen, Z.; Yang, K.; Zhao, L.; Li, Y. Comprehensive comparison of multiple quantitative near-infrared spectroscopy models for Aspergillus flavus contamination detection in peanut. J. Sci. Food Agric. 2019, 99, 5671–5679. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Peng, Y.; Li, L.; Ye, S.; Hong, S. Near-Infrared spectroscopy combined with machine learning methods for distinguishment of the storage years of rice. Infrared Phys. Technol. 2023, 133, 104835. [Google Scholar] [CrossRef]
- Lange, J.; Komissarov, L.; Lang, R.; Enkelmann, D.D.; Anelli, A. Automatic solid form classification in pharmaceutical drug development. arXiv 2024, arXiv:2411.03308. [Google Scholar]
- Wang, H.; Xie, Y.; Li, D.; Deng, H.; Zhao, Y.; Xin, M.; Lin, J. Rapid Identification of X-ray Diffraction Patterns Based on Very Limited Data by Interpretable Convolutional Neural Networks. J. Chem. Inf. Model. 2020, 60, 2004–2011. [Google Scholar] [CrossRef]
- Schuetzke, J.; Benedix, A.; Mikut, R.; Reischl, M. Siamese Networks for 1D Signal Identification. In Proceedings of the 30 Workshop Computational Intelligence, Berlin, Germany, 26–27 November 2020; KIT Scientific Publishing: Eggenstein-Leopoldshafen, Germany, 2020; pp. 17–31. [Google Scholar] [CrossRef]
- Gündüz, H.A.; Binder, M.; To, X.Y.; Mreches, R.; Bischl, B.; McHardy, A.C.; Münch, P.C.; Rezaei, M. A self-supervised deep learning method for data-efficient training in genomics. Commun. Biol. 2023, 6, 928. [Google Scholar] [CrossRef]
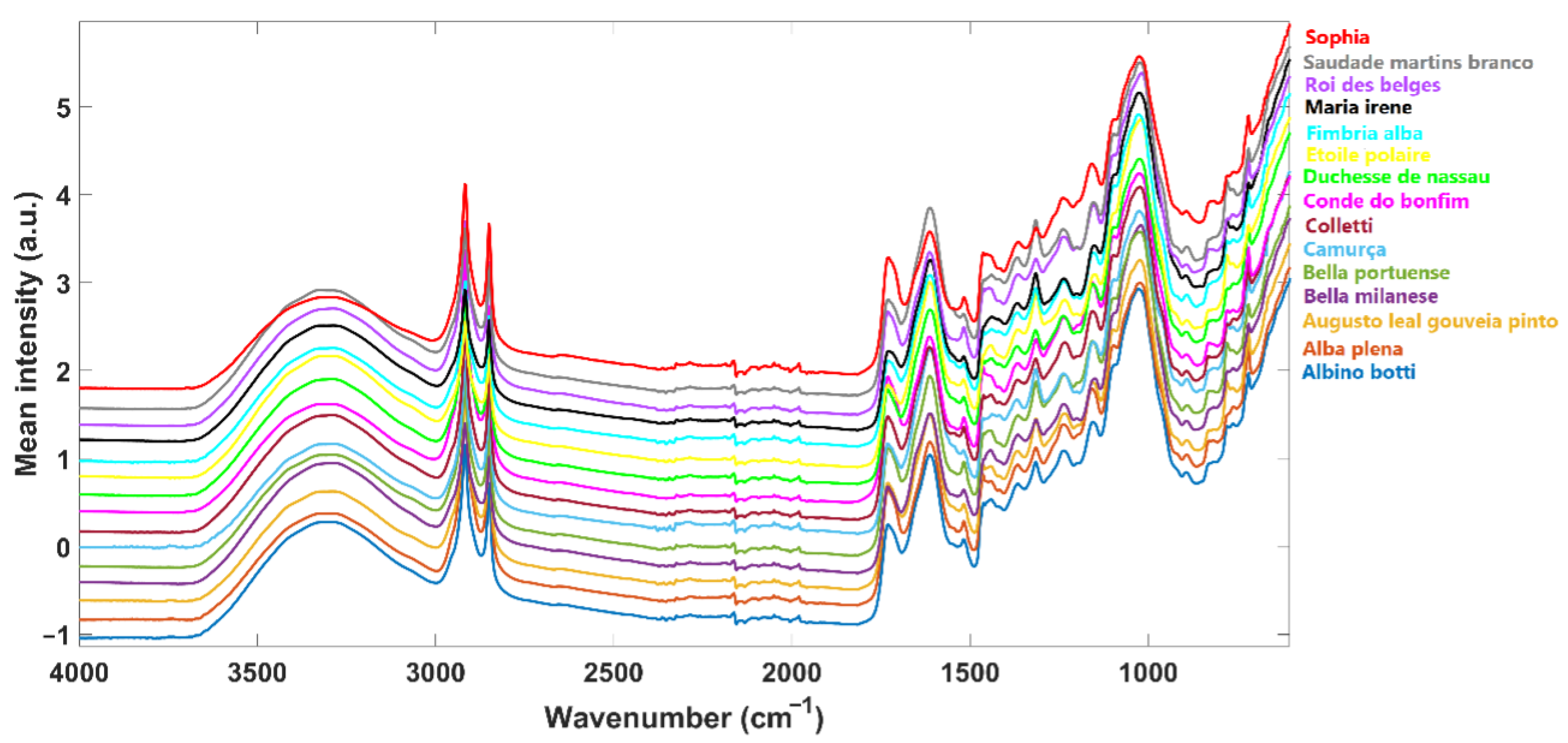
| Cultivar | N° of Plants | Collecting Local |
|---|---|---|
| Albino botti | 1 | VMP |
| 1 | JBP | |
| Alba plena | 1 | VMP |
| 1 | JBP | |
| Augusto leal gouveia pinto | 2 | VMP |
| 3 | JBP | |
| Bella Milanese | 2 | JBP |
| Bella portuense | 2 | JBP |
| Camurça | 2 | VMP |
| Colletti | 2 | VMP |
| 1 | JBP | |
| Conde do bonfim | 3 | JBP |
| Duchesse de nassau | 3 | JBP |
| Etoile polaire | 2 | JBP |
| Fimbria alba | 2 | VMP |
| Maria irene | 2 | JBP |
| Roi des belges | 2 | JBP |
| Saudade martins branco | 4 | VMP |
| Sophia | 2 | JBP |
| Classifier | Hyperparameters |
|---|---|
| AdaBoost Classifier | Default parameters (n_estimators = 50, learning_rate = 1.0, algorithm = “SAMME”) |
| Bagging Classifier | Default parameters (n_estimators = 10) |
| Decision Tree Classifier | Default parameters (Max_depth=5) |
| Gaussian NB | Default parameters |
| Quadratic Discriminant Analysis | Default parameters |
| K Nearest Neighbors Classifier | Default parameters (n_neighbors = 5) |
| Linear Discriminant Analysis | Default parameters |
| Logistic Regression | Default parameters (solver: “lbfgs” + max_iter = 1000) |
| Support-Vector Machines | Default parameters (kernel = “linear”, max_iter = 1000, and C = 1.0) |
| Support-Vector Machines | (kernel = “RBF”, C = 1.0, gamma = ‘scale’, probability = True) |
| NIR | FTIR-ATR | |
|---|---|---|
| Approach 1 | 81.3% | 77.5% |
| Approach 2 | 94.3% | 100.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, P.M.; Sousa, C. Machine Learning-Based Spectral Analyses for Camellia japonica Cultivar Identification. Molecules 2025, 30, 546. https://doi.org/10.3390/molecules30030546
Rodrigues PM, Sousa C. Machine Learning-Based Spectral Analyses for Camellia japonica Cultivar Identification. Molecules. 2025; 30(3):546. https://doi.org/10.3390/molecules30030546
Chicago/Turabian StyleRodrigues, Pedro Miguel, and Clara Sousa. 2025. "Machine Learning-Based Spectral Analyses for Camellia japonica Cultivar Identification" Molecules 30, no. 3: 546. https://doi.org/10.3390/molecules30030546
APA StyleRodrigues, P. M., & Sousa, C. (2025). Machine Learning-Based Spectral Analyses for Camellia japonica Cultivar Identification. Molecules, 30(3), 546. https://doi.org/10.3390/molecules30030546








