Unlocking Antimicrobial Peptides: In Silico Proteolysis and Artificial Intelligence-Driven Discovery from Cnidarian Omics
Abstract
1. Introduction
| ID | Species | Subphylum | Uniprot | Length (AA) | Targets | Reference |
|---|---|---|---|---|---|---|
| Aurelin | Aurelia aurita | Medusozoa | Q0MWV8 | 84 | Gram+ and Gram− Bacteria | [28] |
| AmAMP1 | Acropora millepora | Anthozoa | P0DUG2 | 117 | Gram+ and Gram− Bacteria | [38] |
| Arminin ** | Hydra vulgaris | Medusozoa | D2XUU4 | 88 | Gram+ and Gram− Bacteria | [39] |
| ATX-II * | Anemonia sulcata | Anthozoa | P01528 | 80 | Micrococcus luteus | [31] |
| Crassicorin-I and Crassicorin-II * | Urticina crassicornis | Anthozoa | A0A1X9QHL1 and P0DUG3 | 79 | Bacillus subtilis, Escherichia coli and Salmonella enterica | [30] |
| Damicornin | Pocillopora damicornis | Anthozoa | F1DFM9 | 107 | Gram+ bacteria and the fungus Fusarium oxysporum | [29] |
| Equinin B | Actinia equina | Anthozoa | n.a. | 72 | Escherichia coli, Micrococcus luteus and Vibrio alginolyticus | [40] |
| Hydramacin-1 | Hydra vulgaris | Medusozoa | B3RFR8 | 84 | Gram+ and Gram− Bacteria | [41] |
| APETx1 * | Anthopleura elegantissima | Anthozoa | P61541 | 42 | Salmonella enterica | [30] |
| ShK * | Stichodactyla helianthus | Anthozoa | P29187 | 35 | Bacillus subtilis, Escherichia coli, Salmonella enterica and Pseudomonas aeruginosa | [30] |
| Kazal2 | Hydra magnipapillata | Medusozoa | B8Y8I5 | 168 | Staphylococcus aureus | [42] |
2. Results
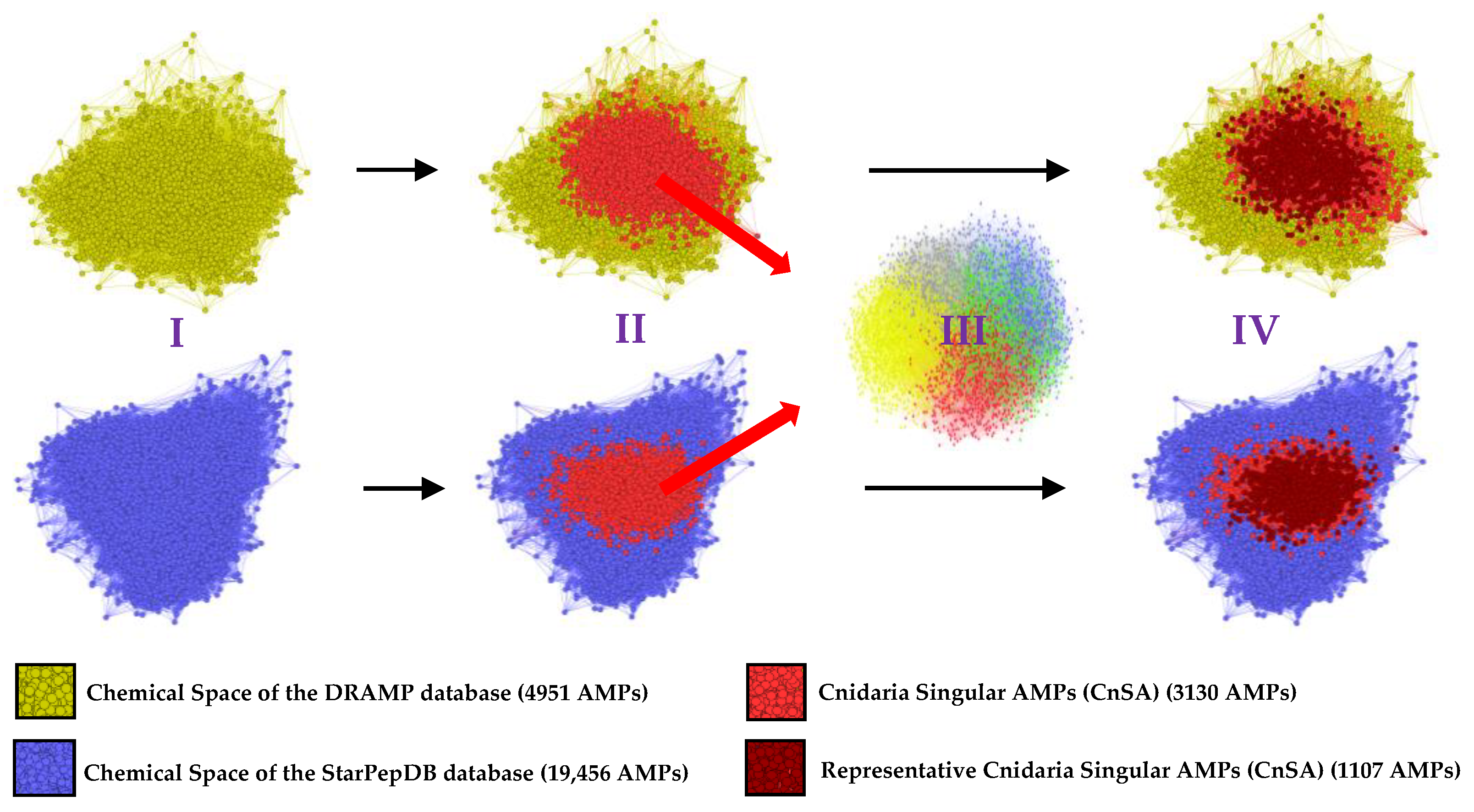
2.1. Cnidaria Databases Reveal Significant Uniqueness
2.2. In Silico Proteolysis of AMP Precursor Datasets with Distinct Proteases Yields Diverse Peptidomes
2.3. Antimicrobial and Toxicity Screening of Virtual Peptidomes Reveals High AMP Diversity
2.4. Physicochemical Properties Indicate the Suitability of the Non-Haemolytic and Non-Toxic AMPs for Targeting Microbial Membranes
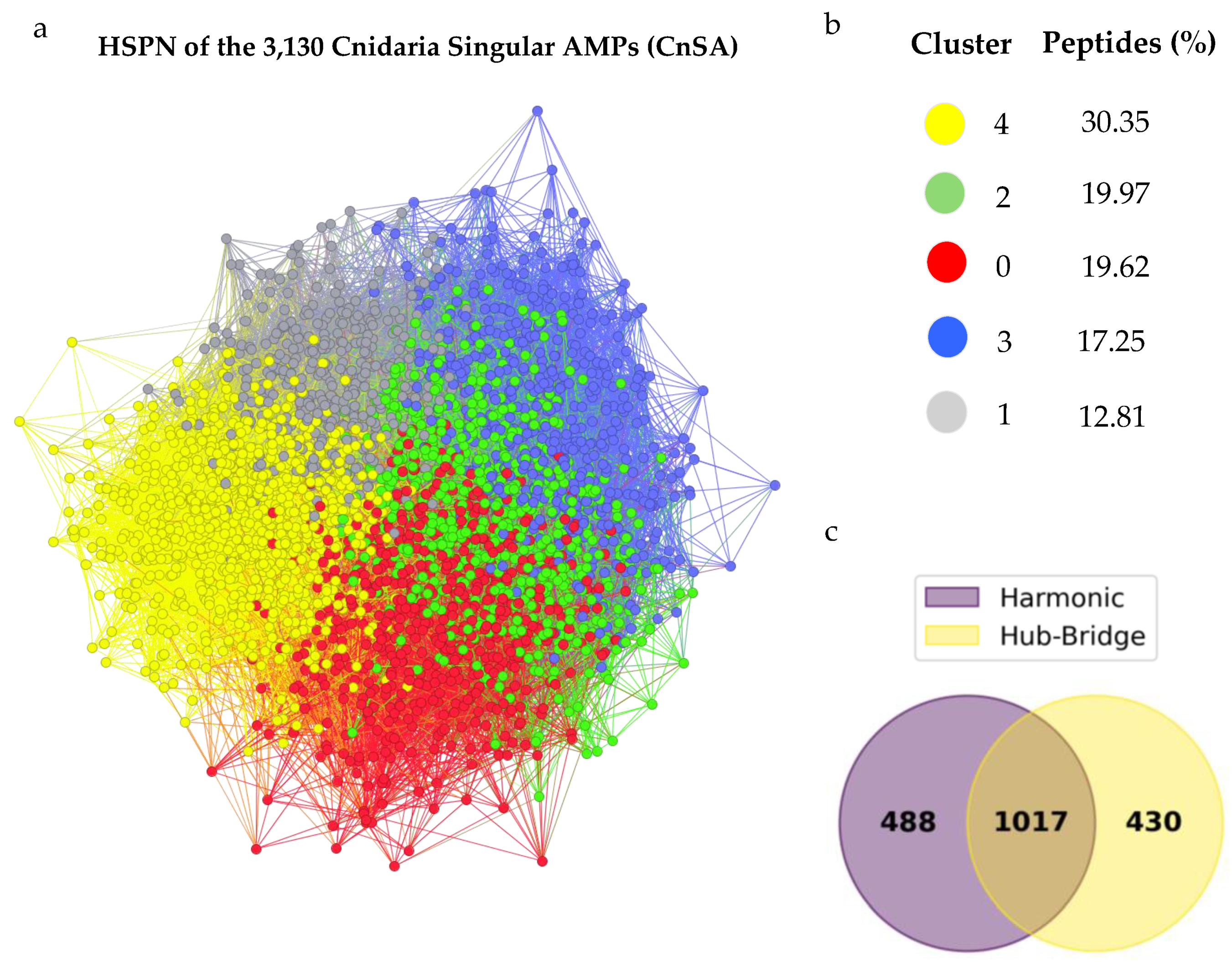
2.5. Cnidaria Singular AMPs (CnSAs) Demonstrate High Internal Sequence Diversity
2.6. Half-Space Proximal Networks (HSPNs) Facilitate the Extraction of Representative CnSA Datasets
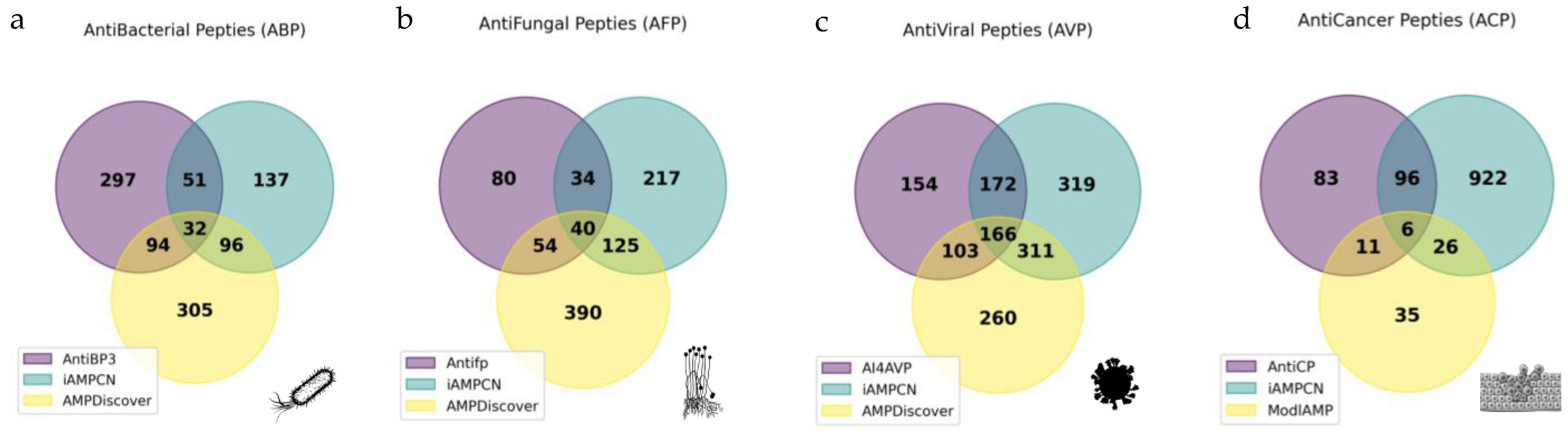
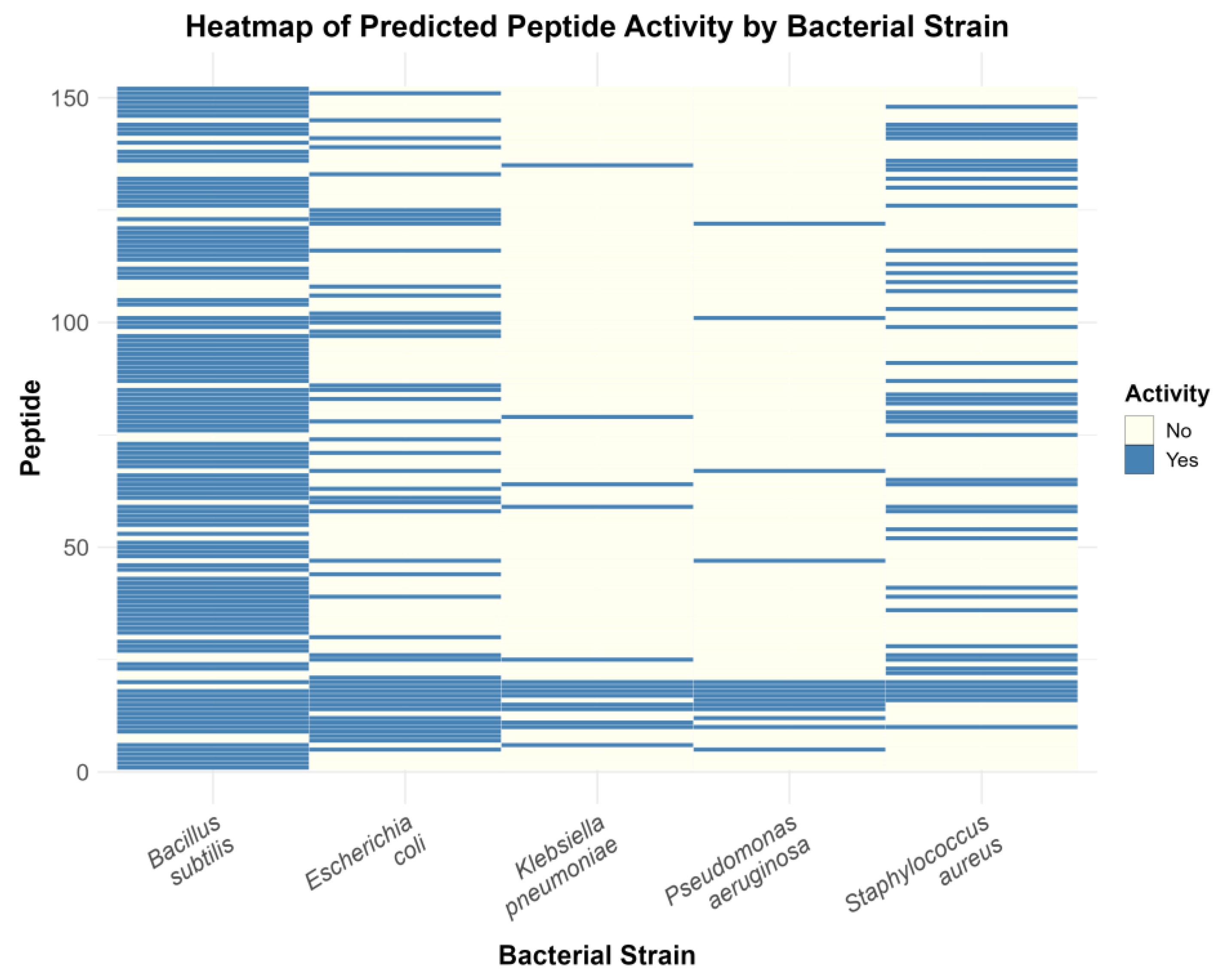
2.7. Strain-Specific Predictions Reveal Novel Candidate Antibacterial Peptides (ABPs)
3. Discussion
4. Materials and Methods
4.1. Gathering of Omics Data from Cnidaria
4.2. Database Construction
4.3. In Silico Proteolysis
4.4. Antimicrobial and Toxicity Screening
4.5. Selection of Cnidaria Singular AMPs (CnSA) Using Complex Network Analyses
4.6. Activity and Strain-Specific Predictions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919. [Google Scholar] [PubMed]
- Aronica, P.G.; Reid, L.M.; Desai, N.; Li, J.; Fox, S.J.; Yadahalli, S.; Essex, J.W.; Verma, C.S. Computational methods and tools in antimicrobial peptide research. J. Chem. Inf. Model. 2021, 61, 3172–3196. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Patocka, J.; Nepovimova, E.; Oleksak, P.; Valis, M.; Wu, W.; Kuca, K. Marine invertebrate peptides: Antimicrobial peptides. Front. Microbiol. 2021, 12, 785085. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Guryanova, S.V.; Balandin, S.V.; Belogurova-Ovchinnikova, O.Y.; Ovchinnikova, T.V. Marine Invertebrate Antimicrobial Peptides and Their Potential as Novel Peptide Antibiotics. Mar. Drugs 2023, 21, 503. [Google Scholar] [CrossRef]
- Wang, S.; Fan, L.; Pan, H.; Li, Y.; Qiu, Y.; Lu, Y. Antimicrobial peptides from marine animals: Sources, structures, mechanisms and the potential for drug development. Front. Mar. Sci. 2023, 9, 1112595. [Google Scholar] [CrossRef]
- Agüero-Chapin, G.; Galpert-Cañizares, D.; Domínguez-Pérez, D.; Marrero-Ponce, Y.; Pérez-Machado, G.; Teijeira, M.; Antunes, A. Emerging computational approaches for antimicrobial peptide discovery. Antibiotics 2022, 11, 936. [Google Scholar] [CrossRef]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Beltran, J.A.; Tellez Ibarra, R.; Guillen-Ramirez, H.A.; Brizuela, C.A. Graph-based data integration from bioactive peptide databases of pharmaceutical interest: Toward an organized collection enabling visual network analysis. Bioinformatics 2019, 35, 4739–4747. [Google Scholar] [CrossRef]
- Kang, X.; Dong, F.; Shi, C.; Liu, S.; Sun, J.; Chen, J.; Li, H.; Xu, H.; Lao, X.; Zheng, H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 2019, 6, 148. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef] [PubMed]
- Ammerman, J.; Fuhrman, J.; Hagström, A.; Azam, F. Bacterioplankton growth in seawater: I. Growth kinetics and cellular characteristics in seawater cultures. Mar. Ecol. Prog. Ser. Oldendorf 1984, 18, 31–39. [Google Scholar] [CrossRef]
- Tincu, J.A.; Taylor, S.W. Antimicrobial peptides from marine invertebrates. Antimicrob. Agents Chemother. 2004, 48, 3645–3654. [Google Scholar] [CrossRef] [PubMed]
- Miyata, T.; Tokunaga, F.; Yoneya, T.; Yoshikawa, K.; Iwanaga, S.; Niwa, M.; Takao, T.; Shimonishi, Y. Antimicrobial peptides, isolated from horseshoe crab hemocytes, tachyplesin II, and polyphemusins I and II: Chemical structures and biological activity. J. Biochem. 1989, 106, 663–668. [Google Scholar] [CrossRef]
- Nakamura, T.; Furunaka, H.; Miyata, T.; Tokunaga, F.; Muta, T.; Iwanaga, S.; Niwa, M.; Takao, T.; Shimonishi, Y. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). Isolation and chemical structure. J. Biol. Chem. 1988, 263, 16709–16713. [Google Scholar] [CrossRef]
- Pan, W.; Liu, X.; Ge, F.; Han, J.; Zheng, T. Perinerin, a novel antimicrobial peptide purified from the clamworm Perinereis aibuhitensis grube and its partial characterization. J. Biochem. 2004, 135, 297–304. [Google Scholar] [CrossRef]
- Xu, G.; Wu, M.; Wang, L.; Zhang, X.; Cao, S.; Liu, M.; Cui, Y. Conformational and dynamics simulation study of antimicrobial peptide hedistin—Heterogeneity of its helix–turn–helix motif. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2009, 1788, 2497–2508. [Google Scholar] [CrossRef][Green Version]
- Mitta, G.; Hubert, F.; Noël, T.; Roch, P. Myticin, a novel cysteine-rich antimicrobial peptide isolated from haemocytes and plasma of the mussel Mytilus galloprovincialis. Eur. J. Biochem. 1999, 265, 71–78. [Google Scholar] [CrossRef]
- Charlet, M.; Chernysh, S.; Philippe, H.; Hetru, C.; Hoffmann, J.A.; Bulet, P. Innate immunity: Isolation of several cysteine-rich antimicrobial peptides from the blood of a mollusc, Mytilus edulis. J. Biol. Chem. 1996, 271, 21808–21813. [Google Scholar] [CrossRef]
- Lee, I.H.; Zhao, C.; Cho, Y.; Harwig, S.S.; Cooper, E.L.; Lehrer, R.I. Clavanins, α-helical antimicrobial peptides from tunicate hemocytes. FEBS Lett. 1997, 400, 158–162. [Google Scholar] [CrossRef]
- Matos, A.; Domínguez-Pérez, D.; Almeida, D.; Agüero-Chapin, G.; Campos, A.; Osório, H.; Vasconcelos, V.; Antunes, A. Shotgun Proteomics of Ascidians Tunic Gives New Insights on Host–Microbe Interactions by Revealing Diverse Antimicrobial Peptides. Mar. Drugs 2020, 18, 362. [Google Scholar] [CrossRef]
- Li, C.; Haug, T.; Styrvold, O.B.; Jørgensen, T.Ø.; Stensvåg, K. Strongylocins, novel antimicrobial peptides from the green sea urchin, Strongylocentrotus droebachiensis. Dev. Comp. Immunol. 2008, 32, 1430–1440. [Google Scholar] [CrossRef]
- Li, C.; Haug, T.; Moe, M.K.; Styrvold, O.B.; Stensvåg, K. Centrocins: Isolation and characterization of novel dimeric antimicrobial peptides from the green sea urchin, Strongylocentrotus droebachiensis. Dev. Comp. Immunol. 2010, 34, 959–968. [Google Scholar] [CrossRef]
- Fautin, D.G. Structural diversity, systematics, and evolution of cnidae. Toxicon 2009, 54, 1054–1064. [Google Scholar] [CrossRef]
- McFadden, C.S.; Quattrini, A.M.; Brugler, M.R.; Cowman, P.F.; Duenas, L.F.; Kitahara, M.V.; Paz-Garcia, D.A.; Reimer, J.D.; Rodriguez, E. Phylogenomics, Origin, and Diversification of Anthozoans (Phylum Cnidaria). Syst. Biol. 2021, 70, 635–647. [Google Scholar] [CrossRef]
- Kayal, E.; Bentlage, B.; Sabrina Pankey, M.; Ohdera, A.H.; Medina, M.; Plachetzki, D.C.; Collins, A.G.; Ryan, J.F. Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol. Biol. 2018, 18, 68. [Google Scholar] [CrossRef]
- Zapata, F.; Goetz, F.E.; Smith, S.A.; Howison, M.; Siebert, S.; Church, S.H.; Sanders, S.M.; Ames, C.L.; McFadden, C.S.; France, S.C.; et al. Phylogenomic Analyses Support Traditional Relationships within Cnidaria. PLoS ONE 2015, 10, e0139068. [Google Scholar] [CrossRef]
- Ovchinnikova, T.V.; Balandin, S.V.; Aleshina, G.M.; Tagaev, A.A.; Leonova, Y.F.; Krasnodembsky, E.D.; Men’shenin, A.V.; Kokryakov, V.N. Aurelin, a novel antimicrobial peptide from jellyfish Aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem. Biophys. Res. Commun. 2006, 348, 514–523. [Google Scholar] [CrossRef]
- Vidal-Dupiol, J.; Ladrière, O.; Destoumieux-Garzon, D.; Sautiere, P.-E.; Meistertzheim, A.-L.; Tambutté, E.; Tambutté, S.; Duval, D.; Fouré, L.; Adjeroud, M. Innate immune responses of a scleractinian coral to vibriosis. J. Biol. Chem. 2011, 286, 22688–22698. [Google Scholar] [CrossRef]
- Kim, C.H.; Lee, Y.J.; Go, H.J.; Oh, H.Y.; Lee, T.K.; Park, J.B.; Park, N.G. Defensin-neurotoxin dyad in a basally branching metazoan sea anemone. FEBS J. 2017, 284, 3320–3338. [Google Scholar] [CrossRef]
- Trapani, M.R.; Parisi, M.G.; Toubiana, M.; Coquet, L.; Jouenne, T.; Roch, P.; Cammarata, M. First evidence of antimicrobial activity of neurotoxin 2 from Anemonia sulcata (Cnidaria). Invertebr. Surviv. J. 2014, 11, 182–191. [Google Scholar]
- Klompen, A.M.L.; Kayal, E.; Collins, A.G.; Cartwright, P. Phylogenetic and Selection Analysis of an Expanded Family of Putatively Pore-Forming Jellyfish Toxins (Cnidaria: Medusozoa). Genome Biol. Evol. 2021, 13, evab081. [Google Scholar] [CrossRef]
- Barroso, R.A.; Ramos, L.; Moreno, H.; Antunes, A. Evolutionary Analysis of Cnidaria Small Cysteine-Rich Proteins (SCRiPs), an Enigmatic Neurotoxin Family from Stony Corals and Sea Anemones (Anthozoa: Hexacorallia). Toxins 2024, 16, 75. [Google Scholar] [CrossRef]
- Mitchell, M.L.; Shafee, T.; Papenfuss, A.T.; Norton, R.S. Evolution of cnidarian trans-defensins: Sequence, structure and exploration of chemical space. Proteins Struct. Funct. Bioinform. 2019, 87, 551–560. [Google Scholar] [CrossRef]
- Leal, E.; Múnera, M.; Suescún-Bolívar, L.P. In silico characterization of Cnidarian’s antimicrobial peptides. Front. Mar. Sci. 2022, 9, 1065717. [Google Scholar] [CrossRef]
- Agüero-Chapin, G.; Domínguez-Pérez, D.; Marrero-Ponce, Y.; Castillo-Mendieta, K.; Antunes, A. Unveiling Encrypted Antimicrobial Peptides from Cephalopods’ Salivary Glands: A Proteolysis-Driven Virtual Approach. ACS Omega 2024, 9, 43353–43367. [Google Scholar] [CrossRef]
- Klimovich, A.; Bosch, T.C. Novel technologies uncover novel ‘anti’-microbial peptides in Hydra shaping the species-specific microbiome. Philos. Trans. R. Soc. B 2024, 379, 20230058. [Google Scholar] [CrossRef]
- Mason, B.; Cooke, I.; Moya, A.; Augustin, R.; Lin, M.-F.; Satoh, N.; Bosch, T.C.; Bourne, D.; Hayward, D.; Andrade, N. AmAMP1 from Acropora millepora and damicornin define a family of coral-specific antimicrobial peptides related to the Shk toxins of sea anemones. Dev. Comp. Immunol. 2021, 114, 103866. [Google Scholar] [CrossRef]
- Augustin, R.; Anton-Erxleben, F.; Jungnickel, S.; Hemmrich, G.; Spudy, B.r.; Podschun, R.; Bosch, T.C. Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob. Agents Chemother. 2009, 53, 5245–5250. [Google Scholar] [CrossRef]
- La Corte, C.; Catania, V.; Dara, M.; Parrinello, D.; Staropoli, M.; Trapani, M.R.; Cammarata, M.; Parisi, M.G. Equinins as Novel Broad-Spectrum Antimicrobial Peptides Isolated from the Cnidarian Actinia equina (Linnaeus, 1758). Mar. Drugs 2024, 22, 172. [Google Scholar] [CrossRef]
- Jung, S.; Dingley, A.J.; Augustin, R.; Anton-Erxleben, F.; Stanisak, M.; Gelhaus, C.; Gutsmann, T.; Hammer, M.U.; Podschun, R.; Bonvin, A.M. Hydramacin-1, structure and antibacterial activity of a protein from the basal metazoan Hydra. J. Biol. Chem. 2009, 284, 1896–1905. [Google Scholar] [CrossRef] [PubMed]
- Augustin, R.; Siebert, S.; Bosch, T.C. Identification of a kazal-type serine protease inhibitor with potent anti-staphylococcal activity as part of Hydra’s innate immune system. Dev. Comp. Immunol. 2009, 33, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Palsson, B. The challenges of in silico biology. Nat. Biotechnol. 2000, 18, 1147–1150. [Google Scholar] [CrossRef]
- Agüero-Chapin, G.; Antunes, A.; Mora, J.R.; Pérez, N.; Contreras-Torres, E.; Valdes-Martini, J.R.; Martinez-Rios, F.; Zambrano, C.H.; Marrero-Ponce, Y. Complex Networks Analyses of Antibiofilm Peptides: An Emerging Tool for Next-Generation Antimicrobials’ Discovery. Antibiotics 2023, 12, 747. [Google Scholar] [CrossRef]
- Romero, M.; Marrero-Ponce, Y.; Rodríguez, H.; Agüero-Chapin, G.; Antunes, A.; Aguilera-Mendoza, L.; Martinez-Rios, F. A novel network science and similarity-searching-based approach for discovering potential tumor-homing peptides from antimicrobials. Antibiotics 2022, 11, 401. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Hernández-Elizárraga, V.H.; Ocharán-Mercado, A.; Olguín-López, N.; Hernández-Matehuala, R.; Caballero-Pérez, J.; Ibarra-Alvarado, C.; Rojas-Molina, A. New Insights into the Toxin Diversity and Antimicrobial Activity of the “Fire Coral” Millepora complanata. Toxins 2022, 14, 206. [Google Scholar] [CrossRef]
- Stabili, L.; Piraino, S.; Rizzo, L. The Mediterranean Zoanthid Parazoanthus axinellae as a Novel Source of Antimicrobial Compounds. J. Mar. Sci. Eng. 2024, 12, 354. [Google Scholar] [CrossRef]
- Fingerhut, L.C.H.W.; Miller, D.J.; Strugnell, J.M.; Daly, N.L.; Cooke, I.R. ampir: An R package for fast genome-wide prediction of antimicrobial peptides. Bioinformatics 2020, 36, 5262–5263. [Google Scholar] [CrossRef]
- Maillet, N. Rapid Peptides Generator: Fast and efficient in silico protein digestion. NAR Genom. Bioinform. 2019, 2, lqz004. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; Heck, A.J. Proteomics beyond trypsin. FEBS J. 2015, 282, 2612–2626. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.; Kudryashova, E.; Grinberg, V.Y.; Grinberg, N.; Burova, T.; Levashov, A. The chemical modification of α-chymotrypsin with both hydrophobic and hydrophilic compounds stabilizes the enzyme against denaturation in water–organic media. Protein Eng. 2001, 14, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, P.J.; Walker, J.M. Proteinase K (EC 3.4. 21.14). Enzym. Mol. Biol. 1993, 305–311. [Google Scholar] [CrossRef]
- Oddo, A.; Hansen, P.R. Hemolytic activity of antimicrobial peptides. In Antimicrobial Peptides; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; pp. 427–435. [Google Scholar]
- Capecchi, A.; Cai, X.; Personne, H.; Köhler, T.; van Delden, C.; Reymond, J.-L. Machine learning designs non-hemolytic antimicrobial peptides. Chem. Sci. 2021, 12, 9221–9232. [Google Scholar] [CrossRef]
- Grafskaia, E.N.; Polina, N.F.; Babenko, V.V.; Kharlampieva, D.D.; Bobrovsky, P.A.; Manuvera, V.A.; Farafonova, T.E.; Anikanov, N.A.; Lazarev, V.N. Discovery of novel antimicrobial peptides: A transcriptomic study of the sea anemone Cnidopus japonicus. J. Bioinform. Comput. Biol. 2018, 16, 1840006. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Ladewig, L.; Gloy, L.; Langfeldt, D.; Pinnow, N.; Weiland-Bräuer, N.; Schmitz, R.A. Antimicrobial peptides originating from expression libraries of Aurelia aurita and Mnemiopsis leidyi prevent biofilm formation of opportunistic pathogens. Microorganisms 2023, 11, 2184. [Google Scholar] [CrossRef]
- Martínez Mondragón, S.B. Actividad Bactericida de Péptidos Antimicrobianos Sintetizados A Partir del Transcriptoma del Cnidario Hydractinia Symbiolongicarpus. Bachelor’s Thesis, Universidad de los Andes, Bogotá, Colombia, 2019. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 19 August 2024).
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Reina, D.; Toral, S.; Johnson, P.; Barrero, F. Improving discovery phase of reactive ad hoc routing protocols using Jaccard distance. J. Supercomput. 2014, 67, 131–152. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Santos-Junior, C.D.; Pan, S.; Zhao, X.-M.; Coelho, L.P. Macrel: Antimicrobial peptide screening in genomes and metagenomes. PeerJ 2020, 8, e10555. [Google Scholar] [CrossRef]
- Li, C.; Sutherland, D.; Hammond, S.A.; Yang, C.; Taho, F.; Bergman, L.; Houston, S.; Warren, R.L.; Wong, T.; Hoang, L.M. AMPlify: Attentive deep learning model for discovery of novel antimicrobial peptides effective against WHO priority pathogens. BMC Genom. 2022, 23, 77. [Google Scholar] [CrossRef]
- Chaudhary, K.; Kumar, R.; Singh, S.; Tuknait, A.; Gautam, A.; Mathur, D.; Anand, P.; Varshney, G.C.; Raghava, G.P. A web server and mobile app for computing hemolytic potency of peptides. Sci. Rep. 2016, 6, 22843. [Google Scholar] [CrossRef]
- Castillo-Mendieta, K.; Agüero-Chapin, G.; Marquez, E.A.; Perez-Castillo, Y.; Barigye, S.J.; Pérez-Cárdenas, M.; Peréz-Giménez, F.; Marrero-Ponce, Y. A New Robust Method for Predicting Hemolytic Toxicity from Peptide Sequence. ChemRxiv 2023. [Google Scholar] [CrossRef]
- Rathore, A.S.; Choudhury, S.; Arora, A.; Tijare, P.; Raghava, G.P. ToxinPred 3.0: An improved method for predicting the toxicity of peptides. Comput. Biol. Med. 2024, 179, 108926. [Google Scholar] [CrossRef]
- Wang, J.-H.; Sung, T.-Y. ToxTeller: Predicting Peptide Toxicity Using Four Different Machine Learning Approaches. ACS Omega 2024, 9, 32116–32123. [Google Scholar] [CrossRef]
- Jiao, S.; Ye, X.; Sakurai, T.; Zou, Q.; Liu, R. Integrated convolution and self-attention for improving peptide toxicity prediction. Bioinformatics 2024, 40, btae297. [Google Scholar] [CrossRef]
- Müller, A.T.; Gabernet, G.; Hiss, J.A.; Schneider, G. modlAMP: Python for antimicrobial peptides. Bioinformatics 2017, 33, 2753–2755. [Google Scholar] [CrossRef] [PubMed]
- Wilcoxon, F. Individual comparisons by ranking methods. In Breakthroughs in Statistics: Methodology and Distribution; Springer: Berlin/Heidelberg, Germany, 1992; pp. 196–202. [Google Scholar]
- Friedman, M. The use of ranks to avoid the assumption of normality implicit in the analysis of variance. J. Am. Stat. Assoc. 1937, 32, 675–701. [Google Scholar] [CrossRef]
- Agüero-Chapin, G.; Galpert, D.; Molina-Ruiz, R.; Ancede-Gallardo, E.; Pérez-Machado, G.; De la Riva, G.A.; Antunes, A. Graph theory-based sequence descriptors as remote homology predictors. Biomolecules 2019, 10, 26. [Google Scholar] [CrossRef]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Tellez-Ibarra, R.; Llorente-Quesada, M.T.; Salgado, J.; Barigye, S.J.; Liu, J. Overlap and diversity in antimicrobial peptide databases: Compiling a non-redundant set of sequences. Bioinformatics 2015, 31, 2553–2559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Garcia-Jacas, C.R.; Chavez, E.; Beltran, J.A.; Guillen-Ramirez, H.A.; Brizuela, C.A. Automatic construction of molecular similarity networks for visual graph mining in chemical space of bioactive peptides: An unsupervised learning approach. Sci. Rep. 2020, 10, 18074. [Google Scholar] [CrossRef]
- Aguilera-Mendoza, L.; Ayala-Ruano, S.; Martinez-Rios, F.; Chavez, E.; García-Jacas, C.R.; Brizuela, C.A.; Marrero-Ponce, Y. StarPep Toolbox: An open-source software to assist chemical space analysis of bioactive peptides and their functions using complex networks. Bioinformatics 2023, 39, btad506. [Google Scholar] [CrossRef]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Boldi, P.; Vigna, S. Axioms for centrality. Internet Math. 2014, 10, 222–262. [Google Scholar] [CrossRef]
- Ghalmane, Z.; Hassouni, M.E.; Cherifi, H. Immunization of networks with non-overlapping community structure. Soc. Netw. Anal. Min. 2019, 9, 45. [Google Scholar] [CrossRef]
- Xu, J.; Li, F.; Li, C.; Guo, X.; Landersdorfer, C.; Shen, H.-H.; Peleg, A.Y.; Li, J.; Imoto, S.; Yao, J. iAMPCN: A deep-learning approach for identifying antimicrobial peptides and their functional activities. Brief. Bioinform. 2023, 24, bbad240. [Google Scholar] [CrossRef]
- Pinacho-Castellanos, S.A.; García-Jacas, C.R.; Gilson, M.K.; Brizuela, C.A. Alignment-free antimicrobial peptide predictors: Improving performance by a thorough analysis of the largest available data set. J. Chem. Inf. Model. 2021, 61, 3141–3157. [Google Scholar] [CrossRef] [PubMed]
- Bajiya, N.; Choudhury, S.; Dhall, A.; Raghava, G.P. AntiBP3: A Method for Predicting Antibacterial Peptides against Gram-Positive/Negative/Variable Bacteria. Antibiotics 2024, 13, 168. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-T.; Sun, Y.-Y.; Wang, C.-T.; Cheng, W.-C.; Lu, I.-H.; Lin, C.-Y.; Chen, S.-H. AI4AVP: An antiviral peptides predictor in deep learning approach with generative adversarial network data augmentation. Bioinform. Adv. 2022, 2, vbac080. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Moriwaki, Y.; Li, C.; Shimizu, K. Prediction of antifungal peptides by deep learning with character embedding. IPSJ Trans. Bioinform. 2019, 12, 21–29. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhagat, D.; Mahalwal, M.; Sharma, N.; Raghava, G.P. AntiCP 2.0: An updated model for predicting anticancer peptides. Brief. Bioinform. 2021, 22, bbaa153. [Google Scholar] [CrossRef]
- Chen, J.; Cheong, H.H.; Siu, S.W. xDeep-AcPEP: Deep learning method for anticancer peptide activity prediction based on convolutional neural network and multitask learning. J. Chem. Inf. Model. 2021, 61, 3789–3803. [Google Scholar] [CrossRef]
- Vishnepolsky, B.; Grigolava, M.; Managadze, G.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M.; Pirtskhalava, M. Comparative analysis of machine learning algorithms on the microbial strain-specific AMP prediction. Brief. Bioinform. 2022, 23, bbac233. [Google Scholar] [CrossRef]
- Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M.; Managadze, G.; Grigolava, M.; Makhatadze, G.I.; Pirtskhalava, M. Predictive model of linear antimicrobial peptides active against gram-negative bacteria. J. Chem. Inf. Model. 2018, 58, 1141–1151. [Google Scholar] [CrossRef]
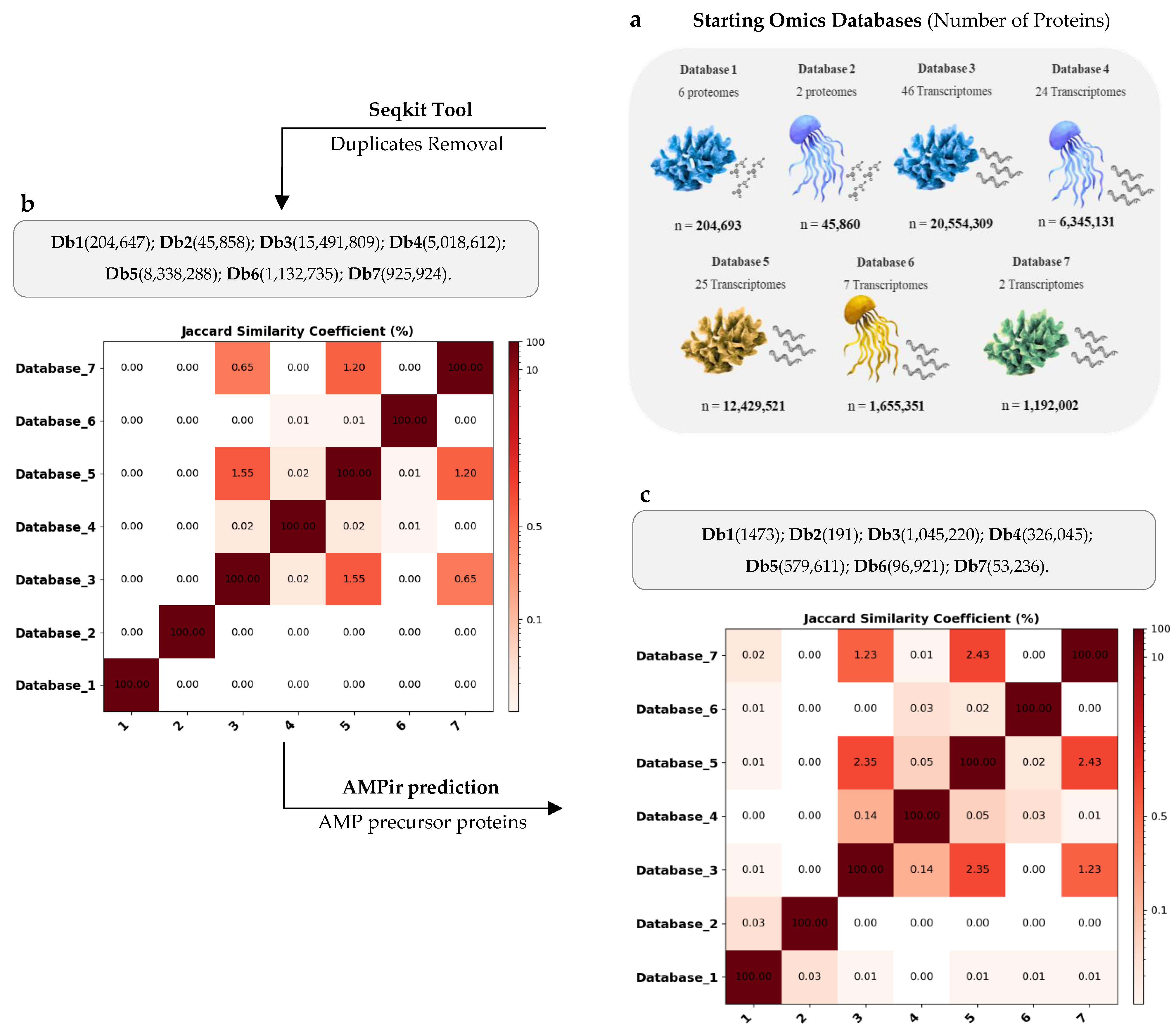
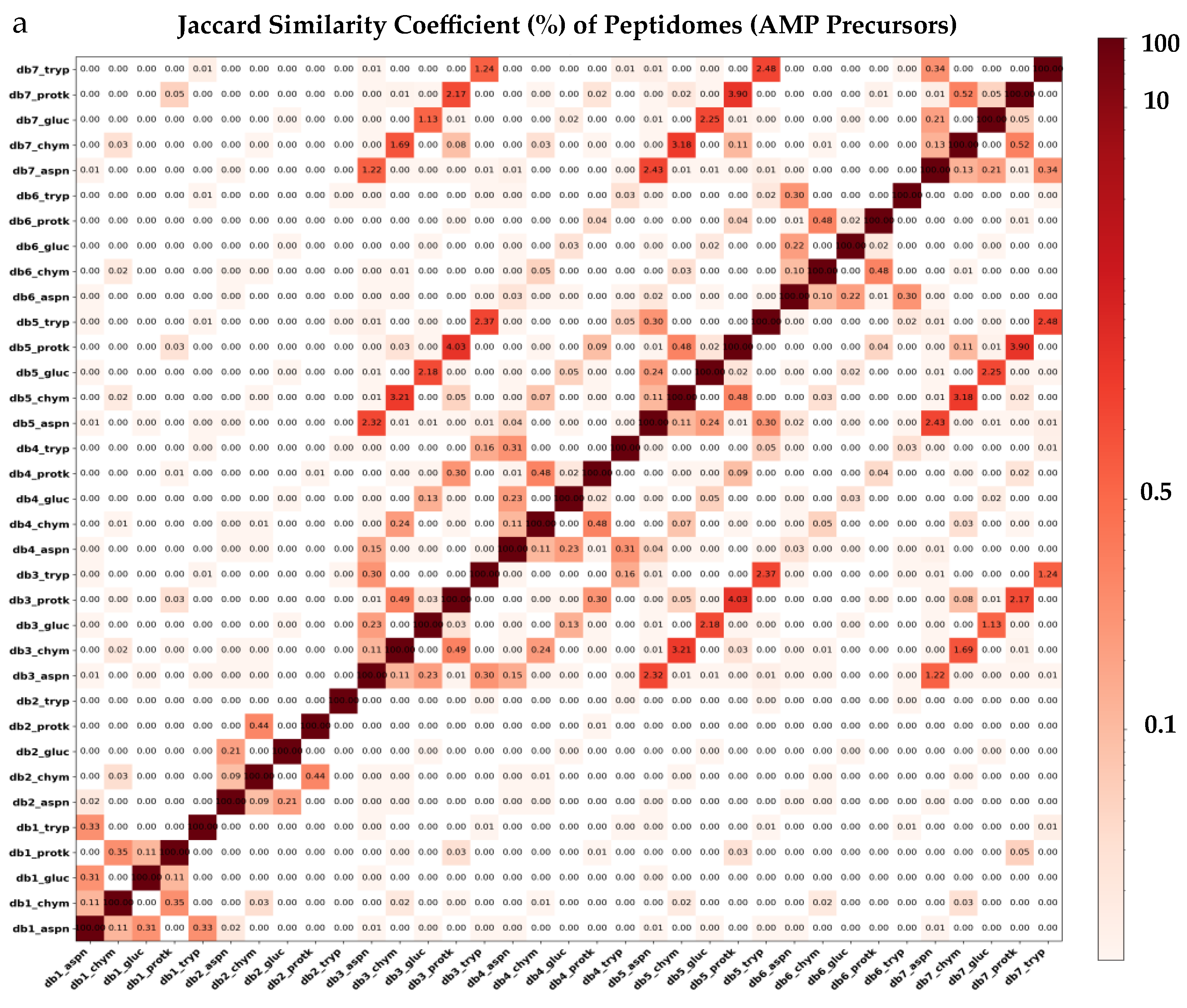
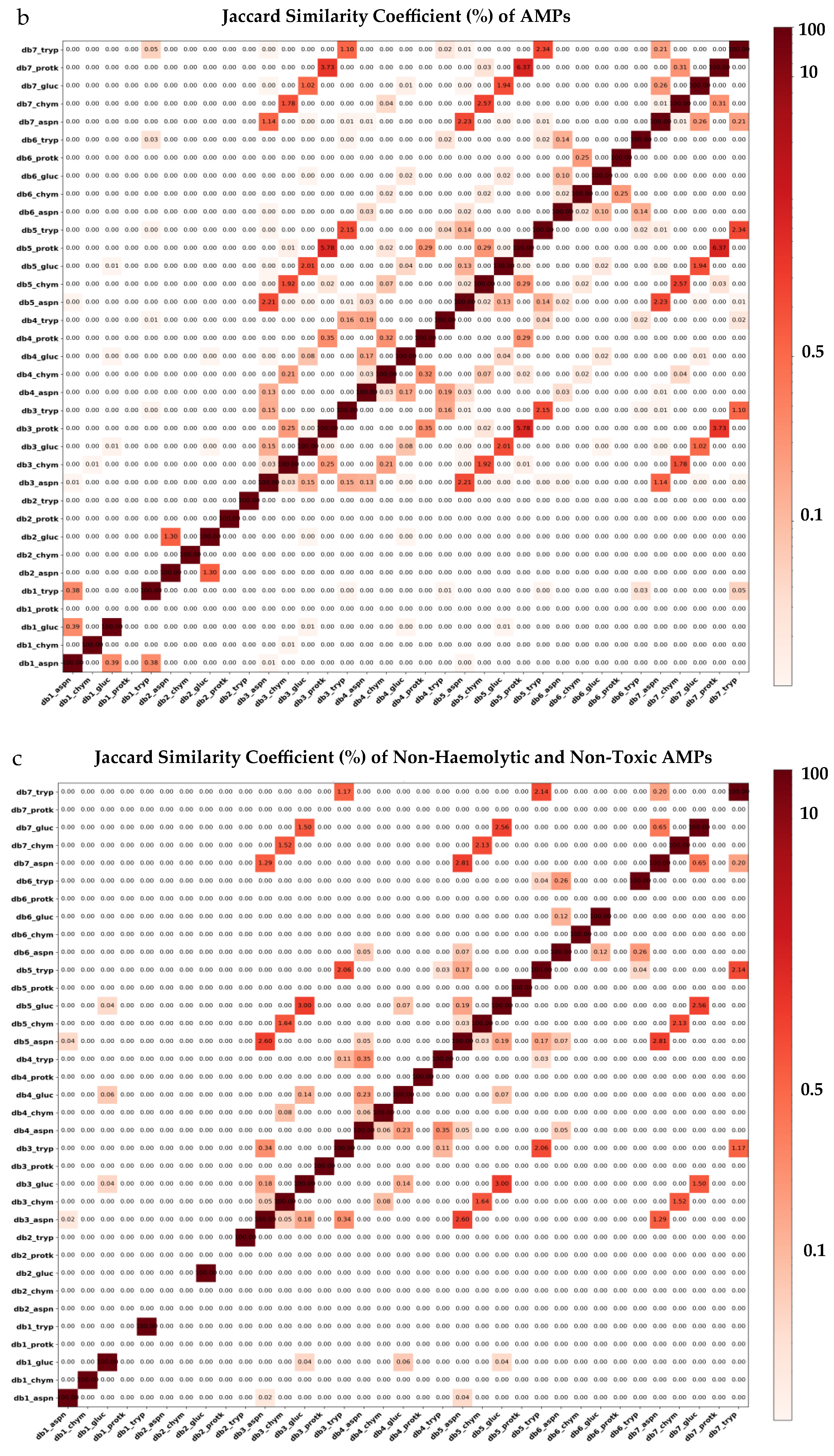

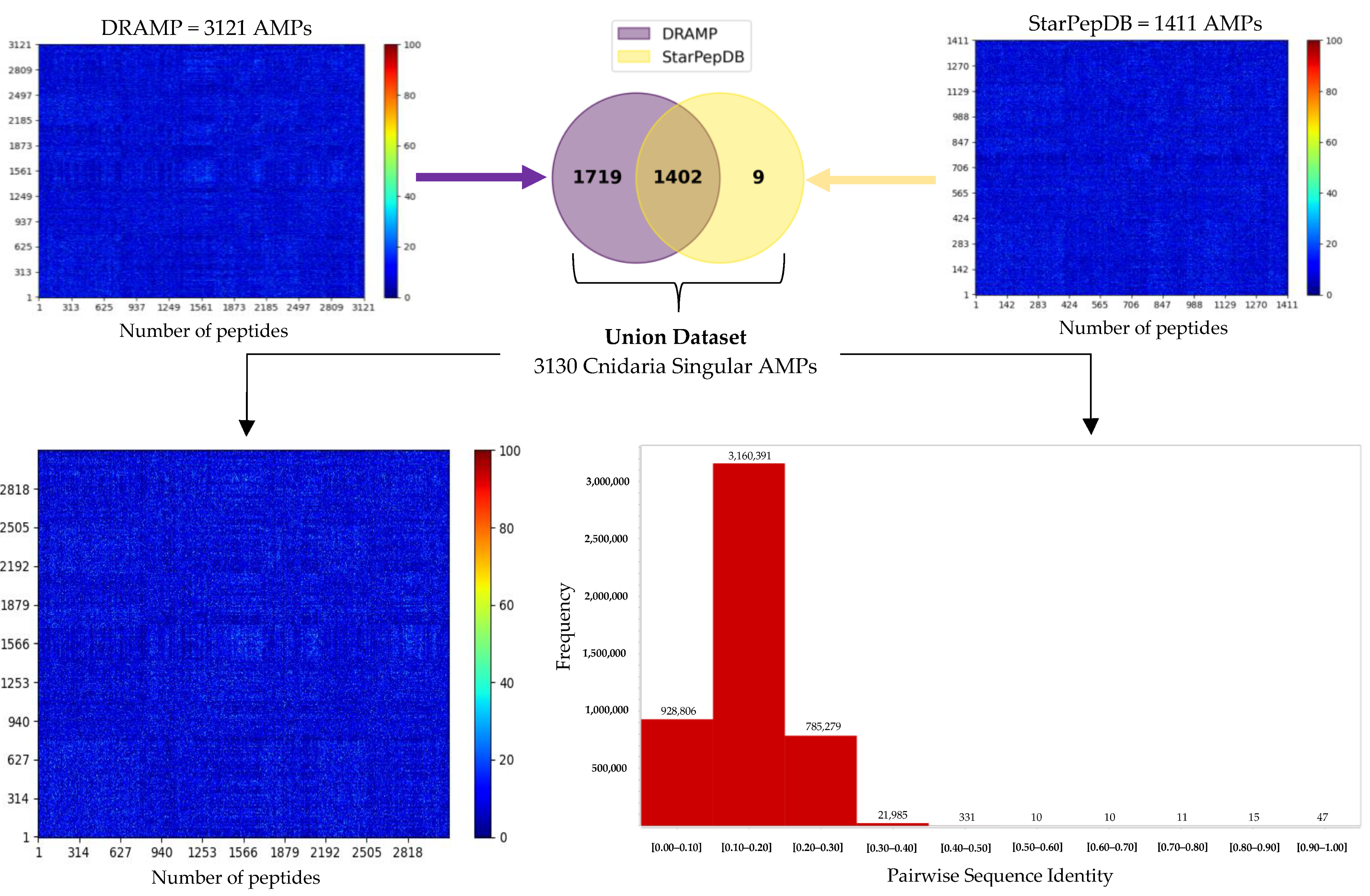

| Proteolysis Protocol | Total Peptides | 6–40 AA Length | Non-Duplicated Peptides | Non-Redundant Peptides | 20 AA Alphabet |
|---|---|---|---|---|---|
| db1_aspn | 15,851 | 8432 | 7980 | 4376 | 4376 |
| db1_chym | 32,418 | 9242 | 8565 | 2696 | 2696 |
| db1_gluc | 15,240 | 8172 | 7649 | 4447 | 4447 |
| db1_protk | 68,118 | 3285 | 2999 | 175 | 175 |
| db1_tryp | 19,198 | 9170 | 8442 | 4318 | 4318 |
| db2_aspn | 2748 | 1422 | 1311 | 698 | 698 |
| db2_chym | 5075 | 1479 | 1338 | 434 | 434 |
| db2_gluc | 2210 | 1261 | 1158 | 744 | 744 |
| db2_protk | 10,692 | 555 | 494 | 25 | 25 |
| db2_tryp | 3122 | 1376 | 1257 | 670 | 670 |
| db3_aspn | 73,527,51 | 4,175,173 | 3,694,840 | 2,115,242 | 1,820,209 |
| db3_chym | 18,999,471 | 3,996,393 | 3,311,893 | 858,127 | 818,622 |
| db3_gluc | 5,438,132 | 3,348,195 | 3,021,505 | 1,947,320 | 1,609,672 |
| db3_protk | 33,464,859 | 1,417,423 | 1,040,671 | 59,408 | 58,164 |
| db3_tryp | 8,467,153 | 4,350,869 | 3,846,043 | 2,082,476 | 1,849,069 |
| db4_aspn | 2,291,447 | 1,292,563 | 1,205,239 | 682,256 | 586,367 |
| db4_chym | 6,080,475 | 1,207,735 | 1,090,192 | 262,041 | 250,336 |
| db4_gluc | 1,702,547 | 1,044,291 | 982,746 | 629,228 | 519,668 |
| db4_protk | 10,512,274 | 417,458 | 357,444 | 17,402 | 17,080 |
| db4_tryp | 2,630,768 | 1,343,898 | 1,252,674 | 673,179 | 595,910 |
| db5_aspn | 4,031,801 | 2,293,354 | 2,038,733 | 1,169,051 | 996,612 |
| db5_chym | 10,705,538 | 2,166,093 | 1,826,084 | 457,572 | 436,127 |
| db5_gluc | 2,983,890 | 1,843,785 | 1,666,856 | 1,080,419 | 883,537 |
| db5_protk | 18,562,675 | 763,345 | 591,295 | 31,353 | 30,722 |
| db5_tryp | 4,665,535 | 2,393,038 | 2,126,340 | 1,151,478 | 1,014,421 |
| db6_aspn | 670,520 | 381,979 | 370,092 | 208,598 | 177,282 |
| db6_chym | 1,831,070 | 348,971 | 333,615 | 72,869 | 69,353 |
| db6_gluc | 482,489 | 300,680 | 292,545 | 190,074 | 153,954 |
| db6_protk | 3,089,109 | 119,570 | 112,026 | 4804 | 4704 |
| db6_tryp | 768,090 | 397,489 | 384,873 | 204,191 | 179,035 |
| db7_aspn | 380,990 | 215,028 | 200,493 | 113,076 | 97,943 |
| db7_chym | 932,823 | 210,478 | 193,912 | 52,098 | 49,536 |
| db7_gluc | 279,267 | 171,878 | 161,719 | 104,066 | 86,699 |
| db7_protk | 1,682,862 | 78,598 | 70,910 | 3998 | 3909 |
| db7_tryp | 424,506 | 222,032 | 207,692 | 112,808 | 100,524 |
| Total Peptides | 12,428,038 | ||||
| Total nr Peptides | 8,278,560 |
| Proteolysis Protocol | Peptidomes | (1) AMP | (2) Non-Haemolytic AMP | (3) Non-Haemolytic and Non-Toxic AMP |
|---|---|---|---|---|
| db1_aspn | 4376 | 214 | 36 | 9 |
| db1_chym | 2696 | 35 | 9 | 3 |
| db1_gluc | 4447 | 298 | 82 | 20 |
| db1_protk | 175 | 0 | 0 | 0 |
| db1_tryp | 4318 | 49 | 8 | 4 |
| Peptides db1 | 16,012 | 596 | 135 | 36 |
| nr Peptides db1 | 10,955 | 537 | 133 | 36 |
| db2_aspn | 698 | 31 | 4 | 0 |
| db2_chym | 434 | 7 | 2 | 0 |
| db2_gluc | 744 | 47 | 15 | 1 |
| db2_protk | 25 | 1 | 0 | 0 |
| db2_tryp | 670 | 11 | 3 | 1 |
| Peptides db2 | 2571 | 97 | 24 | 2 |
| nr Peptides db2 | 1788 | 90 | 24 | 2 |
| db3_aspn | 1,820,209 | 134,007 | 21,138 | 5055 |
| db3_chym | 818,622 | 14,534 | 3665 | 946 |
| db3_gluc | 1,609,672 | 78,250 | 26,331 | 4974 |
| db3_protk | 58,164 | 456 | 96 | 9 |
| db3_tryp | 1,849,069 | 38,029 | 12,366 | 4023 |
| Peptides db3 | 6,155,736 | 265,276 | 63,596 | 15,007 |
| nr Peptides db3 | 4,229,977 | 244,894 | 62,423 | 14,816 |
| db4_aspn | 586,367 | 42,756 | 6531 | 1486 |
| db4_chym | 250,336 | 4260 | 1033 | 269 |
| db4_gluc | 519,668 | 24,700 | 8248 | 1586 |
| db4_protk | 17,080 | 111 | 22 | 3 |
| db4_tryp | 595,910 | 12,479 | 4080 | 1356 |
| Peptides db4 | 1,969,361 | 84,306 | 19,950 | 4700 |
| nr Peptides db4 | 1,357,350 | 77,857 | 19,583 | 4638 |
| db5_aspn | 996,612 | 72,434 | 11,325 | 2633 |
| db5_chym | 436,127 | 3929 | 934 | 234 |
| db5_gluc | 883,537 | 40,721 | 13,585 | 2640 |
| db5_protk | 30,722 | 239 | 56 | 11 |
| db5_tryp | 1,014,421 | 20,694 | 6565 | 2134 |
| Peptides db5 | 3,361,419 | 138,017 | 32,465 | 7652 |
| nr Peptides db5 | 2,316,742 | 127,952 | 31,909 | 7580 |
| db6_aspn | 177,282 | 12,861 | 1907 | 389 |
| db6_chym | 69,353 | 1162 | 292 | 70 |
| db6_gluc | 153,954 | 6591 | 2300 | 416 |
| db6_protk | 4704 | 21 | 2 | 0 |
| db6_tryp | 179,035 | 3800 | 1204 | 375 |
| Peptides db6 | 584,328 | 24,435 | 5705 | 1250 |
| nr Peptides db6 | 404,314 | 22,683 | 5585 | 1237 |
| db7_aspn | 97,943 | 6914 | 1138 | 299 |
| db7_chym | 49,536 | 937 | 254 | 54 |
| db7_gluc | 86,699 | 4444 | 1486 | 320 |
| db7_protk | 3909 | 45 | 7 | 0 |
| db7_tryp | 100,524 | 2029 | 643 | 208 |
| Peptides db7 | 338,611 | 14,369 | 3528 | 881 |
| nr Peptides db7 | 230,192 | 13,177 | 3456 | 867 |
| Total Peptides | 12,428,038 | 527,096 | 125,403 | 29,528 |
| Total nr Peptides | 8,278,560 | 473,747 | 119,531 | 28,279 |
| Species | Subphylum | Class (Order) | Candidate ABP Sequence | Rank | Predicted Antimicrobial Activity |
|---|---|---|---|---|---|
| Ctenactis echinata | Anth. | Hexacorallia (Scleractinia) | CGVWQYRQGNSLYVQVISRPKKSGFRFR | I | B. subtilis |
| Galaxea fascicularis | Anth. | Hexacorallia (Scleractinia) | DLFFRFVNYLGNQYNQLGWWKKVRSSGSRG | I | B. subtilis |
| Favites colemani | Anth. | Hexacorallia (Scleractinia) | DRFGKEEKQWPFVPWQWPVRRNVLLRRQR | I | B. subtilis |
| Catalaphyllia jardinei * | Anth. | Hexacorallia (Scleractinia) | GAWSGAKRYGTGQRHISSNSSLFRKWGND | I | B. subtilis |
| Fimbriaphyllia ancora | Anth. | Hexacorallia (Scleractinia) | VFPRFRSIFSPGVTRGLRAVSSLSKD | I | B. subtilis, E. coli, P. aeruginosa |
| Alveopora japonica | Anth. | Hexacorallia (Scleractinia) | CRKQVYKPPLQFSGLSSSSFLSYLVKRFNTQQRGSFWR | II | B. subtilis, K. pneumoniae |
| Heliopora coerulea | Anth. | Octocorallia (Scleralcyonacea) | PMKAWITGIAANRGTKGGSAKCAVGLFKSRVKD | II | E. coli |
| Protopalythoa variabilis | Anth. | Hexacorallia (Zoantharia) | QPRLIFFGSTSSFRAPHGQQKQVHKFAAKVQCCK | II | E. coli |
| Acropora millepora | Anth. | Hexacorallia (Scleractinia) | RGQWQINKRTGSKSCARLKTTGAPHMASGWQVWK | II | B. subtilis, E. coli |
| Goniopora lobata * | Anth. | Hexacorallia (Scleractinia) | RGRKLCLPWTFWLGSRTVIQGRCTQPASASGSKGPQRRF | II | B. subtilis, E. coli, K. pneumoniae, P. aeruginosa, S. aureus |
| Chironex fleckeri * | Med. | Cubozoa (Chirodropida) | RWRNVNGWGKSKKKNANGSHIGLWLTGGGG | II | B. subtilis, E. coli, K. pneumoniae |
| Fimbriaphyllia ancora | Anth. | Hexacorallia (Scleractinia) | TLNIPVAGGTKSTAGMWRRCWNGAVPSRTPSKRFG | II | B. subtilis, E. coli, P. aeruginosa |
| Alveopora japonica | Anth. | Hexacorallia (Scleractinia) | YYWNPRLRPGLQVSCSHGSCKTSLAFGRLLKSKD | II | B. subtilis |
| Ricordea yuma | Anth. | Hexacorallia (Corallimorpharia) | CRSNRTQQWGLGSYIRILGRASVVTLKQPL | III | B. subtilis, E. coli, K. pneumoniae, P. aeruginosa |
| Montipora digitata | Anth. | Hexacorallia (Scleractinia) | CSMRPISSSWLRFSKKIWSTSAR | III | B. subtilis, E. coli, K. pneumoniae, P. aeruginosa |
| Phyllodiscus semoni * | Anth. | Hexacorallia (Actiniaria) | CWTWVATPTFAHGMVQVWRASQRVRSRLTN | III | B. subtilis, E. coli, P. aeruginosa, S. aureus |
| Polymyces wellsi | Anth. | Hexacorallia (Scleractinia) | NISFNSSASGRSLFGHFGRFRTLSWLRGWGG | III | B. subtilis, E. coli, K. pneumoniae, P. aeruginosa, S. aureus |
| Goniopora norfolkensis | Anth. | Hexacorallia (Scleractinia) | RPAISGAVTISGKFQKAWGSVHKPLNRCRSSLWGGG | III | B. subtilis, E. coli, K. pneumoniae, P. aeruginosa, S. aureus |
| Goniopora norfolkensis | Anth. | Hexacorallia (Scleractinia) | SGLRKSRMMKWPLSTGGRWSRGGLVA | III | E. coli, K. pneumoniae, P. aeruginosa, S. aureus |
| Galaxea fascicularis | Anth. | Hexacorallia (Scleractinia) | YPKPSLANWTRSSGTSIKGKLWLTGRHPHLRAGSG | III | E. coli, K. pneumoniae, P. aeruginosa, S. aureus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barroso, R.A.; Agüero-Chapin, G.; Sousa, R.; Marrero-Ponce, Y.; Antunes, A. Unlocking Antimicrobial Peptides: In Silico Proteolysis and Artificial Intelligence-Driven Discovery from Cnidarian Omics. Molecules 2025, 30, 550. https://doi.org/10.3390/molecules30030550
Barroso RA, Agüero-Chapin G, Sousa R, Marrero-Ponce Y, Antunes A. Unlocking Antimicrobial Peptides: In Silico Proteolysis and Artificial Intelligence-Driven Discovery from Cnidarian Omics. Molecules. 2025; 30(3):550. https://doi.org/10.3390/molecules30030550
Chicago/Turabian StyleBarroso, Ricardo Alexandre, Guillermin Agüero-Chapin, Rita Sousa, Yovani Marrero-Ponce, and Agostinho Antunes. 2025. "Unlocking Antimicrobial Peptides: In Silico Proteolysis and Artificial Intelligence-Driven Discovery from Cnidarian Omics" Molecules 30, no. 3: 550. https://doi.org/10.3390/molecules30030550
APA StyleBarroso, R. A., Agüero-Chapin, G., Sousa, R., Marrero-Ponce, Y., & Antunes, A. (2025). Unlocking Antimicrobial Peptides: In Silico Proteolysis and Artificial Intelligence-Driven Discovery from Cnidarian Omics. Molecules, 30(3), 550. https://doi.org/10.3390/molecules30030550













