Volatile Compositional Profile, Antioxidant Properties, and Molecular Docking of Ethanolic Extracts from Philodendron heleniae
Abstract
1. Introduction
2. Results
2.1. Phytochemical Screening
2.2. Total Phenolics and Flavonoids Content
2.3. Antioxidant Assay
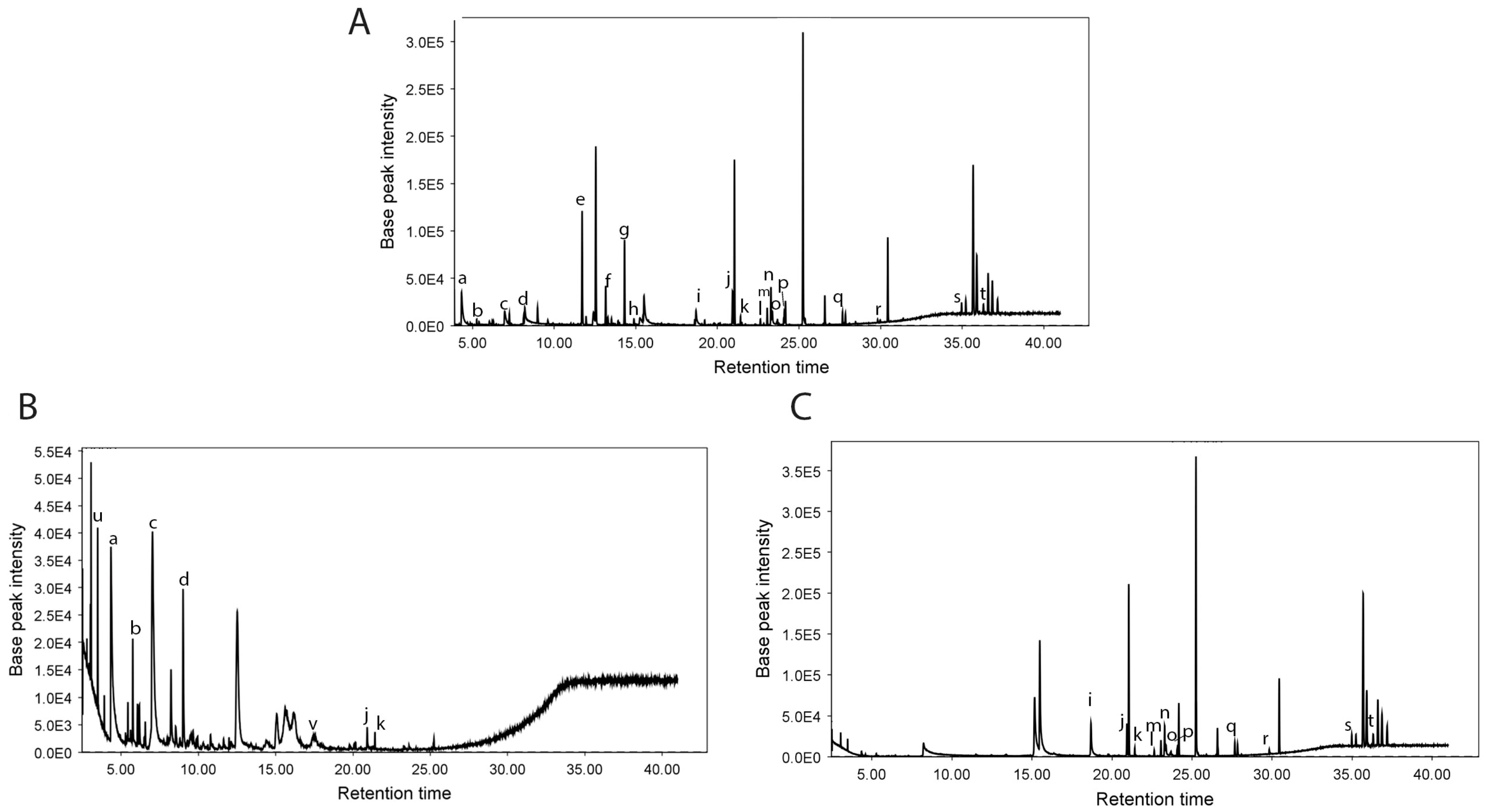
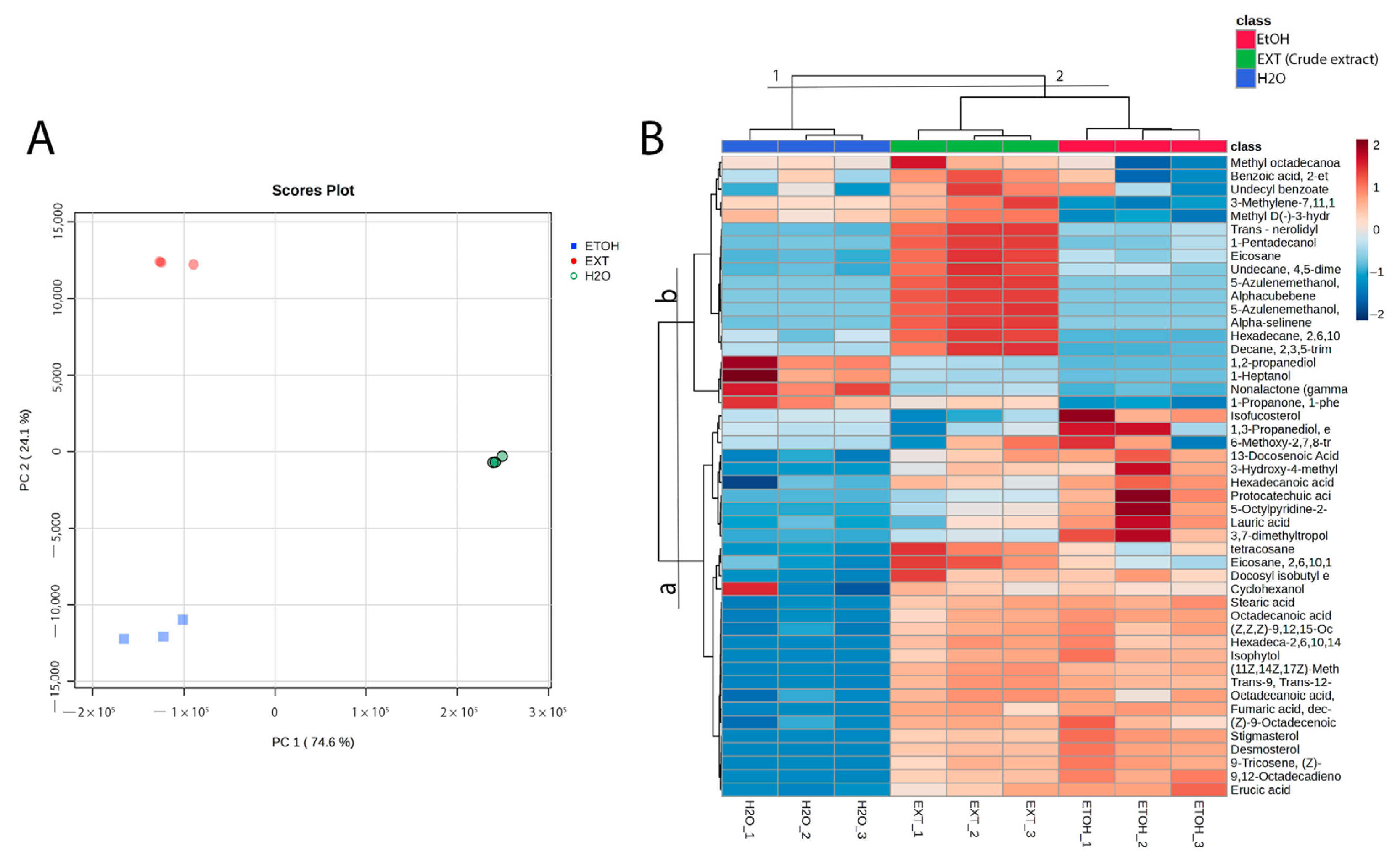
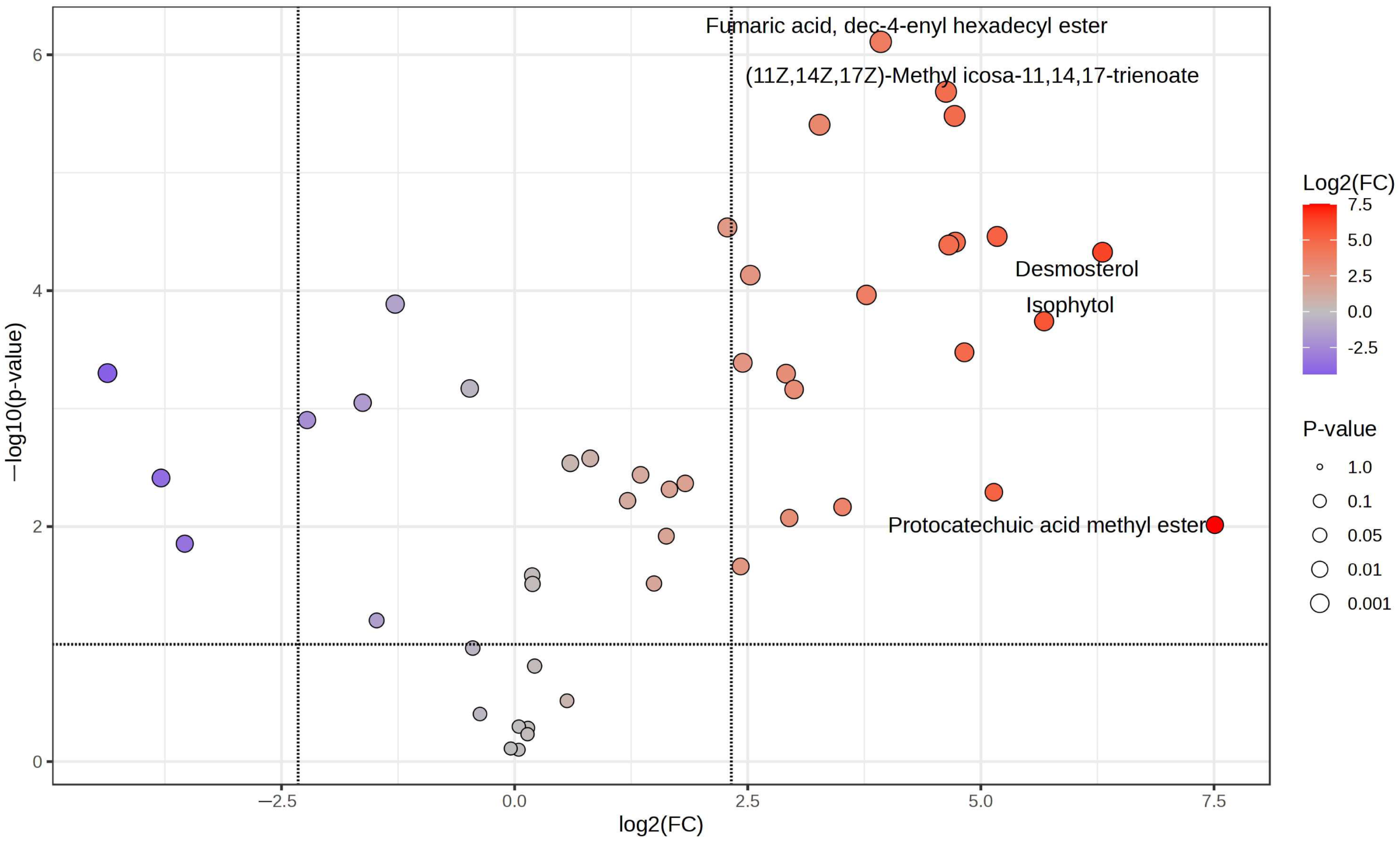
2.4. GC-MS Analysis
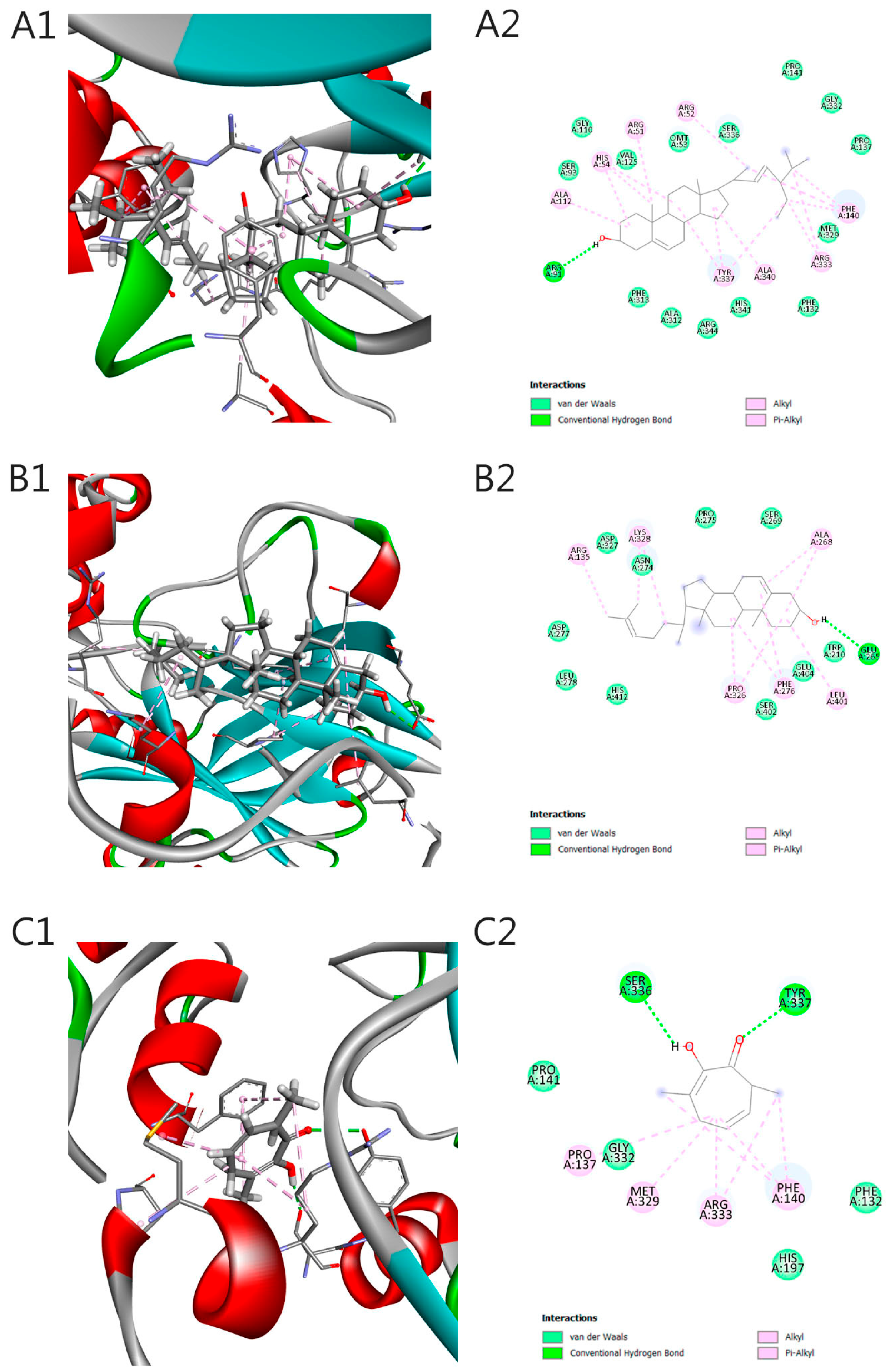
2.5. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Preparation of P. heleniae Crude Extracts
4.3. Phytochemical Analysis
4.4. Total Phenol Content
4.5. Total Flavonoid Content
4.6. Antioxidant Activity Evaluation
4.6.1. ABTS Radical Scavenging Assay
4.6.2. DPPH Radical Scavenging Assay
4.7. GC-MS Characterization
4.7.1. Sample Preparation
4.7.2. Metabolomic Data Acquisition and Metabolite Profiling
4.8. Computational Molecular Docking Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Halliwell, B. Antioxidants in human health and disease. Annu. Rev. Nutr. 1996, 16, 33–50. [Google Scholar] [CrossRef]
- Flieger, J.; Flieger, W.; Baj, J.; Maciejewski, R. Antioxidants: Classification, Natural Sources, Activity/Capacity Measurements, and Usefulness for the Synthesis of Nanoparticles. Materials 2021, 14, 4135. [Google Scholar] [CrossRef]
- Zehiroglu, C.; Sarikaya, O. The importance of antioxidants and place in today’s scientific and technological studies. J. Food Sci. Technol. 2019, 56, 4757–4774. [Google Scholar] [CrossRef]
- Alsulami, F.; Shaheed, S. Role of Natural Antioxidants in Cancer. Cancer Treat. Res. 2024, 191, 95–117. [Google Scholar] [CrossRef]
- Vengatesan, M.; Rambabu, K.; Banat, F.; Mittal, V. Natural antioxidants-based edible active food packaging: An overview of current advancements. Food Biosci. 2021, 43, 101251. [Google Scholar] [CrossRef]
- Deshmukh, R.; Gaikwad, K. Natural antimicrobial and antioxidant compounds for active food packaging applications. Biomass Convers. Biorefin. 2024, 14, 4419–4440. [Google Scholar] [CrossRef]
- Kuai, L.; Liu, F.; Chiou, B.-S.; Avena-Bustillos, R.; McHugh, T.; Zhong, F. Controlled release of antioxidants from active food packaging: A review. Food Hydrocoll. 2021, 120, 106992. [Google Scholar] [CrossRef]
- David, E.; Kopac, J. Survey on antioxidants used as additives to improve biodiesel’s stability to degradation through oxidation. Molecules 2023, 28, 7765. [Google Scholar] [CrossRef]
- Papas, A. Diet and Antioxidant Status. Food Chem. Toxicol. 1999, 37, 999–1007. [Google Scholar] [CrossRef]
- Uzombah, T. The Implications of replacing synthetic antioxidants with natural ones in the food systems. In Natural food Additives; IntechOpen: London, UK, 2022; pp. 1–15. [Google Scholar] [CrossRef]
- Akbarirad, H.; Gohari, A.; Kazemeini, S.; Mousavi, K. An overview on some of important sources of natural antioxidants. Int. Food Res. J. 2016, 23, 928–933. Available online: http://ifrj.upm.edu.my/23%20(03)%202016/(3).pdf (accessed on 22 January 2025).
- Chandran, R.; Abrahamse, H. Identifying plant-based natural medicine against oxidative stress and neurodegenerative disorders. Oxid. Med. Cell Longev. 2020, 2020, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pimm, S.L.; Jenkins, C.N.; Abell, R.; Brooks, T.M.; Gittleman, J.L.; Joppa, L.N.; Raven, P.H.; Roberts, C.M.; Sexton, J.O. The biodiversity of species and their rates of extinction, distribution, and protection background rates of species extinctions. Science 2014, 344, 1246752. [Google Scholar] [CrossRef]
- Sarkar, B.; Rana, N.; Singh, C.; Singh, A. Medicinal herbal remedies in neurodegenerative diseases: An update on antioxidant potential. Naunyn Schmiedebergs Arch. Pharmacol. 2024, 397, 5483–5511. [Google Scholar] [CrossRef]
- Noriega, P.; Guerrini, A.; Sacchetti, G.; Grandini, A.; Ankuash, E.; Manfredini, S. Chemical composition and biological activity of five essential oils from the Ecuadorian Amazon rain forest. Molecules 2019, 24, 1637. [Google Scholar] [CrossRef] [PubMed]
- Mestanza-Ramón, C.; Henkanaththegedara, S.M.; Vásconez Duchicela, P.; Vargas Tierras, Y.; Sánchez Capa, M.; Constante Mejía, D.; Jimenez Gutierrez, M.; Charco Guamán, M.; Mestanza Ramón, P. In-situ and ex-situ biodiversity conservation in ecuador: A review of policies, actions and challenges. Diversity 2020, 12, 315. [Google Scholar] [CrossRef]
- López, V. Consecuencias de la disminución de la flora endémica del Mundo, Ecuador y la Amazonia. ConcienciaDigital 2018, 1, 53–63. [Google Scholar] [CrossRef]
- Giovannini, P.; Howes, M.J.R. Medicinal plants used to treat snakebite in Central America: Review and assessment of scientific evidence. J. Ethnopharmacol. 2017, 199, 240–256. [Google Scholar] [CrossRef]
- Croat, T.; Grayum, M. A new species of Philodendron subgenus Philodendron (Araceae) from Costa Rica. Aroideana 2022, 45, 34–46. [Google Scholar]
- Bogner, J.; Bunting, G. A new Philodendron Species (Araceae) from Ecuador. Willdenowia 1983, 13, 183–185. [Google Scholar]
- Ballesteros, J.; Bracco, F.; Cerna, M.; Finzi, P.V.; Vidari, G. Ethnobotanical research at the Kutukú scientific station, Morona-Santiago, Ecuador. Biomed. Res. Int. 2016, 2016, 9105746. [Google Scholar] [CrossRef]
- Instituto Nacional de Biodiversidad. Banco Nacional de Datos de Biodiversidad de Ecuador. Available online: https://bndb.sisbioecuador.bio/bndb/collections/list.php?usethes=1&taxa=13565 (accessed on 22 January 2025).
- Caballero-Serrano, V.; McLaren, B.; Carrasco, J.C.; Alday, J.G.; Fiallos, L.; Amigo, J.; Onaindia, M. Traditional ecological knowledge and medicinal plant diversity in Ecuadorian amazon home gardens. Glob. Ecol. Conserv. 2019, 17, e00524. [Google Scholar] [CrossRef]
- Da Costa Guimarães, N.; Freitas-de-Sousa, L.A.; de Souza, M.C.; de Almeida, P.D.; Dos-Santos, M.C.; Nunez, C.V.; de Oliveira, R.B.; Mourão, R.H.; de Moura, V.M. Evaluation of the anti-snakebite, antimicrobial and antioxidant potential of Philodendron megalophyllum Schott (Araceae), traditionally used in accidents caused by snakes in the western region of Pará, Brazil. Toxicon 2020, 184, 99–108. [Google Scholar] [CrossRef]
- Soares, A.M.; Ticli, F.K.; Marcussi, S.; Lourenco, M.V.; Januario, A.H.; Sampaio, S.V.; Giglio, J.R.; Lomonte, B.; Pereira, P.S. Medicinal Plants with inhibitory properties against snake venoms. Curr. Med. Chem. 2005, 12, 2625–2641. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, J.; Jiménez, S.L.; Gómez, I.C.; Rey, J.P.; Henao, A.M.; Marín, D.M.; Romero, J.O.; Alarcón, J.C. Snakebites and ethnobotany in the Eastern region of Antioquia, Colombia—The traditional use of plants. J. Ethnopharmacol. 2013, 146, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Joffard, N.; Legendre, L.; Gibernau, M.; Pascal, L. Differential Accumulation of Volatile Organic compounds by leaves and roots of two guianese Philodendron Species, P. fragrantissimum Kunth and P. melinonii Brongn. Chem. Biodivers. 2017, 14, e1600415. [Google Scholar] [CrossRef]
- Castellar, A.; Oliveira, D.R.; Leitão, S.G.; Bizzo, H.R.; Soares, M.D.; Kinupp, V.F.; Veiga-Junior, V.F. Essential oil from Philodendron fragrantissimum, an aromatic Araceae from Amazonia, Brazil. J. Essent. Oil Res. 2013, 25, 194–197. [Google Scholar] [CrossRef]
- Scapinello, J.; Müller, L.G.; Schindler, M.S.; Anzollin, G.S.; Siebel, A.M.; Boligon, A.A.; Niero, R.; Saraiva, T.E.; Maus, N.P.; Betti, A.H.; et al. Antinociceptive and anti-inflammatory activities of Philodendron bipinnatifidum Schott ex Endl (Araceae). J. Ethnopharmacol. 2019, 236, 21–30. [Google Scholar] [CrossRef]
- Summer, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Ebrahim, H.; Deyab, N.; Shaheen, B.; Gabr, A.; Allam, N. In vitro: The extraordinary enhancement in rutin accumulation and antioxidant activity in Philodendron ‘Imperial Red’ plantlets using Ti-Mo-Ni-O nanotubes as a novel elicitor. BioTech 2024, 13, 24. [Google Scholar] [CrossRef]
- Sun, W.; Shahrajabian, M. Therapeutic potential of phenolic compounds in medicinal plants—Natural health products for human health. Molecules 2023, 28, 1845. [Google Scholar] [CrossRef]
- Mutha, R.; Tatiya, A.; Surana, S. Flavonoids as natural phenolic compounds and their role in therapeutics: An overview. Future J. Pharm. Sci. 2021, 7, 1–3. [Google Scholar] [CrossRef]
- Alotaibi, B.; Ijaz, M.; Buabeid, M.; Kharaba, Z.; Yaseen, H.; Murtaza, G. Therapeutic effects and safe uses of plant-derived polyphenolic compounds in cardiovascular diseases: A review. Drug Des. Devel Ther. 2021, 15, 4713–4732. [Google Scholar] [CrossRef]
- Rajasekaran, S.; Rajasekar, N.; Sivanantham, A. Therapeutic potential of plant-derived tannins in non-malignant respiratory diseases. J. Nutr. Biochem. 2021, 94, 108632. [Google Scholar] [CrossRef] [PubMed]
- De Moura Santos Mota, N.; Brandão, M.M.; de Andrade Royo, V.; de Melo Sacramento, V.; Simões, M.O.; Ferreira, L.C.; Menezes, E.V.; de Oliveira, D.A.; de Melo Junior, A.F. Bioactive activities and phytochemical prospection of Philodendron adamantinum Schott (Araceae): An endemic species from the Espinhaço Range. Rev. Unimontes Científica 2024, 26, 2. [Google Scholar] [CrossRef]
- Ingle, K.; Deshmukh, A.; Padole, D.; Dudhare, M.; Moharil, M.; Khelurkar, V. Phytochemicals: Extraction methods, identification and detection of bioactive compounds from plant extracts. J. Pharmacogn. Phytochem. 2017, 6, 32–36. [Google Scholar]
- Arnao, M.; Cano, A.; Acosta, M. Methods to measure the antioxidant activity in plant material. A comparative discussion. Free Radic. Res. 1999, 31 (Suppl. S1), 89–96. [Google Scholar] [CrossRef]
- Sasidharan, S.; Chen, Y.; Saravanan, D.; Sundram, K.M.; Latha, L.Y. Extraction, isolation and characterization of bioactive compounds from plant´s extrcats. Afr. J. Tradit. Complement. Altern. Med. 2011, 8, 1–10. [Google Scholar]
- Abubakar, A.; Haque, M. Preparation of medicinal plants: Basic extraction and fractionation procedures for experimental purposes. J. Pharm. Bioallied Sci. 2020, 12, 1–10. [Google Scholar] [CrossRef]
- Arzola-Rodríguez, S.; Muñoz-Castellanos, L.; López-Camarillo, C.; Salas, E. Phenolipids, Amphipilic Phenolic Antioxidants with Modified Properties and Their Spectrum of Applications in Development: A Review. Biomolecules 2022, 12, 1897. [Google Scholar] [CrossRef]
- Rajauria, G. In-vitro antioxidant properties of lipophilic antioxidant compounds from 3 brown seaweed. Antioxidants 2019, 8, 596. [Google Scholar] [CrossRef]
- Alliance, D.; Santos, C.B.; Barbosa, L.M.; Gomes, J.S.; Lobato, C.C.; Viana, J.C.; Ramos, R.D.; Ferreira, R.M.; Souto, R.N. Chemical study, predictions in silico and larvicide activity of the essential oil of root Philodendron deflexum Poe. J. Comput. Theor. Nanosci. 2017, 14, 3330–3337. [Google Scholar] [CrossRef]
- De Andrade Santiago, J.; das Graças Cardoso, M.; da Silva Figueiredo, A.C.; de Moraes, J.C.; de Assis, F.A.; Teixeira, M.L.; Santiago, W.D.; Sales, T.A.; Camargo, K.C.; Nelson, D.L. Chemical characterization and application of the essential oils from Chenopodium ambrosioides and Philodendron bipinnatifidum in the control of Diabrotica speciosa (Coleoptera: Chrysomelidae). Am. J. Plant Sci. 2014, 5, 3994–4002. [Google Scholar] [CrossRef]
- Francisco, V.; Andrade-Neto, M.; Pouliquen, Y.; Lucien, V. Chemical composition of the essential oil from roots of Philodendron acutatum Schott. J. Essent. Oil Res. 2002, 14, 172–174. [Google Scholar] [CrossRef]
- Nainwal, P. Review on Philodendron species-plant seeking for validation of its therapeutic approaches. J. Pharm. Sci. Res. 2019, 11, 2003–2006. [Google Scholar]
- Pant, P.; Pandey, S.; Dall’Acqua, S. The influence of environmental conditions on secondary metabolites in medicinal plants: A literature review. Chem Biodivers. 2021, 18, e2100345. [Google Scholar] [CrossRef]
- Shahraki, S.; Samareh, H.; Saeidifar, M. Catalase inhibition by two Schiff base derivatives. Kinetics, thermodynamic and molecular docking studies. J. Mol. Liq. 2019, 287, 111003. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, J.; Zhu, Y.; Zhang, Y. Interaction of chromium (III) or chromium (VI) with catalase and its effect on the structure and function of catalase: An in vitro study. Food Chem. 2018, 244, 378–385. [Google Scholar] [CrossRef]
- Husin, F.; Khalid, N.; Misran, E.; Hasham, R.; Hamid, M.; Ya’akob, H. In silico molecular docking study and in vitro evaluation of antioxidant activity in Kacip Fatimah. Mater. Today Proc. 2024, 110, 197–206. [Google Scholar] [CrossRef]
- Rana, S.; Dixit, S.; Mittal, A. In silico target identification and validation for antioxidant and anti-inflammatory activity of selective phytochemicals. Braz. Arch. Biol. Technol. 2019, 62, e19190048. [Google Scholar] [CrossRef]
- Bakrim, S.; Benkhaira, N.; Bourais, I.; Benali, T.; Lee, L.H.; El Omari, N.; Sheikh, R.A.; Goh, K.W.; Ming, L.C.; Bouyahya, A. Health benefits and pharmacological properties of stigmasterol. Antioxidants 2022, 11, 1912. [Google Scholar] [CrossRef]
- Haque, N.; Hannan, A.; Dash, R.; Choi, S.M.; Moon, I. The potential LXRβ agonist stigmasterol protects against hypoxia/reoxygenation injury by modulating mitophagy in primary hippocampal neurons. Phytomedicine 2021, 81, 153415. [Google Scholar] [CrossRef]
- Obisesan, A.; Adebiyi, O.; Olopade, J.; Olayemi, F. Neuroprotective effect of Grewia carpinifolia extract against Behavioral Impairments induced by vanadium. Physiology 2024, 39, 432. [Google Scholar] [CrossRef]
- Harborne, J. Phytochemical Methods: A Guide to Modern Techniques of Plant Analysis, 1st ed.; Chapman and Hall: London, UK, 1973. [Google Scholar]
- Ayoola, G.A.; Coker, H.A.; Adesegun, S.A.; Adepoju-Bello, A.A.; Obaweya, K.; Ezennia, E.C.; Atangbayila, T.O. Phytochemical screening and antioxidant activities of some selected medicinal plants used for malaria therapy in southwestern Nigeria. Trop. J. Pharm. Res. 2008, 7, 1019–1024. [Google Scholar] [CrossRef]
- Waterman, P.; Mole, S. Analysis of Phenolic Plant Metabolites. In Ecological Methods and Concepts; Blackwell Scientific Publications: Oxford, UK, 1994; pp. 77–99. [Google Scholar]
- Chang, C.; Yang, M.; Wen, H.; Chern, J. Estimation of total flavonoid content in propolis by two complementary colometric methods. J. Food Drug Anal. 2020, 10, 3. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Sanner, M.F. PYTHON: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef]
- O’Boyle, N.; Banck, M.; James, C.; Morley, C.; Vandermeersch, T.; Hutchison, G. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Hanwell, M.; Curtis, D.; Lonie, D.; Vandermeersch, T.; Zurek, E.; Hutchison, G. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Biovia, D.S. Discovery Studio Modeling Environment; Dassault Systèmes: San Diego, CA, USA, 2023. [Google Scholar]

| Specialized Metabolites | Ethanolic Extract |
|---|---|
| Alkaloids | − * |
| Terpenoids | ++ * |
| Taninns | +++ * |
| Quinones | ++ * |
| Phenolic compounds | +++ * |
| Flavonoids | ++ * |
| Specialized Metabolites | Content (μg/mg Extract) | Limit of Quantification (LOQ) |
|---|---|---|
| Total phenolics (μg GAE/mg extract) | 574.7± 0.9 * | 3.2 |
| Total flavonoids (μg QE/mg extract) | 2.5 ± 0.2 * | 0.3 |
| TEAC (mg/gdw) * | ||
|---|---|---|
| Sample | ABTS | DPPH |
| Ethanolic extract | 1.03 ± 0.02 | 0.67 ± 0.06 |
| IL * | RT (min) * | Score * | LTPRI Exp * | LTPRI Lit * | Identified Metabolite Name | Super Class |
|---|---|---|---|---|---|---|
| 2 | 2.785 | 84.5 | 1084.56 | 1083 | 1-Heptanol | Lipids and lipid-like molecules |
| 2 | 2.941 | 72.8 | 1093.84 | 1077.25 | 1,2 Propanediol | Organic oxygen compounds |
| 2 | 3.48 | 98.1 | 1125.93 | 1121.68 | Cyclohexanol | Organic oxygen compounds |
| 2 | 4.339 | 85.7 | 1177.11 | 1176 | Propiophenone | Organic oxygen compounds |
| 2 | 5.205 | 76.4 | 1228.69 | 1266 | 3,7-dimethyltropolone | Terpenoids |
| 2 | 5.269 | 82 | 1232.52 | 1230 | 4,5-Dimethylundecane | Hydrocarbons |
| 2 | 7.074 | 75.2 | 1335.86 | 1312 | Methyl 3-hydroxyhexadecanoate | Lipids and lipid-like molecules |
| 2 | 7.78 | 71.1 | 1373.57 | 1365 | Gamma-nonalactone | Organoheterocyclic compounds |
| 2 | 8.25 | 96.2 | 1398.66 | 1405 | 2,3,5-Trimethyldecane | Hydrocarbons |
| 2 | 11.482 | 79.5 | 1571.38 | 1555 | 3-Hydroxy-4-methylbenzoic acid | Alkaloids and derivatives |
| 3 | 11.722 | 84.6 | 1584.42 | 1351 | alpha-Cubebene | Lipids and lipid-like molecules |
| 3 | 13.295 | 80.9 | 1672.35 | 1771 | 1-Pentadecanol | Lipids and lipid-like molecules |
| 2 | 13.301 | 96.5 | 1672.67 | 1667 | 5-Azulenemethanol, 1,2,3,3a,4,5,6,7-octahydro-.alpha.,.alpha.,3,8-tetramethyl-,[3S-(3.alpha.,3a.beta.,5.alpha.)]- | Terpenoids |
| 3 | 13.369 | 78 | 1676.51 | 1569 | Lauric acid | Fatty Acids |
| 2 | 13.743 | 74.5 | 1697.58 | 1688 | Alpha-selinene | Lipids and lipid-like molecules |
| 2 | 14.319 | 97.7 | 1731.55 | 1727 | 5-Azulenemethanol, 1,2,3,4,5,6,7,8-octahydro-.alpha.,.alpha.,3,8-tetramethyl-, acetate, [3S-(3.alpha.,5.alpha.,8.alpha.)]- | Terpenoids |
| 2 | 14.895 | 90.6 | 1765.58 | 1756.38 | Trans-Nerolidyl formate | Organic acids and derivatives |
| 3 | 24.105 | 85.6 | 2354.59 | 2192 | Phytane | Lipids and lipid-like molecules |
| 2 | 17.575 | 72 | 1931.45 | 1953 | 5-Octyl-2-pyridinecarboxylic acid | Alkaloids and derivatives |
| 2 | 17.973 | 89.1 | 1957.26 | 1967 | Hexadecanoic acid | Alkaloids and derivatives |
| 2 | 18.618 | 96.4 | 1999.06 | 2005 | Eicosane | Hydrocarbons |
| 3 | 18.697 | 70.2 | 2004.42 | 1688 | Protocatechuic acid methyl ester | Phenolic compounds |
| 3 | 19.226 | 72.2 | 2040.46 | 1844 | 3-Methylene-7,11,15-Trimethyl-1-Hexadecene | Lipids and lipid-like molecules |
| 3 | 20.079 | 82.8 | 2098.55 | 1786 | Undecyl benzoate | Benzenoids |
| 3 | 20.174 | 83.7 | 2105.28 | 1735 | Benzoic acid, 2-ethylhexyl ester | Benzenoids |
| 2 | 20.465 | 74.1 | 2125.89 | 2128 | Methyl octadecanoate | Lipids and lipid-like molecules |
| 2 | 20.928 | 81.5 | 2158.73 | 2172 | Octadecanoic acid | Alkaloids and derivatives |
| 2 | 21.423 | 99.9 | 2193.82 | 2195 | Octadecanoic acid, ethyl ester | Alkaloids and derivatives |
| 2 | 21.49 | 87.5 | 2198.62 | 2210 | 2,6,10,14,18-Pentamethyleicosane | Organic compounds |
| 3 | 22.33 | 78.8 | 2248.85 | 2546 | Erucic acid | Alkaloids and derivatives |
| 2 | 22.641 | 92.9 | 2267.37 | 2275 | Muscalure | Lipids and lipid-like molecules |
| 3 | 23.057 | 77.8 | 2292.17 | 1950 | Isophytol | Lipids and lipid-like molecules |
| 3 | 23.287 | 82.8 | 2305.84 | 1980 | Trans-9, Trans-12-Octadecadienoic Acid Methyl Ester | Lipids and lipid-like molecules |
| 2 | 22.375 | 95.5 | 2311.13 | 2305 | (11Z,14Z,17Z)-Methyl icosa-11,14,17-trienoate | Lipids and lipid-like molecules |
| 2 | 23.637 | 96.5 | 2326.7 | 2321.57 | Stearic acid | Lipids and lipid-like molecules |
| 3 | 23.754 | 75.1 | 2333.7 | 2172 | (Z)-9-Octadecenoic Acid Methyl Ester | Lipids and lipid-like molecules |
| 3 | 23.774 | 81 | 2334.89 | 2101 | (Z,Z,Z)-9,12,15-Octadecatrienoic Acid Methyl Ester | Lipids and lipid-like molecules |
| 3 | 24.105 | 85.6 | 2354.59 | 2192 | Hexadeca-2,6,10,14-tetraen-1-ol, 3,7,11,16-tetramethyl | Lipids and lipid-like molecules |
| 2 | 24.143 | 77.4 | 2356.88 | 2371 | tetracosane | Hydrocarbons |
| 2 | 27.67 | 88.8 | 2566.96 | 2626.26 | 1,3-Propanediol, eicosyl ethyl ether | Organic oxygen compounds |
| 2 | 29.826 | 97.3 | 2695.39 | 2700 | 9,12-Octadecadienoic acid (Z,Z)-, 2,3-dihydroxypropyl ester | Lipids and lipid-like molecules |
| 3 | 29.855 | 74.9 | 2697.12 | 2697 | Fumaric acid, dec-4-enyl hexadecyl ester | Organic acids and derivatives |
| 3 | 30.288 | 70.3 | 2722.89 | 2500 | 13-Docosenoic Acid Methyl Ester | Lipids and lipid-like molecules |
| 3 | 31.922 | 71.6 | 2820.23 | 2626 | Docosyl isobutyl ether | Organic oxygen compounds |
| 2 | 34.496 | 88.5 | 2973.58 | 2984 | 6-Methoxy-2,7,8-trimethyl-2-(4,8,12-trimethyltridecyl)chroman | Phenolic compounds |
| 3 | 34.974 | 73.7 | 3001.07 | 3125 | Desmosterol | Steroids and steroid derivatives |
| 3 | 36.308 | 76.1 | 3081.54 | 3248 | Stigmasterol | Steroids and steroid derivatives |
| 3 | 37.444 | 70.7 | 3149.18 | 3343 | Isofucosterol | Steroids and steroid derivatives |
| Code | Ligand | Pubchem ID | Binding Energy (kcal/mol) | Ki (µM) |
|---|---|---|---|---|
| A | Stigmasterol | 5280794 | −11.0 | 8.58 nM |
| B | Desmosterol | 439577 | −8.3 | 0.82 µM |
| C | 3,7-dimethyltropolone | 13403206 | −7.0 | 7.36 µM |
| D | 3-Hydroxy-4-methylbenzoic acid | 68512 | −6.9 | 8.7 µM |
| E | Isophytol | 10453 | −6.5 | 17.11 µM |
| F | Protocatechuic acid methyl ester | 287064 | −6 | 39.8 µM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ochoa-Ocampo, M.; Espinosa de los Monteros-Silva, N.; Pastuña-Fasso, J.V.; Sacoto, J.D.; Peñuela-Mora, M.C.; Casanola-Martin, G.; Almeida, J.R.; Diéguez-Santana, K.; Mogollón, N.G.S. Volatile Compositional Profile, Antioxidant Properties, and Molecular Docking of Ethanolic Extracts from Philodendron heleniae. Molecules 2025, 30, 1366. https://doi.org/10.3390/molecules30061366
Ochoa-Ocampo M, Espinosa de los Monteros-Silva N, Pastuña-Fasso JV, Sacoto JD, Peñuela-Mora MC, Casanola-Martin G, Almeida JR, Diéguez-Santana K, Mogollón NGS. Volatile Compositional Profile, Antioxidant Properties, and Molecular Docking of Ethanolic Extracts from Philodendron heleniae. Molecules. 2025; 30(6):1366. https://doi.org/10.3390/molecules30061366
Chicago/Turabian StyleOchoa-Ocampo, Melanie, Nina Espinosa de los Monteros-Silva, Jefferson V. Pastuña-Fasso, Juan Diego Sacoto, María Cristina Peñuela-Mora, Gerardo Casanola-Martin, José R. Almeida, Karel Diéguez-Santana, and Noroska G. S. Mogollón. 2025. "Volatile Compositional Profile, Antioxidant Properties, and Molecular Docking of Ethanolic Extracts from Philodendron heleniae" Molecules 30, no. 6: 1366. https://doi.org/10.3390/molecules30061366
APA StyleOchoa-Ocampo, M., Espinosa de los Monteros-Silva, N., Pastuña-Fasso, J. V., Sacoto, J. D., Peñuela-Mora, M. C., Casanola-Martin, G., Almeida, J. R., Diéguez-Santana, K., & Mogollón, N. G. S. (2025). Volatile Compositional Profile, Antioxidant Properties, and Molecular Docking of Ethanolic Extracts from Philodendron heleniae. Molecules, 30(6), 1366. https://doi.org/10.3390/molecules30061366







