Structural Disorder within Paramyxoviral Nucleoproteins and Phosphoproteins in Their Free and Bound Forms: From Predictions to Experimental Assessment
Abstract
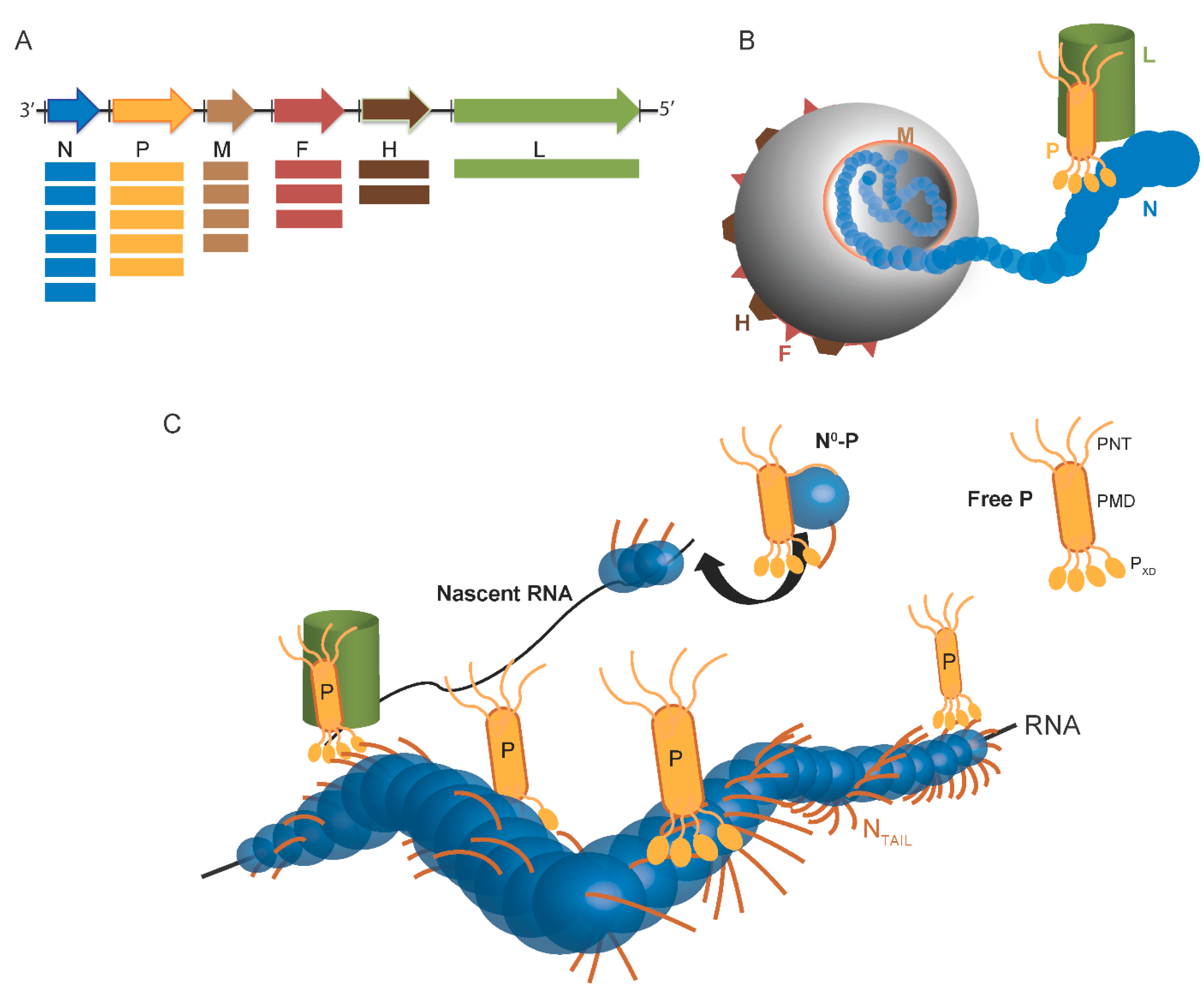
:1. Overview of the Replicative Complex of Paramyxoviruses
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Eaton, B.T.; Mackenzie, J.S.; Wang, L.F. Henipaviruses. In Fields Virology, 5th ed.; Fields, B.N., Knipe, D.M., Howley, P.M., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 2007; pp. 1587–1600. [Google Scholar]
- Lamb, R.A.; Parks, G.D. Paramyxoviridae: The viruses and their replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 1450–1497. [Google Scholar]
- Wang, L.F.; Yu, M.; Hansson, E.; Pritchard, L.I.; Shiell, B.; Michalski, W.P.; Eaton, B.T. The exceptionally large genome of Hendra virus: Support for creation of a new genus within the family Paramyxoviridae. J. Virol. 2000, 74, 9972–9979. [Google Scholar] [CrossRef] [PubMed]
- Halpin, K.; Bankamp, B.; Harcourt, B.H.; Bellini, W.J.; Rota, P.A. Nipah virus conforms to the rule of six in a minigenome replication assay. J. Gen. Virol. 2004, 85, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Kolakofsky, D. Paramyxoviridae: The viruses and their replication. In Fields Virology, 4th ed.; Fields, B.N., Knipe, D.M., Howley, P.M., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 2001; pp. 1305–1340. [Google Scholar]
- Bhella, D.; Ralph, A.; Murphy, L.B.; Yeo, R.P. Significant differences in nucleocapsid morphology within the Paramyxoviridae. J. Gen. Virol. 2002, 83, 1831–1839. [Google Scholar] [PubMed]
- Tan, W.S.; Ong, S.T.; Eshaghi, M.; Foo, S.S.; Yusoff, K. Solubility, immunogenicity and physical properties of the nucleocapsid protein of nipah virus produced in Escherichia coli. J. Med. Virol. 2004, 73, 105–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, M.; Cattaneo, R.; Spielhofer, P.; Orvell, C.; Norrby, E.; Messerli, M.; Perriard, J.C.; Billeter, M.A. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology 1991, 185, 299–308. [Google Scholar] [CrossRef]
- Spehner, D.; Drillien, R.; Howley, P.M. The assembly of the measles virus nucleoprotein into nucleocapsid-like particles is modulated by the phosphoprotein. Virology 1997, 232, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Albertini, A.A.V.; Schoehn, G.; Ruigrok, R.W. Structures impliquées dans la réplication et la transcription des virus à arn non segmentés de sens négatif. Virologie 2005, 9, 83–92. (In French) [Google Scholar]
- Blocquel, D.; Bourhis, J.M.; Eléouët, J.F.; Gerlier, D.; Habchi, J.; Jamin, M.; Longhi, S.; Yabukarski, F. Transcription et réplication des mononégavirales: Une machine moléculaire originale. Virologie 2012, 16, 225–257. (In French) [Google Scholar]
- Lamb, R.A.; Krug, R.M. Orthomyxoviridae: The viruses and their replication. In Fields Virology, 4th ed.; Fields, B.N., Knipe, D.M., Howley, P.M., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 2001; pp. 1487–1531. [Google Scholar]
- Longhi, S.; Canard, B. Mécanismes de transcription et de réplication des Paramyxoviridae. Virologie 1999, 3, 227–240. (In French) [Google Scholar]
- Roux, L. Dans le génome des paramyxovirinae, les promoteurs et leurs activités sont façonnés par la «règle de six». Virologie 2005, 9, 19–34. (In French) [Google Scholar]
- Morin, B.; Rahmeh, A.A.; Whelan, S.P. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J. 2012, 31, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Banerjee, A.K. Phosphoprotein, P of human parainfluenza virus type 3 prevents self-association of RNA-dependent RNA polymerase, L. Virology 2009, 383, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, M.; Shaila, M.S. Recombinant L and P protein complex of Rinderpest virus catalyses mRNA synthesis in vitro. Virus Res. 2008, 135, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T.; Kobayashi, M.; Iwama, M.; Mizumoto, K. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 2005, 280, 4429–4435. [Google Scholar] [CrossRef] [PubMed]
- Noton, S.L.; Deflube, L.R.; Tremaglio, C.Z.; Fearns, R. The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS Pathog. 2012, 8, e1002980. [Google Scholar] [CrossRef] [PubMed]
- Fix, J.; Galloux, M.; Blondot, M.L.; Eleouet, J.F. The insertion of fluorescent proteins in a variable region of respiratory syncytial virus L polymerase results in fluorescent and functional enzymes but with reduced activities. Open Virol. J. 2011, 5, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Bourhis, J.M.; Longhi, S. Measles virus nucleoprotein: Structural organization and functional role of the intrinsically disordered C-terminal domain. In Measles Virus Nucleoprotein; Longhi, S., Ed.; Nova Publishers Inc.: Hauppage, NY, USA, 2007; pp. 1–35. [Google Scholar]
- Longhi, S. Measles Virus Nucleoprotein; Nova Publishers Inc.: Hauppage, NY, USA, 2007. [Google Scholar]
- Longhi, S. Nucleocapsid structure and function. Curr. Top. Microbiol. Immunol. 2009, 329, 103–128. [Google Scholar] [PubMed]
- Longhi, S.; Oglesbee, M. Structural disorder within the measles virus nucleoprotein and phosphoprotein. Protein Pept. Lett. 2010, 17, 961–978. [Google Scholar] [CrossRef] [PubMed]
- Longhi, S. Structural disorder within the measles virus nucleoprotein and phosphoprotein: Functional implications for transcription and replication. In Negative Strand RNA Virus; Luo, M., Ed.; World Scientific Publishing: Singapore, Singapore, 2011; pp. 95–125. [Google Scholar]
- Habchi, J.; Longhi, S. Structural disorder within paramyxovirus nucleoproteins and phosphoproteins. Mol. Biosyst. 2012, 8, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Habchi, J.; Mamelli, L.; Longhi, S. Structural disorder within the nucleoprotein and phosphoprotein from measles, nipah and hendra viruses. In Flexible Viruses: Structural Disorder in Viral Proteins; Uversky, V.N., Longhi, S., Eds.; John Wiley and Sons: Hoboken, NJ, USA, 2012; pp. 47–94. [Google Scholar]
- Tompa, P.; Fuxreiter, M. Fuzzy complexes: Polymorphism and structural disorder in protein–protein interactions. Trends Biochem. Sci. 2008, 33, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing protein intrinsic disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habchi, J.; Mamelli, L.; Darbon, H.; Longhi, S. Structural disorder within Henipavirus nucleoprotein and phosphoprotein: From predictions to experimental assessment. PLoS ONE 2010, 5, e11684. [Google Scholar] [CrossRef] [PubMed]
- Karlin, D.; Ferron, F.; Canard, B.; Longhi, S. Structural disorder and modular organization in Paramyxovirinae N and P. J. Gen. Virol. 2003, 84, 3239–3252. [Google Scholar] [CrossRef] [PubMed]
- Karlin, D.; Longhi, S.; Receveur, V.; Canard, B. The N-terminal domain of the phosphoprotein of morbilliviruses belongs to the natively unfolded class of proteins. Virology 2002, 296, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Bourhis, J.; Johansson, K.; Receveur-Bréchot, V.; Oldfield, C.J.; Dunker, A.K.; Canard, B.; Longhi, S. The C-terminal domain of measles virus nucleoprotein belongs to the class of intrinsically disordered proteins that fold upon binding to their physiological partner. Virus Res. 2004, 99, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Houben, K.; Marion, D.; Tarbouriech, N.; Ruigrok, R.W.; Blanchard, L. Interaction of the C-terminal domains of sendai virus N and P proteins: Comparison of polymerase-nucleocapsid interactions within the Paramyxovirus family. J. Virol. 2007, 81, 6807–6816. [Google Scholar] [CrossRef] [PubMed]
- Johansson, K.; Bourhis, J.M.; Campanacci, V.; Cambillau, C.; Canard, B.; Longhi, S. Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J. Biol. Chem. 2003, 278, 44567–44573. [Google Scholar] [CrossRef] [PubMed]
- Kingston, R.L.; Hamel, D.J.; Gay, L.S.; Dahlquist, F.W.; Matthews, B.W. Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc. Natl. Acad. Sci. USA 2004, 101, 8301–8306. [Google Scholar] [CrossRef] [PubMed]
- Kingston, R.L.; Walter, A.B.; Gay, L.S. Characterization of nucleocapsid binding by the measles and the mumps virus phosphoprotein. J. Virol. 2004, 78, 8630–8640. [Google Scholar] [CrossRef] [PubMed]
- Bernado, P.; Blanchard, L.; Timmins, P.; Marion, D.; Ruigrok, R.W.; Blackledge, M. A structural model for unfolded proteins from residual dipolar couplings and small-angle X-ray scattering. Proc. Natl. Acad. Sci. USA 2005, 102, 17002–17007. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, L.; Tarbouriech, N.; Blackledge, M.; Timmins, P.; Burmeister, W.P.; Ruigrok, R.W.; Marion, D. Structure and dynamics of the nucleocapsid-binding domain of the sendai virus phosphoprotein in solution. Virology 2004, 319, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Houben, K.; Blanchard, L.; Blackledge, M.; Marion, D. Intrinsic dynamics of the partly unstructured PX domain from the sendai virus RNA polymerase cofactor P. Biophys. J. 2007, 93, 2830–2844. [Google Scholar] [CrossRef] [PubMed]
- Communie, G.; Crepin, T.; Maurin, D.; Jensen, M.R.; Blackledge, M.; Ruigrok, R.W. Structure of the tetramerization domain of measles virus phosphoprotein. J. Virol. 2013, 87, 7166–7169. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.M.; Sun, D.; Schmitt, A.P.; He, B. Phosphorylation of paramyxovirus phosphoprotein and its role in viral gene expression. Future Microbiol. 2010, 5, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Gerard, F.C.; Ribeiro Ede, A., Jr.; Leyrat, C.; Ivanov, I.; Blondel, D.; Longhi, S.; Ruigrok, R.W.; Jamin, M. Modular organization of rabies virus phosphoprotein. J. Mol. Biol. 2009, 388, 978–996. [Google Scholar] [CrossRef] [PubMed]
- Leyrat, C.; Gerard, F.C.; de Almeida Ribeiro, E., Jr.; Ivanov, I.; Ruigrok, R.W.; Jamin, M. Structural disorder in proteins of the rhabdoviridae replication complex. Protein Pept. Lett. 2010, 17, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Ringkjøbing Jensen, M.; Communie, G.; Ribeiro, E.D., Jr.; Martinez, N.; Desfosses, A.; Salmon, L.; Mollica, L.; Gabel, F.; Jamin, M.; Longhi, S.; et al. Intrinsic disorder in measles virus nucleocapsids. Proc. Natl. Acad. Sci. USA 2011, 108, 9839–9844. [Google Scholar]
- Ruigrok, R.W.; Crepin, T.; Kolakofsky, D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr. Opin. Microbiol. 2011, 14, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Warnes, A.; Fooks, A.R.; Dowsett, A.B.; Wilkinson, G.W.; Stephenson, J.R. Expression of the measles virus nucleoprotein gene in Escherichia coli and assembly of nucleocapsid-like structures. Gene 1995, 160, 173–178. [Google Scholar] [CrossRef]
- Bhella, D. Measles virus nucleocapsid structure, conformational flexibility and the rule of six. In Measles Virus Nucleoprotein; Longhi, S., Ed.; Nova Publishers Inc.: Hauppage, NY, USA, 2007. [Google Scholar]
- Eshaghi, M.; Tan, W.S.; Ong, S.T.; Yusoff, K. Purification and characterization of Nipah virus nucleocapsid protein produced in insect cells. J. Clin. Microbiol. 2005, 43, 3172–3177. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.R.; Bernado, P.; Houben, K.; Blanchard, L.; Marion, D.; Ruigrok, R.W.; Blackledge, M. Structural disorder within sendai virus nucleoprotein and phosphoprotein: Insight into the structural basis of molecular recognition. Protein Pept. Lett. 2010, 17, 952–960. [Google Scholar] [CrossRef] [PubMed]
- Heggeness, M.H.; Scheid, A.; Choppin, P.W. Conformation of the helical nucleocapsids of paramyxoviruses and vesicular stomatitis virus: Reversible coiling and uncoiling induced by changes in salt concentration. Proc. Natl. Acad. Sci. USA 1980, 77, 2631–2635. [Google Scholar] [CrossRef] [PubMed]
- Heggeness, M.H.; Scheid, A.; Choppin, P.W. The relationship of conformational changes in the sendai virus nucleocapsid to proteolytic cleavage of the NP polypeptide. Virology 1981, 114, 555–562. [Google Scholar] [CrossRef]
- Karlin, D.; Longhi, S.; Canard, B. Substitution of two residues in the measles virus nucleoprotein results in an impaired self-association. Virology 2002, 302, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Bankamp, B.; Horikami, S.M.; Thompson, P.D.; Huber, M.; Billeter, M.; Moyer, S.A. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 1996, 216, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Liston, P.; Batal, R.; DiFlumeri, C.; Briedis, D.J. Protein interaction domains of the measles virus nucleocapsid protein (NP). Arch. Virol. 1997, 142, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Longhi, S.; Receveur-Brechot, V.; Karlin, D.; Johansson, K.; Darbon, H.; Bhella, D.; Yeo, R.; Finet, S.; Canard, B. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 2003, 278, 18638–18648. [Google Scholar] [CrossRef] [PubMed]
- Bhella, D.; Ralph, A.; Yeo, R.P. Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J. Mol. Biol. 2004, 340, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Schoehn, G.; Mavrakis, M.; Albertini, A.; Wade, R.; Hoenger, A.; Ruigrok, R.W. The 12Å structure of trypsin-treated measles virus N–RNA. J. Mol. Biol. 2004, 339, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Tawar, R.G.; Duquerroy, S.; Vonrhein, C.; Varela, P.F.; Damier-Piolle, L.; Castagné, N.; MacLellan, K.; Bedouelle, H.; Bricogne, G.; Bhella, D.; et al. 3D structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 2009, 326, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Alayyoubi, M.; Leser, G.P.; Kors, C.A.; Lamb, R.A. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex. Proc. Natl. Acad. Sci. USA 2015, 112, E1792–E1799. [Google Scholar] [CrossRef] [PubMed]
- Desfosses, A.; Goret, G.; Farias Estrozi, L.; Ruigrok, R.W.; Gutsche, I. Nucleoprotein-RNA orientation in the measles virus nucleocapsid by three-dimensional electron microscopy. J. Virol. 2011, 85, 1391–1395. [Google Scholar] [CrossRef] [PubMed]
- Communie, G.; Habchi, J.; Yabukarski, F.; Blocquel, D.; Schneider, R.; Tarbouriech, N.; Papageorgiou, N.; Ruigrok, R.W.; Jamin, M.; Ringkjøbing-Jensen, M.; et al. Atomic resolution description of the interaction between the nucleoprotein and phosphoprotein of hendra virus. PLoS Pathog. 2013, 9, e1003631. [Google Scholar] [CrossRef] [PubMed]
- Baronti, L.; Erales, J.; Habchi, J.; Felli, I.C.; Pierattelli, R.; Longhi, S. Dynamics of the intrinsically disordered C-terminal domain of the nipah virus nucleoprotein and interaction with the X domain of the phosphoprotein as unveiled by NMR spectroscopy. ChemBioChem 2015, 16, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Gutsche, I.; Desfosses, A.; Effantin, G.; Ling, W.L.; Haupt, M.; Ruigrok, R.W.; Sachse, C.; Schoehn, G. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science 2015, 348, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Yabukarski, F.; Lawrence, P.; Tarbouriech, N.; Bourhis, J.M.; Delaforge, E.; Jensen, M.R.; Ruigrok, R.W.; Blackledge, M.; Volchkov, V.; Jamin, M. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat. Struct. Mol. Biol. 2014, 21, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Garner, E.; Romero, P.; Dunker, A.K.; Brown, C.; Obradovic, Z. Predicting binding regions within disordered proteins. Genome Inform. 1999, 10, 41–50. [Google Scholar]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled folding and binding with α-helix-forming molecular recognition elements. Biochemistry 2005, 44, 12454–12470. [Google Scholar] [CrossRef] [PubMed]
- Mohan, A.; Oldfield, C.J.; Radivojac, P.; Vacic, V.; Cortese, M.S.; Dunker, A.K.; Uversky, V.N. Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 2006, 362, 1043–1059. [Google Scholar] [CrossRef] [PubMed]
- Vacic, V.; Oldfield, C.J.; Mohan, A.; Radivojac, P.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Characterization of molecular recognition features, morfs, and their binding partners. J. Proteome Res. 2007, 6, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Kawano, M.; Tsurudome, M.; Nishio, M.; Ito, M.; Ohgimoto, S.; Suga, S.; Komada, H.; Ito, Y. Binding of the V proteins to the nucleocapsid proteins of human parainfluenza type 2 virus. Med. Microbiol. Immunol. 1996, 185, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Karlin, D.; Belshaw, R. Detecting remote sequence homology in disordered proteins: Discovery of conserved motifs in the N-termini of Mononegavirales phosphoproteins. PLoS ONE 2012, 7, e31719. [Google Scholar] [CrossRef] [PubMed]
- Leyrat, C.; Jensen, M.R.; Ribeiro, E.A., Jr.; Gerard, F.C.; Ruigrok, R.W.; Blackledge, M.; Jamin, M. The N0-binding region of the vesicular stomatitis virus phosphoprotein is globally disordered but contains transient α-helices. Protein Sci. 2011, 20, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Leyrat, C.; Yabukarski, F.; Tarbouriech, N.; Ribeiro, E.A., Jr.; Jensen, M.R.; Blackledge, M.; Ruigrok, R.W.; Jamin, M. Structure of the vesicular stomatitis virus N0-P complex. PLoS Pathog. 2011, 7, e1002248. [Google Scholar] [CrossRef] [PubMed]
- Sweetman, D.A.; Miskin, J.; Baron, M.D. Rinderpest virus C and V proteins interact with the major (L) component of the viral polymerase. Virology 2001, 281, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Zhang, G.; Yang, X.; Zhang, S.; Chen, L.; Yan, Q.; Xu, M.; Banerjee, A.K.; Chen, M. Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production. Cell Host Microbe 2014, 15, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Blocquel, D.; Beltrandi, M.; Erales, J.; Barbier, P.; Longhi, S. Biochemical and structural studies of the oligomerization domainof the Nipah virus phosphoprotein: evidence for an elongated coiled-coil homotrimer. Virology 2013, 446, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Beltrandi, M.; Blocquel, D.; Erales, J.; Barbier, P.; Cavalli, A.; Longhi, S. Insights into the coiled-coil organization of the Hendra virus phosphoprotein from combined biochemical and SAXS studies. Virology 2015, 477, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Blocquel, D.; Habchi, J.; Durand, E.; Sevajol, M.; Ferron, F.; Erales, J.; Papageorgiou, N.; Longhi, S. Coiled-coil deformations in crystal structures: The measles virus phosphoprotein multimerization domain as an illustrative example. Acta Cryst. D 2014, 70, 1589–1603. [Google Scholar] [CrossRef] [PubMed]
- Blocquel, D.; Habchi, J.; Gruet, A.; Blangy, S.; Longhi, S. Compaction and binding properties of the intrinsically disordered C-terminal domain of henipavirus nucleoprotein as unveiled by deletion studies. Mol. Biosyst. 2012, 8, 392–410. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P. Structure and Function of Intrinsically Disordered Proteins; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2010. [Google Scholar]
- Hazy, E.; Tompa, P. Limitations of induced folding in molecular recognition by intrinsically disordered proteins. Chemphyschem 2009, 10, 1415–1419. [Google Scholar] [CrossRef] [PubMed]
- Martinho, M.; Habchi, J.; El Habre, Z.; Nesme, L.; Guigliarelli, B.; Belle, V.; Longhi, S. Assessing induced folding within the intrinsically disordered C-terminal domain of the Henipavirus nucleoproteins by site directed spin labeling EPR spectroscopy. J. Biomol. Struct. Dyn. 2013, 31, 453–471. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chu, X.; Longhi, S.; Roche, P.; Han, W.; Wang, E.; Wang, J. Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein. Proc. Natl. Acad. Sci. USA 2013, 110, E3743–E3752. [Google Scholar] [CrossRef] [PubMed]
- Morin, B.; Bourhis, J.M.; Belle, V.; Woudstra, M.; Carrière, F.; Guigliarelli, B.; Fournel, A.; Longhi, S. Assessing induced folding of an intrinsically disordered protein by site-directed spin-labeling epr spectroscopy. J. Phys. Chem. B 2006, 110, 20596–20608. [Google Scholar] [CrossRef] [PubMed]
- Belle, V.; Rouger, S.; Costanzo, S.; Liquiere, E.; Strancar, J.; Guigliarelli, B.; Fournel, A.; Longhi, S. Mapping α-helical induced folding within the intrinsically disordered C-terminal domain of the measles virus nucleoprotein by site-directed spin-labeling EPR spectroscopy. Proteins Struct. Funct. Bioinform. 2008, 73, 973–988. [Google Scholar] [CrossRef] [PubMed]
- Gely, S.; Lowry, D.F.; Bernard, C.; Ringkjobing-Jensen, M.; Blackledge, M.; Costanzo, S.; Darbon, H.; Daughdrill, G.W.; Longhi, S. Solution structure of the C-terminal X domain of the measles virus phosphoprotein and interaction with the intrinsically disordered C-terminal domain of the nucleoprotein. J. Mol. Recognit. 2010, 23, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.R.; Houben, K.; Lescop, E.; Blanchard, L.; Ruigrok, R.W.; Blackledge, M. Quantitative conformational analysis of partially folded proteins from residual dipolar couplings: Application to the molecular recognition element of sendai virus nucleoprotein. J. Am. Chem. Soc. 2008, 130, 8055–8061. [Google Scholar] [CrossRef] [PubMed]
- Serrano, L.; Fersht, A.R. Capping and α-helix stability. Nature 1989, 342, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Habchi, J.; Blangy, S.; Mamelli, L.; Ringkjobing Jensen, M.; Blackledge, M.; Darbon, H.; Oglesbee, M.; Shu, Y.; Longhi, S. Characterization of the interactions between the nucleoprotein and the phosphoprotein of henipaviruses. J. Biol. Chem. 2011, 286, 13583–13602. [Google Scholar] [CrossRef] [PubMed]
- Bourhis, J.M.; Receveur-Bréchot, V.; Oglesbee, M.; Zhang, X.; Buccellato, M.; Darbon, H.; Canard, B.; Finet, S.; Longhi, S. The intrinsically disordered C-terminal domain of the measles virus nucleoprotein interacts with the C-terminal domain of the phosphoprotein via two distinct sites and remains predominantly unfolded. Protein Sci. 2005, 14, 1975–1992. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Glendening, C.; Linke, H.; Parks, C.L.; Brooks, C.; Udem, S.A.; Oglesbee, M. Identification and characterization of a regulatory domain on the carboxyl terminus of the measles virus nucleocapsid protein. J. Virol. 2002, 76, 8737–8746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bourhis, J.M.; Longhi, S.; Carsillo, T.; Buccellato, M.; Morin, B.; Canard, B.; Oglesbee, M. Hsp72 recognizes a P binding motif in the measles virus N protein C-terminus. Virology 2005, 337, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Couturier, M.; Buccellato, M.; Costanzo, S.; Bourhis, J.M.; Shu, Y.; Nicaise, M.; Desmadril, M.; Flaudrops, C.; Longhi, S.; Oglesbee, M. High affinity binding between Hsp70 and the C-terminal domain of the measles virus nucleoprotein requires an Hsp40 co-chaperone. J. Mol. Recognit. 2010, 23, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Tenoever, B.R.; Servant, M.J.; Grandvaux, N.; Lin, R.; Hiscott, J. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 2002, 76, 3659–3669. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Bourhis, J.M.; Chamontin, C.; Soriano, C.; Villet, S.; Costanzo, S.; Couturier, M.; Belle, V.; Fournel, A.; Darbon, H.; et al. The interaction between the measles virus nucleoprotein and the interferon regulator factor 3 relies on a specific cellular environment. Virol. J. 2009, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Laine, D.; Bourhis, J.; Longhi, S.; Flacher, M.; Cassard, L.; Canard, B.; Sautès-Fridman, C.; Rabourdin-Combe, C.; Valentin, H. Measles virus nucleoprotein induces cell proliferation arrest and apoptosis through NTAIL/NR and NCORE/FCγRIIB1 interactions, respectively. J. Gen. Virol. 2005, 86, 1771–1784. [Google Scholar] [CrossRef] [PubMed]
- Laine, D.; Trescol-Biémont, M.; Longhi, S.; Libeau, G.; Marie, J.; Vidalain, P.; Azocar, O.; Diallo, A.; Canard, B.; Rabourdin-Combe, C.; et al. Measles virus (MV) nucleoprotein binds to a novel cell surface receptor distinct from FcγRII via its C-terminal domain: Role in MV-induced immunosuppression. J. Virol. 2003, 77, 11332–11346. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Masuda, M.; Miura, R.; Yoneda, M.; Kai, C. Morbillivirus nucleoprotein possesses a novel nuclear localization signal and a CRM1-independent nuclear export signal. Virology 2006, 352, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Takeda, M.; Shirogane, Y.; Nakatsu, Y.; Nakamura, T.; Yanagi, Y. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J. Virol. 2009, 83, 10374–10383. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Yoneda, M.; Ikeda, F.; Sugai, A.; Sato, H.; Kai, C. Peroxiredoxin 1 is required for efficient transcription and replication of measles virus. J. Virol. 2011, 85, 2247–2253. [Google Scholar] [CrossRef] [PubMed]
- De, B.P.; Banerjee, A.K. Involvement of actin microfilaments in the transcription/replication of human parainfluenza virus type 3: Possible role of actin in other viruses. Microsc. Res. Tech. 1999, 47, 114–123. [Google Scholar] [CrossRef]
- Moyer, S.A.; Baker, S.C.; Horikami, S.M. Host cell proteins required for measles virus reproduction. J. Gen. Virol. 1990, 71, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Krumm, S.A.; Takeda, M.; Plemper, R.K. The measles virus nucleocapsid protein tail domain is dispensable for viral polymerase recruitment and activity. J. Biol. Chem. 2013, 288, 29943–29953. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Cortay, J.C.; Gerlier, D. Measles virus protein interactions in yeast: New findings and caveats. Virus Res. 2003, 98, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Liston, P.; DiFlumeri, C.; Briedis, D.J. Protein interactions entered into by the measles virus P, V, and C proteins. Virus Res. 1995, 38, 241–259. [Google Scholar] [CrossRef]
- Curran, J.; Pelet, T.; Kolakofsky, D. An acidic activation-like domain of the sendai virus P protein is required for RNA synthesis and encapsidation. Virology 1994, 202, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Curran, J.; Marq, J.B.; Kolakofsky, D. An N-terminal domain of the sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 1995, 69, 849–855. [Google Scholar] [PubMed]
- Tarbouriech, N.; Curran, J.; Ruigrok, R.W.; Burmeister, W.P. Tetrameric coiled coil domain of sendai virus phosphoprotein. Nat. Struct. Biol. 2000, 7, 777–781. [Google Scholar] [PubMed]
- Rahaman, A.; Srinivasan, N.; Shamala, N.; Shaila, M.S. Phosphoprotein of the rinderpest virus forms a tetramer through a coiled coil region important for biological function. A structural insight. J. Biol. Chem. 2004, 279, 23606–23614. [Google Scholar] [CrossRef] [PubMed]
- Llorente, M.T.; Barreno-Garcia, B.; Calero, M.; Camafeita, E.; Lopez, J.A.; Longhi, S.; Ferron, F.; Varela, P.F.; Melero, J.A. Structural analysis of the human respiratory syncitial virus phosphoprotein: Characterization of an α-helical domain involved in oligomerization. J. Gen. Virol. 2006, 87, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.; Green, T.J.; Purushotham, S.; Deivanayagam, C.; Bedwell, G.J.; Prevelige, P.E.; Luo, M. Structural and functional characterization of the mumps virus phosphoprotein. J. Virol. 2013, 87, 7558–7568. [Google Scholar] [CrossRef] [PubMed]
- Leyrat, C.; Renner, M.; Harlos, K.; Grimes, J.M. Solution and crystallographic structures of the central region of the phosphoprotein from human metapneumovirus. PLoS ONE 2013, 8, e80371. [Google Scholar] [CrossRef] [PubMed]
- Bruhn-Johannsen, J.F.; Barnett, K.; Bibby, J.; Thomas, J.; Keegan, R.; Rigden, D.; Bornholdt, Z.A.; Saphire, E.O. Crystal structure of the Nipah virus phosphoprotein tetramerization domain. J. Virol. 2014, 88, 758–762. [Google Scholar] [CrossRef] [PubMed]
- Dutta, K.; Alexandrov, A.; Huang, H.; Pascal, S.M. pH-induced folding of an apoptotic coiled coil. Protein Sci. 2001, 10, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Lupas, A.N.; Gruber, M. The structure of α-helical coiled coils. Adv. Protein Chem. 2005, 70, 37–78. [Google Scholar] [PubMed]
- Oshaben, K.M.; Salari, R.; McCaslin, D.R.; Chong, L.T.; Horne, W.S. The native GCN4 leucine-zipper domain does not uniquely specify a dimeric oligomerization state. Biochemistry 2012, 51, 9581–9591. [Google Scholar] [CrossRef] [PubMed]
- Curran, J. A role for the sendai virus p protein trimer in RNA synthesis. J. Virol. 1998, 72, 4274–4280. [Google Scholar] [PubMed]
- Curran, J.; Boeck, R.; Lin-Marq, N.; Lupas, A.; Kolakofsky, D. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology 1995, 214, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Kingston, R.L.; Gay, L.S.; Baase, W.S.; Matthews, B.W. Structure of the nucleocapsid-binding domain from the mumps virus polymerase; an example of protein folding induced by crystallization. J. Mol. Biol. 2008, 379, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Yegambaram, K.; Bulloch, E.M.; Kingston, R.L. Protein domain definition should allow for conditional disorder. Protein Sci. 2013, 22, 1502–1518. [Google Scholar] [CrossRef] [PubMed]
- D’Urzo, A.; Konijnenberg, A.; Rossetti, G.; Habchi, J.; Li, J.; Carloni, P.; Sobott, F.; Longhi, S.; Grandori, R. Molecular basis for structural heterogeneity of an intrinsically disordered protein bound to a partner by combined ESI-IM-MS and modeling. J. Am. Soc. Mass Spectrom. 2015, 26, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Delano, W.L. The pymol molecular graphics system. Proteins Struct. Funct. Bioinform. 2002, 30, 442–454. [Google Scholar]
- Erales, J.; Beltrandi, M.; Roche, J.; Maté, M.; Longhi, S. Insights into the hendra virus NTAIL-XD complex: Evidence for a parallel organization of the helical more at the XD surface stabilized by a combination of hydrophobic and polar interactions. Biochim. Biophys. Acta 2015, 1854, 1038–1053. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; Haran, G.; Zhou, H.X. Fundamental aspects of protein-protein association kinetics. Chem. Rev. 2009, 109, 839–860. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Yuwen, T.; Zhu, F.; Skrynnikov, N.R. Role of electrostatic interactions in binding of peptides and intrinsically disordered proteins to their folded targets. 1. NMR and MD characterization of the complex between the c-Crk N-SH3 domain and the peptide sos. Biochemistry 2014, 53, 6473–6495. [Google Scholar] [CrossRef] [PubMed]
- Dosnon, M.; Bonetti, D.; Morrone, A.; Erales, J.; di Silvio, E.; Longhi, S.; Gianni, S. Demonstration of a folding after binding mechanism in the recognition between the measles virus NTAIL and X domains. ACS Chem. Biol. 2015, 10, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Kavalenka, A.; Urbancic, I.; Belle, V.; Rouger, S.; Costanzo, S.; Kure, S.; Fournel, A.; Longhi, S.; Guigliarelli, B.; Strancar, J. Conformational analysis of the partially disordered measles virus NTAIL-XD complex by SDSL EPR spectroscopy. Biophys. J. 2010, 98, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Belle, V.; Rouger, S.; Costanzo, S.; Longhi, S.; Fournel, A. Site-directed spin labeling EPR spectroscopy. In Instrumental Analysis of Intrinsically Disordered Proteins: Assessing Structure and Conformation; Uversky, V.N., Longhi, S., Eds.; John Wiley and Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Meszaros, B.; Tompa, P.; Simon, I.; Dosztanyi, Z. Molecular principles of the interactions of disordered proteins. J. Mol. Biol. 2007, 372, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.D.; Ma, B.; Kumar, S.; Wolfson, H.; Nussinov, R. Protein folding: Binding of conformationally fluctuating building blocks via population selection. Crit. Rev. Biochem. Mol. Biol. 2001, 36, 399–433. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; Ma, B.; Sham, Y.Y.; Kumar, S.; Nussinov, R. Structured disorder and conformational selection. Proteins Struct. Funct. Bioinform. 2001, 44, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Espinoza-Fonseca, L.M. Reconciling binding mechanisms of intrinsically disordered proteins. Biochem. Biophys. Res. Commun. 2009, 382, 479–482. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, B.A.; Portman, J.J.; Wolynes, P.G. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc. Natl. Acad. Sci. USA 2000, 97, 8868–8873. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.; Pickar, A.; Qiu, S.; Tsao, J.; Rodenburg, C.; Dokland, T.; Elson, A.; He, B.; Luo, M. Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc. Natl. Acad. Sci. USA 2014, 111, 15208–15213. [Google Scholar] [CrossRef] [PubMed]
- Oglesbee, M.; Ringler, S.; Krakowka, S. Interaction of canine distemper virus nucleocapsid variants with 70K heat-shock proteins. J. Gen. Virol. 1990, 71, 1585–1590. [Google Scholar] [CrossRef] [PubMed]
- Oglesbee, M.; Tatalick, L.; Rice, J.; Krakowka, S. Isolation and characterization of canine distemper virus nucleocapsid variants. J. Gen. Virol. 1989, 70, 2409–2419. [Google Scholar] [CrossRef] [PubMed]
- Brunel, J.; Chopy, D.; Dosnon, M.; Bloyet, L.M.; Devaux, P.; Urzua, E.; Cattaneo, R.; Longhi, S.; Gerlier, D. Sequence of events in measles virus replication: Role of phosphoprotein-nucleocapsid interactions. J. Virol. 2014, 88, 10851–10863. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Habchi, J.; Costanzo, S.; Padilla, A.; Brunel, J.; Gerlier, D.; Oglesbee, M.; Longhi, S. Plasticity in structural and functional interactions between the phosphoprotein and nucleoprotein of measles virus. J. Biol. Chem. 2012, 287, 11951–11967. [Google Scholar] [CrossRef] [PubMed]
- Fuxreiter, M.; Tompa, P. Fuzzy interactome: The limitations of models in molecular biology. Trends Biochem. Sci. 2009, 34, 3. [Google Scholar] [CrossRef]
- Fuxreiter, M. Fuzziness: Linking regulation to protein dynamics. Mol. Biosyst. 2012, 8, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Longhi, S. The measles virus NTAIL-XD complex: An illustrative example of fuzziness. Adv. Exp. Med. Biol. 2012, 725, 126–141. [Google Scholar] [PubMed]
- Carsillo, T.; Zhang, X.; Vasconcelos, D.; Niewiesk, S.; Oglesbee, M. A single codon in the nucleocapsid protein C terminus contributes to in vitro and in vivo fitness of edmonston measles virus. J. Virol. 2006, 80, 2904–2912. [Google Scholar] [CrossRef] [PubMed]
- Oglesbee, M. Nucleocapsid protein interactions with the major inducible heat shock protein. In Measles Virus Nucleoprotein; Longhi, S., Ed.; Nova Publishers Inc.: Hauppage, NY, USA, 2007; pp. 53–98. [Google Scholar]
- Carsillo, T.; Traylor, Z.; Choi, C.; Niewiesk, S.; Oglesbee, M. Hsp72, a host determinant of measles virus neurovirulence. J. Virol. 2006, 80, 11031–11039. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J. Proteome Res. 2007, 6, 1917–1932. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, K.; Sato, H.; Inoue, Y.; Watanabe, A.; Yoneda, M.; Ikeda, F.; Fujita, K.; Fukuda, H.; Takamura, C.; Kozuka-Hata, H.; et al. Phosphorylation of measles virus nucleoprotein upregulates the transcriptional activity of minigenomic RNA. Proteomics 2008, 8, 1871–1879. [Google Scholar] [CrossRef] [PubMed]
- Sugai, A.; Sato, H.; Yoneda, M.; Kai, C. Phosphorylation of measles virus nucleoprotein affects viral growth by changing gene expression and genomic RNA stability. J. Virol. 2013, 87, 11684–11692. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Sato, H.; Hagiwara, K.; Watanabe, A.; Sugai, A.; Ikeda, F.; Kozuka-Hata, H.; Oyama, M.; Yoneda, M.; Kai, C. Determination of a phosphorylation site in Nipah virus nucleoprotein and its involvement in virus transcription. J. Gen. Virol. 2011, 92, 2133–2141. [Google Scholar] [CrossRef] [PubMed]
- Gruet, A.; Longhi, S.; Bignon, C. One-step generation of error-prone PCR libraries using Gateway® technology. Microb. Cell Fact. 2012, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Pancsa, R.; Fuxreiter, M. Interactions via intrinsically disordered regions: What kind of motifs? IUBMB Life 2012, 64, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.G.; Magliery, T.J.; Regan, L. Detecting protein–protein interactions with GFP-fragment reassembly. Nat. Methods 2004, 1, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Gruet, A.; Dosnon, M.; Vassena, A.; Lombard, V.; Gerlier, D.; Bignon, C.; Longhi, S. Dissecting partner recognition by an intrinsically disordered protein using descriptive random mutagenesis. J. Mol. Biol. 2013, 425, 3495–3509. [Google Scholar] [CrossRef] [PubMed]
- Sebolt-Leopold, J.S.; English, J.M. Mechanisms of drug inhibition of signalling molecules. Nature 2006, 441, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Lagerstrom, M.C.; Schioth, H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008, 7, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Betzi, S.; Restouin, A.; Opi, S.; Arold, S.T.; Parrot, I.; Guerlesquin, F.; Morelli, X.; Collette, Y. Protein protein interaction inhibition (2P2I) combining high throughput and virtual screening: Application to the HIV-1 Nef protein. Proc. Natl. Acad. Sci. USA 2007, 104, 19256–19261. [Google Scholar] [CrossRef] [PubMed]
- Galloux, M.; Gabiane, G.; Sourimant, J.; Richard, C.A.; England, P.; Moudjou, M.; Aumont-Nicaise, M.; Fix, J.; Rameix-Welti, M.A.; Eleouet, J.F. Identification and characterization of the binding site of the respiratory syncytial virus phosphoprotein to RNA-free nucleoprotein. J. Virol. 2015, 89, 3484–3496. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Legall, T.; Oldfield, C.J.; Mueller, J.P.; Van, Y.Y.; Romero, P.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006, 24, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Targeting intrinsically disordered proteins in neurodegenerative and protein dysfunction diseases: Another illustration of the D2 concept. Expert Rev. Proteomics 2010, 7, 543–564. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Uversky, V.N. Drugs for “protein clouds”: Targeting intrinsically disordered transcription factors. Curr. Opin. Pharmacol. 2010, 10, 782–788. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins and novel strategies for drug discovery. Expert Opin. Drug Discov. 2012, 7, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Lo Conte, L.; Chothia, C.; Janin, J. The atomic structure of protein–protein recognition sites. J. Mol. Biol. 1999, 285, 2177–2198. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, K.; Tsai, C.J.; Nussinov, R. Analysis of ordered and disordered protein complexes reveals structural features discriminating between stable and unstable monomers. J. Mol. Biol. 2004, 341, 1327–1341. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Vassilev, L.T. Targeting the p53-MDM2 interaction to treat cancer. Br. J. Cancer 2004, 91, 1415–1419. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T. Small-molecule antagonists of p53-MDM2 binding: Research tools and potential therapeutics. Cell Cycle 2004, 3, 419–421. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [PubMed]
- Dey, S.; Pal, A.; Chakrabarti, P.; Janin, J. The subunit interfaces of weakly associated homodimeric proteins. J. Mol. Biol. 2010, 398, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Bourgeas, R.; Basse, M.J.; Morelli, X.; Roche, P. Atomic analysis of protein–protein interfaces with known inhibitors: The 2P2I database. PLoS ONE 2010, 5, e9598. [Google Scholar] [CrossRef] [PubMed]
- Jordan, I.K.; Sutter, B.A.; McClure, M.A. Molecular evolution of the Paramyxoviridae and Rhabdoviridae multiple-protein-encoding P gene. Mol. Biol. Evol. 2000, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Narechania, A.; Terai, M.; Burk, R.D. Overlapping reading frames in closely related human papillomaviruses result in modular rates of selection within E2. J. Gen. Virol. 2005, 86, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Rancurel, C.; Khosravi, M.; Dunker, K.A.; Romero, P.R.; Karlin, D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J. Virol. 2009, 83, 10719–10736. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, E.; Tompa, P.; Liliom, K.; Kalmar, L. Dual coding in alternative reading frames correlates with intrinsic protein disorder. Proc. Natl. Acad. Sci. USA 2010, 107, 5429–5434. [Google Scholar] [CrossRef] [PubMed]
- Bourhis, J.M.; Canard, B.; Longhi, S. Structural disorder within the replicative complex of measles virus: Functional implications. Virology 2006, 344, 94–110. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Blocquel, D.; Habchi, J.; Uversky, A.V.; Kurgan, L.; Uversky, V.N.; Longhi, S. Structural disorder in viral proteins. Chem. Rev. 2014, 114, 6880–6911. [Google Scholar] [CrossRef] [PubMed]
- Tokuriki, N.; Oldfield, C.J.; Uversky, V.N.; Berezovsky, I.N.; Tawfik, D.S. Do viral proteins possess unique biophysical features? Trends Biochem. Sci. 2009, 34, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Williams, R.W.; Oldfield, C.J.; Goh, G.K.; Dunker, A.K.; Uversky, V.N. Viral disorder or disordered viruses: Do viral proteins possess unique features? Protein Pept. Lett. 2010, 17, 932–951. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; van Roey, K.; Weatheritt, R.J.; Toedt, G.; Uyar, B.; Altenberg, B.; Budd, A.; Diella, F.; Dinkel, H.; Gibson, T.J. Attributes of short linear motifs. Mol. Biosyst. 2012, 8, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; Trave, G.; Gibson, T.J. How viruses hijack cell regulation. Trends Biochem. Sci. 2011, 36, 159–169. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habchi, J.; Longhi, S. Structural Disorder within Paramyxoviral Nucleoproteins and Phosphoproteins in Their Free and Bound Forms: From Predictions to Experimental Assessment. Int. J. Mol. Sci. 2015, 16, 15688-15726. https://doi.org/10.3390/ijms160715688
Habchi J, Longhi S. Structural Disorder within Paramyxoviral Nucleoproteins and Phosphoproteins in Their Free and Bound Forms: From Predictions to Experimental Assessment. International Journal of Molecular Sciences. 2015; 16(7):15688-15726. https://doi.org/10.3390/ijms160715688
Chicago/Turabian StyleHabchi, Johnny, and Sonia Longhi. 2015. "Structural Disorder within Paramyxoviral Nucleoproteins and Phosphoproteins in Their Free and Bound Forms: From Predictions to Experimental Assessment" International Journal of Molecular Sciences 16, no. 7: 15688-15726. https://doi.org/10.3390/ijms160715688