CHARMM Force Field Parameterization of Peroxisome Proliferator-Activated Receptor γ Ligands
Abstract
:1. Introduction
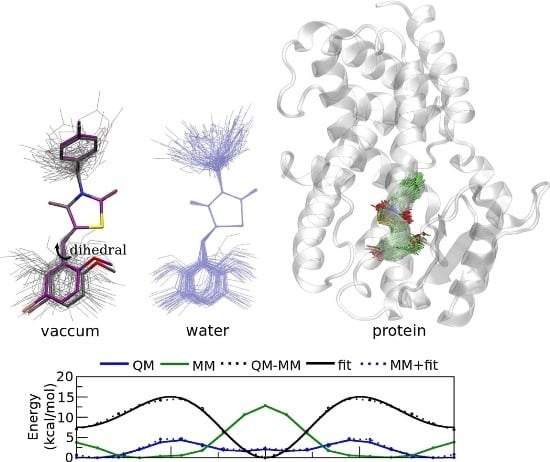
2. Results and Discussion
3. Materials and Methods
3.1. Parameterization of the PPARγ Ligands SR1664 and GQ16
3.2. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lehrke, M.; Lazar, M.A. The many faces of PPARγ. Cell 2005, 123, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Willson, T.M.; Brown, P.J.; Sternbach, D.D.; Henke, B.R. The PPARs: From orphan receptors to drug discovery. J. Med. Chem. 2000, 43, 527–550. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARγ signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Francis, G.A.; Fayard, E.; Picard, F.; Auwerx, J. Nuclear receptors and the control of metabolism. Annu. Rev. Physiol. 2003, 65, 261–311. [Google Scholar] [CrossRef] [PubMed]
- Desvergne, B.; Wahli, W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr. Rev. 1999, 20, 649–688. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Banks, A.S.; Kamenecka, T.M.; Busby, S.A.; Chalmers, M.J.; Kumar, N.; Kuruvilla, D.S.; Shin, Y.; He, Y.; Bruning, J.B.; et al. Antidiabetic actions of a non-agonist PPARγ ligand blocking Cdk5-mediated phosphorylation. Nature 2011, 477, 477–481. [Google Scholar] [PubMed]
- Choi, S.; Kim, E.S.; Koh, M.; Lee, S.; Lim, D.; Yang, Y.R.; Jang, H.; Seo, K.; Min, S.; Lee, I.H.; et al. A novel non-agonist peroxisome proliferator-activated receptor (PPAR γ) ligand UHC1 blocks PPARγ phosphorylation by cyclin-dependent kinase 5 ( CDK5 ) and improves insulin sensitivity. J. Biol. Chem. 2014, 289, 26618–26629. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Banks, A.S.; Estall, J.L.; Kajimura, S.; Boström, P.; Laznik, D.; Ruas, J.L.; Chalmers, M.J.; Kamenecka, T.M.; Blüher, M.; et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARγ by CDK. Nature 2010, 466, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Banks, A.S.; Mcallister, F.E.; Camporez, P.G.; Zushin, P.H.; Jurczak, M.J.; Laznik-Bogoslavski, D.; Shulman, G.I.; Gygi, S.P.; Spiegelman, B.M. An ERK/CDK5 axis controls the diabetogenic actions of PPARγ. Nature 2015, 517, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Shu, Y.; Niu, Z.; Zheng, W.; Wu, H.; Lu, Y.; Shen, P. PPARγ1 phosphorylation enhances proliferation and drug resistance in human fibrosarcoma cells. Exp. Cell Res. 2014, 322, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Bruning, J.B.; Chalmers, M.J.; Prasad, S.; Busby, S.A.; Kamenecka, T.M.; He, Y.; Nettles, K.W.; Griffin, P.R. Partial agonists activate PPARγ using a helix 12 independent mechanism. Structure 2007, 15, 1258–1271. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.S.; Chalmers, M.J.; Novick, S.; Kuruvilla, D.S.; Chang, M.R.; Kamenecka, T.M.; Rance, M.; Johnson, B.A.; Burris, T.P.; Griffin, P.R.; et al. Ligand and receptor dynamics contribute to the mechanism of graded PPARγ agonism. Structure 2012, 20, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Kroker, A.J.; Bruning, J.B. Review of the structural and dynamic mechanisms of PPARγ partial agonism. PPAR Res. 2015, 2015, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nolte, R.T.; Wisely, G.B.; Westin, S.; Cobb, J.E.; Lambert, M.H.; Kurokawa, R.; Rosenfeld, M.G.; Willson, T.M.; Glass, C.K.; Milburn, M. V ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature 1998, 395, 137–143. [Google Scholar] [PubMed]
- Garcia-Vallvé, S.; Guasch, L.; Tomas-Hernández, S.; del Bas, J.M.; Ollendorff, V.; Arola, L.; Pujadas, G.; Mulero Abellan, M. Peroxisome proliferator-activated receptor γ (PPARγ) and ligand choreography: Newcomers take the stage. J. Med. Chem. 2015, 58, 5381–5394. [Google Scholar] [CrossRef] [PubMed]
- Waku, T.; Shiraki, T.; Oyama, T.; Fujimoto, Y.; Maebara, K.; Kamiya, N.; Jingami, H.; Morikawa, K. Structural insight into PPARγ activation through covalent modification with endogenous fatty acids. J. Mol. Biol. 2009, 385, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.J.; Tomkinson, N.C.O.; Villeneuve, M.S.; Blanchard, S.G.; Willson, T.M. Differential activity of rosiglitazone enantiomers at PPARγ. Bioorg. Med. Chem. Lett. 1998, 8, 3657–3658. [Google Scholar] [CrossRef]
- Lehmann, J.M.; Moore, L.B.; Smith-Oliver, T.A.; Wilkison, W.O.; Willson, T.M.; Kliewer, S.A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR γ). J. Biol. Chem. 1995, 270, 12953–12956. [Google Scholar] [CrossRef] [PubMed]
- Soccio, R.E.; Chen, E.R.; Lazar, M.A. Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metab. 2014, 20, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Mourão, R.H.; Silva, T.G.; Soares, A.L.M.; Vieira, E.S.; Santos, J.N.; Lima, M.C.A.; Lima, V.L.M.; Galdino, S.L.; Barbe, J.; Pitta, I.R. Synthesis and biological activity of novel acridinylidene and benzylidene thiazolidinediones. Eur. J. Med. Chem. 2005, 40, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lambert, M.H.; Xu, H.E. Activation of nuclear receptors: A perspective from structural genomics. Structure 2003, 11, 741–746. [Google Scholar] [CrossRef]
- Amato, A.A.; Rajagopalan, S.; Lin, J.Z.; Carvalho, B.M.; Figueira, A.C.M.; Lu, J.; Ayers, S.D.; Mottin, M.; Silveira, R.L.; Souza, P.C.T.; et al. GQ-16, a novel peroxisome proliferator-activated receptor γ (PPARγ) ligand, promotes insulin sensitization without weight gain. J. Biol. Chem. 2012, 287, 28169–28179. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.S.; de Lima, C.L.; Royer, C.; Silva, J.B.; Oliveira, F.C.B.; Christ, C.G.; Pereira, S.A.; Bao, S.N.; Lima, M.C.A.; Pitta, M.G.R.; et al. GQ-16, a TZD-derived partial PPARγ agonist, induces the expression of thermogenesis-related genes in brown fat and visceral white fat and decreases visceral adiposity in obese and hyperglycemic mice. PLoS ONE 2016, 11, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Chandra, V.; Rastinejad, F. Structural overview of the nuclear receptor superfamily: Insights into physiology and therapeutics. Annu. Rev. Physiol. 2010, 72, 247–272. [Google Scholar] [CrossRef] [PubMed]
- Marciano, D.P.; Kuruvilla, D.S.; Boregowda, S.V.; Asteian, A.; Hughes, T.S.; Garcia-Ordonez, R.; Corzo, C.A.; Khan, T.M.; Novick, S.J.; Park, H.; et al. Pharmacological repression of PPARγ promotes osteogenesis. Nat. Commun. 2015, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y. Human PPARγ ligand binding dmain in complex with SR. 2016; in press. [Google Scholar]
- Itoh, T.; Fairall, L.; Amin, K.; Inaba, Y.; Szanto, A.; Balint, B.; Nagy, L.; Yamamoto, K.; Schwabe, J. Structural basis for the activation of PPARγ by oxidized fatty acids. Nat. Struct. Mol. Biol. 2008, 15, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Liberato, M.V.; Nascimento, A.S.; Ayers, S.D.; Lin, J.Z.; Cvoro, A.; Silveira, R.L.; Martínez, L.; Souza, P.C.; Saidemberg, D.; Deng, T.; et al. Medium chain fatty acids are selective peroxisome proliferator activated receptor (PPAR) γ activators and pan-PPAR partial agonists. PLoS ONE 2012, 7, e36297. [Google Scholar] [CrossRef] [Green Version]
- Martínez, L.; Webb, P.; Polikarpov, I.; Skaf, M.S. Molecular dynamics simulations of ligand dissociation from thyroid hormone receptors: Evidence of the likeliest escape pathway and its implications for the design of novel ligands. J. Med. Chem. 2006, 49, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Martínez, L.; Sonoda, M.T.; Webb, P.; Baxter, J.D.; Skaf, M.S.; Polikarpov, I. Molecular dynamics simulations reveal multiple pathways of ligand dissociation from thyroid hormone receptors. Biophys. J. 2005, 89, 2011–2023. [Google Scholar] [CrossRef] [PubMed]
- Martínez, L.; Nascimento, A.S.; Nunes, F.M.; Phillips, K.; Aparício, R.; Dias, S.M.G.; Figueira, A.C.M.; Lin, J.H.; Nguyen, P.; Apriletti, J.W.; et al. Gaining ligand selectivity in thyroid hormone receptors via entropy. Proc. Natl. Acad. Sci. USA 2009, 106, 20717–20722. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, A.; Souza, P.C.T.; Muniz, J.R.C.; Ricci, C.G.; Ayers, S.D.; Parekh, N.M.; Godoy, A.S. Molecular mechanism of peroxisome proliferator-activated receptor α activation by WY14643: A new mode of ligand recognition and receptor stabilization. J. Mol. Biol. 2013, 425, 2878–2893. [Google Scholar] [CrossRef] [PubMed]
- Mottin, M.; Souza, P.C.T.; Skaf, M.S. Molecular recognition of PPARγ by kinase CDK5/p25: Insights from a combination of protein–protein docking and adaptive biasing force simulations. J. Phys. Chem. B 2015, 119, 8330–8339. [Google Scholar] [CrossRef] [PubMed]
- Souza, P.C.T.; Puhl, A.C.; Martínez, L.; Aparício, R.; Nascimento, A.S.; Figueira, A.C.M.; Nguyen, P.; Webb, P.; Skaf, M.; Polikarpov, I. Identification of a new hormone-binding site on the surface of thyroid hormone receptor. Mol. Endocrinol. 2014, 28, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Fratev, F.; Tsakovska, I.; Sharif, M.A.; Mihaylova, E.; Pajeva, I. Structural and dynamical insight into PPARγ antagonism: In silico study of the ligand-receptor interactions of non-covalent antagonists. Int. J. Mol. Sci. 2015, 16, 15405–15424. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, L.; Zhao, X.; Sun, X. Molecular recognition of agonist and antagonist for peroxisome proliferator-activated receptor-α studied by molecular dynamics simulations. Int. J. Mol. Sci. 2014, 15, 8743–8752. [Google Scholar] [CrossRef] [PubMed]
- Hansson, A.; Souza, P.C.T.; Silveira, R.L.; Skaf, M.S.; Marti, L. CHARMM force field parameterization of rosiglitazone. Int. J. Quantum Chem. 2011, 111, 1346–1354. [Google Scholar] [CrossRef] [Green Version]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field (CGenFF): A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [PubMed]
- Vanommeslaeghe, K.; Mackerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. Chem. Inf. Model. 2012, 52, 3144–3154. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Raman, E.P.; Mackerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D., Jr.; Bashford, D.; Bellott, M.; Dunbrack, R.L., Jr.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 5647, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, J.Á.; Manzanaro, S.; Martín, M.J.; de Quesada, T.G.; Reymundo, I.; Luengo, S.M.; Gago, F. Synthesis, activity, and molecular modeling studies of novel human aldose reductase inhibitors based on a marine natural product. J. Med. Chem. 2003, 46, 5208–5221. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Montgomery, J.A.; Vreven, T.; Kudin, K.N.; Burant, J.C.; et al. Gaussian 03, revision C. 2004. Available online: http://www.citeulike.org/group/7862/article/3740703 (accessed on 20 September 2016).
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Besler, B.H.; Merz, K.M.; Kollman, P.A. Atomic charges derived from semiempirical methods. J. Comput. Chem. 1990, 11, 431–439. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Mackerell, A.J.; Feig, M.; Brooks, C. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004, 25, 1400–1415. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of N-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Leach, A.R. Molecular Modelling: Principles and Applications, 2nd ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2001. [Google Scholar]






| Dihedral Angle | kφn (kcal/mol) | n | δ (°) | |
|---|---|---|---|---|
| T1 | C13 N2 C20 C21 | 0.1795 | 1 | 0.0 |
| 0.3623 | 3 | 0.0 | ||
| 0.4022 | 4 | 0.0 | ||
| T2 | N1 C9 C10 C11 | 0.1000 | 1 | 180.0 |
| 0.7100 | 2 | 180.0 | ||
| T3 | C5 C7 N1 C9 | 1.5000 | 1 | 180.0 |
| 0.1900 | 2 | 0.0 | ||
| 0.2000 | 3 | 0.0 | ||
| 0.7000 | 4 | 0.0 | ||
| Dihedral Angle | kφn (kcal/mol) | n | δ (°) | |
|---|---|---|---|---|
| T1 | C11 N1 C12 C13 | −0.3572 | 4 | 180.0 |
| T2 | C6 C7 C8 C9 | 2.8584 | 1 | 0.0 |
| 5.6498 | 2 | 180.0 | ||
| 0.8824 | 3 | 0.0 | ||
| T3 | S1 C9 C8 C7 | 2.4869 | 1 | 0.0 |
| 10.3926 | 2 | 180.0 | ||
| 0.6072 | 3 | 0.0 | ||
| 0.8350 | 4 | 0.0 | ||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mottin, M.; Souza, P.C.T.; Ricci, C.G.; Skaf, M.S. CHARMM Force Field Parameterization of Peroxisome Proliferator-Activated Receptor γ Ligands. Int. J. Mol. Sci. 2017, 18, 15. https://doi.org/10.3390/ijms18010015
Mottin M, Souza PCT, Ricci CG, Skaf MS. CHARMM Force Field Parameterization of Peroxisome Proliferator-Activated Receptor γ Ligands. International Journal of Molecular Sciences. 2017; 18(1):15. https://doi.org/10.3390/ijms18010015
Chicago/Turabian StyleMottin, Melina, Paulo C. T. Souza, Clarisse G. Ricci, and Munir S. Skaf. 2017. "CHARMM Force Field Parameterization of Peroxisome Proliferator-Activated Receptor γ Ligands" International Journal of Molecular Sciences 18, no. 1: 15. https://doi.org/10.3390/ijms18010015
APA StyleMottin, M., Souza, P. C. T., Ricci, C. G., & Skaf, M. S. (2017). CHARMM Force Field Parameterization of Peroxisome Proliferator-Activated Receptor γ Ligands. International Journal of Molecular Sciences, 18(1), 15. https://doi.org/10.3390/ijms18010015