Increased Plasma Circulating Cell-Free DNA Could Be a Potential Marker for Oral Cancer
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
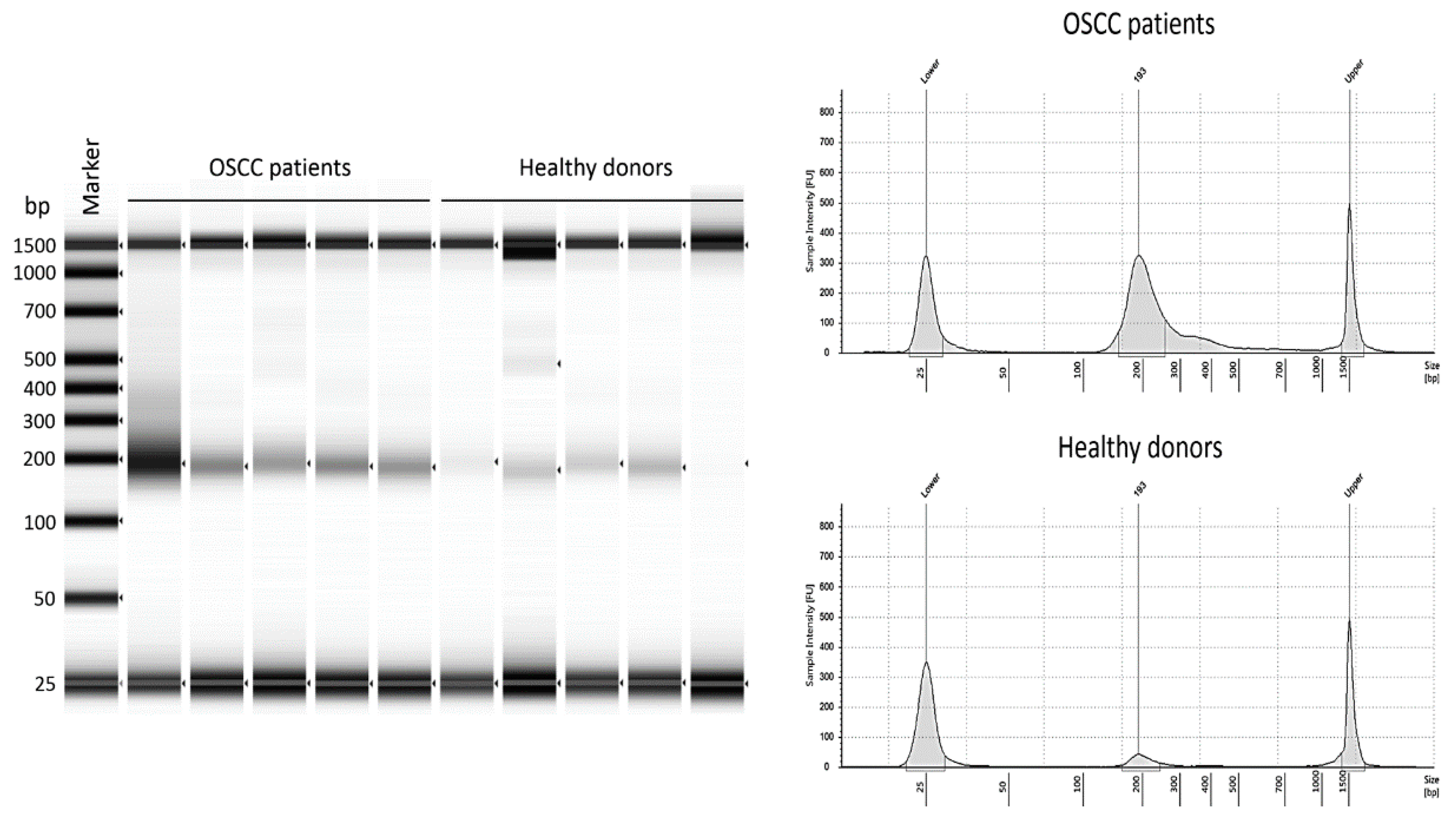
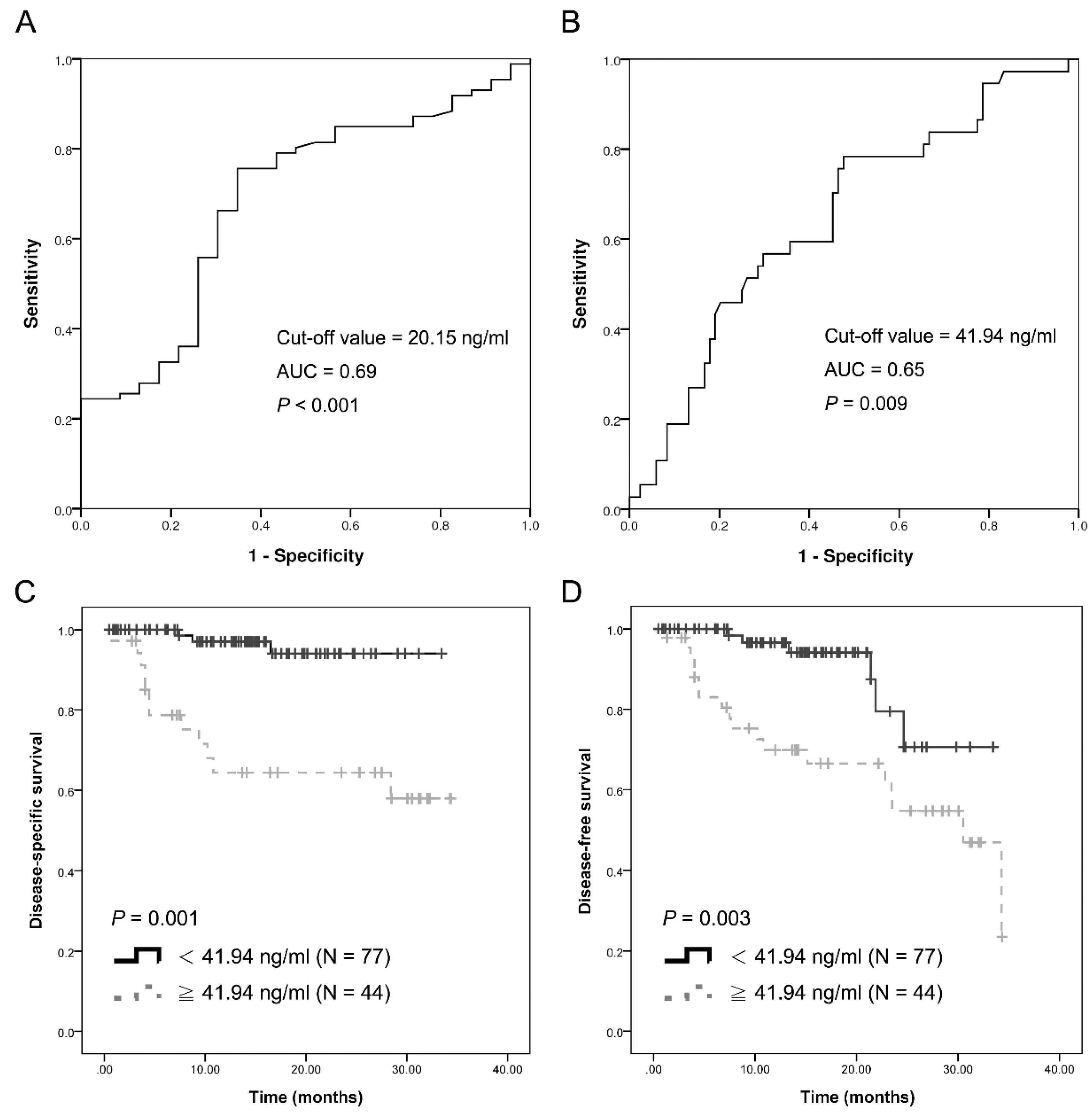
2.2. Plasma cfDNA as a Potential Diagnostic Marker
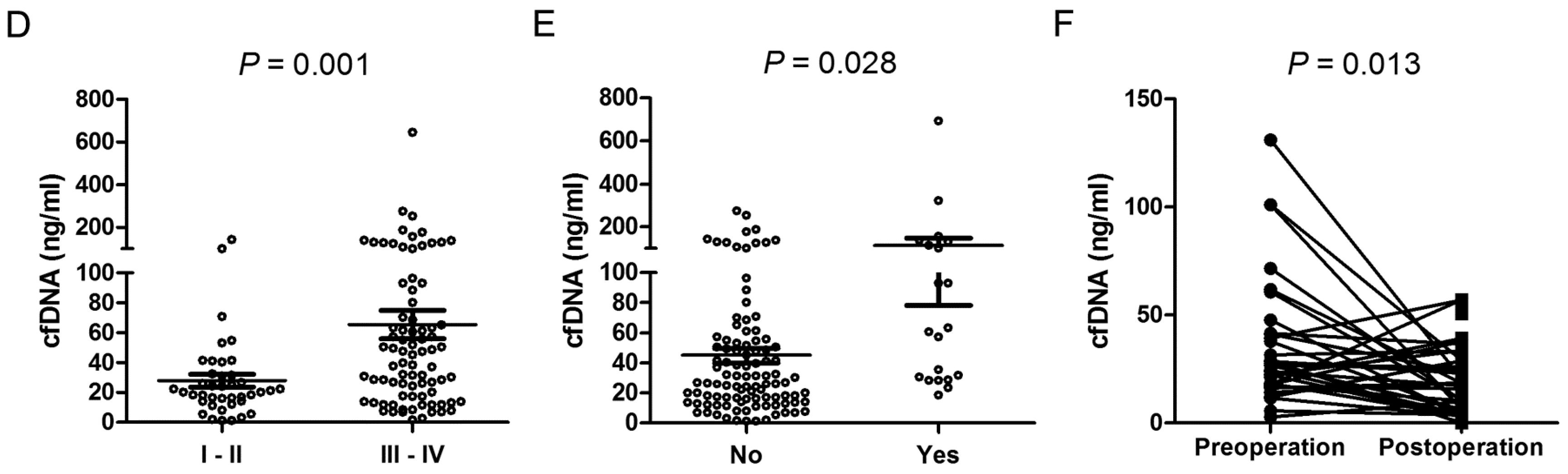
2.3. cfDNA Level as an Independent Factor of Cervical Lymph Node Metastasis in OSCC
2.4. Decrease of cfDNA in Patients’ Plasma after the Resection of Oral Primary Tumors
2.5. Association between cfDNA Levels and Survival of Patients with OSCC
3. Discussion
4. Materials and Methods
4.1. Plasma Samples
4.2. cfDNA Extraction
4.3. Plasma DNA Quantification
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Shieh, T.M.; Lin, S.C.; Liu, C.J.; Chang, S.S.; Ku, T.H.; Chang, K.W. Association of expression aberrances and genetic polymorphisms of lysyl oxidase with areca-associated oral tumorigenesis. Clin. Cancer Res. 2007, 13, 4378–4385. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.C.; Liu, C.J.; Ko, S.Y.; Chang, H.C.; Liu, T.Y.; Chang, K.W. Copy number amplification of 3q26-27 oncogenes in microdissected oral squamous cell carcinoma and oral brushed samples from areca chewers. J. Pathol. 2005, 206, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.W.; Liu, C.J.; Chu, T.H.; Cheng, H.W.; Hung, P.S.; Hu, W.Y.; Lin, S.C. Association between high miR-211 microRNA expression and the poor prognosis of oral carcinoma. J. Dent. Res. 2008, 87, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Chang, K.W.; Lin, S.C.; Cheng, H.W. Presurgical serum levels of matrix metalloproteinase-9 and vascular endothelial growth factor in oral squamous cell carcinoma. Oral Oncol. 2009, 45, 920–925. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.S.; Liu, X.B.; Wong, B.Y.; Ng, R.W.; Yuen, A.P.; Wei, W.I. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin. Cancer Res. 2008, 14, 2588–2592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braakhuis, B.J.; Tabor, M.P.; Leemans, C.R.; van der Waal, I.; Snow, G.B.; Brakenhoff, R.H. Second primary tumors and field cancerization in oral and oropharyngeal cancer: Molecular techniques provide new insights and definitions. Head Neck 2002, 24, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Kulasinghe, A.; Kenny, L.; Punyadeera, C. The development of a liquid biopsy for head and neck cancers. Oral Oncol. 2016, 61, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Kidess, E.; Jeffrey, S.S. Circulating tumor cells versus tumor-derived cell-free DNA: Rivals or partners in cancer care in the era of single-cell analysis? Genome Med. 2013, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Spellman, P.T.; Gray, J.W. Detecting cancer by monitoring circulating tumor DNA. Nat. Med. 2014, 20, 474–475. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed]
- Ulivi, P.; Silvestrini, R. Role of quantitative and qualitative characteristics of free circulating DNA in the management of patients with non-small cell lung cancer. Cell. Oncol. 2013, 36, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Sidransky, D. Emerging molecular markers of cancer. Nat. Rev. Cancer 2002, 2, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Couraud, S.; Vaca-Paniagua, F.; Villar, S.; Oliver, J.; Schuster, T.; Blanche, H.; Girard, N.; Tredaniel, J.; Guilleminault, L.; Gervais, R.; et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: A proof-of-concept study from BioCAST/IFCT-1002. Clin. Cancer Res. 2014, 20, 4613–4624. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Fleischhacker, M.; Rabien, A. Cell-free DNA in the blood as a solid tumor biomarker--a critical appraisal of the literature. Clin. Chim. Acta 2010, 411, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, A.M.; Rutkowski, T.; Fiszer-Kierzkowska, A.; Malusecka, E.; Skladowski, K. Assessment of the total cfDNA and HPV16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral Oncol. 2016, 54, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.J.; Chen, L.C.; Chien, K.Y.; Chen, Y.J.; Chang, J.T.; Wang, H.M.; Liao, C.T.; Chen, I.H. Oral cancer plasma tumor marker identified with bead-based affinity-fractionated proteomic technology. Clin. Chem. 2005, 51, 2236–2244. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, D.; Yamamoto, N.; Takagi, R.; Katakura, A.; Mizoe, J.E.; Shibahara, T. Detection of tumor DNA in plasma using whole genome amplification. Bull. Tokyo Dent. Coll. 2006, 47, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Olmo, D.C.; Gutierrez-Gonzalez, L.; Samos, J.; Picazo, M.G.; Atienzar, M.; Garcia-Olmo, D. Surgery and hematogenous dissemination: Comparison between the detection of circulating tumor cells and of tumor DNA in plasma before and after tumor resection in rats. Ann. Surg. Oncol. 2006, 13, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Bijian, K.; Mlynarek, A.M.; Balys, R.L.; Jie, S.; Xu, Y.; Hier, M.P.; Black, M.J.; Di Falco, M.R.; LaBoissiere, S.; Alaoui-Jamali, M.A. Serum proteomic approach for the identification of serum biomarkers contributed by oral squamous cell carcinoma and host tissue microenvironment. J. Proteome Res. 2009, 8, 2173–2185. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Kao, S.Y.; Tu, H.F.; Tsai, M.M.; Chang, K.W.; Lin, S.C. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010, 16, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.C.; Chang, K.W.; Chou, F.C.; Cheng, C.Y.; Liu, C.J. Association of pretreatment thrombocytosis with disease progression and survival in oral squamous cell carcinoma. Oral Oncol. 2007, 43, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Tissot, C.; Toffart, A.C.; Villar, S.; Souquet, P.J.; Merle, P.; Moro-Sibilot, D.; Perol, M.; Zavadil, J.; Brambilla, C.; Olivier, M.; et al. Circulating free DNA concentration is an independent prognostic biomarker in lung cancer. Eur. Respir. J. 2015, 46, 1773–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, A.; Zhang, X.; Zhou, S.L.; Cao, Y.; Huang, X.W.; Fan, J.; Yang, X.R.; Zhou, J. Plasma Circulating Cell-free DNA Integrity as a Promising Biomarker for Diagnosis and Surveillance in Patients with Hepatocellular Carcinoma. J. Cancer 2016, 7, 1798–1803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murtaza, M.; Dawson, S.J.; Tsui, D.W.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.F.; Kingsbury, Z.; Wong, A.S.; et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013, 497, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Shukla, D.; Kale, A.D.; Hallikerimath, S.; Yerramalla, V.; Subbiah, V. Can quantifying free-circulating DNA be a diagnostic and prognostic marker in oral epithelial dysplasia and oral squamous cell carcinoma? J. Oral Maxillofac. Surg. 2013, 71, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Coulet, F.; Blons, H.; Cabelguenne, A.; Lecomte, T.; Lacourreye, O.; Brasnu, D.; Beaune, P.; Zucman, J.; Laurent-Puig, P. Detection of plasma tumor DNA in head and neck squamous cell carcinoma by microsatellite typing and p53 mutation analysis. Cancer Res. 2000, 60, 707–711. [Google Scholar] [PubMed]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar] [PubMed]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Stroun, M.; Anker, P.; Lyautey, J.; Lederrey, C.; Maurice, P.A. Isolation and characterization of DNA from the plasma of cancer patients. Eur. J. Cancer Clin. Oncol. 1987, 23, 707–712. [Google Scholar] [CrossRef]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.C.; Weiss, J.; Hudson, C.; Christophi, C.; Cebon, J.; Behren, A.; Dobrovic, A. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci. Rep. 2015, 5, 11198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautschi, O.; Bigosch, C.; Huegli, B.; Jermann, M.; Marx, A.; Chasse, E.; Ratschiller, D.; Weder, W.; Joerger, M.; Betticher, D.C.; et al. Circulating deoxyribonucleic Acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J. Clin. Oncol. 2004, 22, 4157–4164. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.K.; Kim, H.Y.; Lee, J.S. Circulating cell-free DNA in plasma of never smokers with advanced lung adenocarcinoma receiving gefitinib or standard chemotherapy as first-line therapy. Clin. Cancer Res. 2011, 17, 5179–5187. [Google Scholar] [CrossRef] [PubMed]
- Basnet, S.; Zhang, Z.Y.; Liao, W.Q.; Li, S.H.; Li, P.S.; Ge, H.Y. The Prognostic Value of Circulating Cell-Free DNA in Colorectal Cancer: A Meta-Analysis. J. Cancer 2016, 7, 1105–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, S.J.; Rosenfeld, N.; Caldas, C. Circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 369, 93–94. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Kallianpur, S.; Mani, A.; Tijare, M.S.; Khan, S.; Jain, M.; Mathur, V.; Ahuja, R.; Saxena, V. Quantification of circulating plasma cell free DNA fragments in patients with oral cancer and precancer. Gulf J. Oncol. 2018, 1, 11–17. [Google Scholar]
- Ocana, A.; Diez-Gonzalez, L.; Garcia-Olmo, D.C.; Templeton, A.J.; Vera-Badillo, F.; Jose Escribano, M.; Serrano-Heras, G.; Corrales-Sanchez, V.; Seruga, B.; Andres-Pretel, F.; et al. Circulating DNA and Survival in Solid Tumors. Cancer Epidemiol. Biomark. Prev. 2016, 25, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Tang, Q.; Cao, X.; Burwinkel, B. Cell-Free Circulating DNA Integrity Based on Peripheral Blood as a Biomarker for Diagnosis of Cancer: A Systematic Review. Cancer Epidemiol. Biomark. Prev. 2017, 26, 1595–1602. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thornton, B.; Basu, C. Real-time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 2011, 39, 145–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Messaoudi, S.; Rolet, F.; Mouliere, F.; Thierry, A.R. Circulating cell free DNA: Preanalytical considerations. Clin. Chim. Acta 2013, 424, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Moraga, D.; Panayotova, N.; Zhou, X.H.; Farmerie, W.G.; Shanker, S. Modified Library Construction Method for 3-5kb Illumina Mate-pair Libraries. J. Biomol. Tech. 2013, 24, S41. [Google Scholar]
- Serrao, E.; Cherepanov, P.; Engelman, A.N. Amplification, Next-generation Sequencing, and Genomic DNA Mapping of Retroviral Integration Sites. J. Vis. Exp. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nygaard, A.D.; Holdgaard, P.C.; Spindler, K.L.; Pallisgaard, N.; Jakobsen, A. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br. J. Cancer 2014, 110, 363–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norton, S.E.; Lechner, J.M.; Williams, T.; Fernando, M.R. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin. Biochem. 2013, 46, 1561–1565. [Google Scholar] [CrossRef] [PubMed]
- Hung, E.C.; Chiu, R.W.; Lo, Y.M. Detection of circulating fetal nucleic acids: A review of methods and applications. J. Clin. Pathol. 2009, 62, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.; Yeung, S.W.; Lui, W.B.; Rainer, T.H.; Lo, Y.M. Effects of preanalytical factors on the molecular size of cell-free DNA in blood. Clin. Chem. 2005, 51, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.; Zhang, J.; Hui, A.B.; Wong, N.; Lau, T.K.; Leung, T.N.; Lo, K.W.; Huang, D.W.; Lo, Y.M. Size distributions of maternal and fetal DNA in maternal plasma. Clin. Chem. 2004, 50, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zimmermann, B.; Rusterholz, C.; Kang, A.; Holzgreve, W.; Hahn, S. Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms. Clin. Chem. 2004, 50, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Guleria, R.; Singh, V.; Bharti, A.C.; Mohan, A.; Das, B.C. Efficacy of circulating plasma DNA as a diagnostic tool for advanced non-small cell lung cancer and its predictive utility for survival and response to chemotherapy. Lung Cancer 2010, 70, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Lou, F.; Ma, Y.; Li, J.; Yang, B.; Chen, W.; Ye, H.; Zhang, J.B.; Zhao, M.Y.; Wu, W.J.; et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci. Rep. 2016, 6, 33519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziegler, A.; Zangemeister-Wittke, U.; Stahel, R.A. Circulating DNA: A new diagnostic gold mine? Cancer Treat. Rev. 2002, 28, 255–271. [Google Scholar] [CrossRef]
- Agostini, M.; Enzo, M.V.; Bedin, C.; Belardinelli, V.; Goldin, E.; Del Bianco, P.; Maschietto, E.; D’Angelo, E.; Izzi, L.; Saccani, A.; et al. Circulating cell-free DNA: A promising marker of regional lymphonode metastasis in breast cancer patients. Cancer Biomark. 2012, 11, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Olmo, D.; Garcia-Olmo, D.C.; Ontanon, J.; Martinez, E. Horizontal transfer of DNA and the “genometastasis hypothesis”. Blood 2000, 95, 724–725. [Google Scholar] [PubMed]
- Pinzani, P.; Salvianti, F.; Pazzagli, M.; Orlando, C. Circulating nucleic acids in cancer and pregnancy. Methods 2010, 50, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Valastyan, S.; Reinhardt, F.; Benaich, N.; Calogrias, D.; Szasz, A.M.; Wang, Z.C.; Brock, J.E.; Richardson, A.L.; Weinberg, R.A. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 2009, 137, 1032–1046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, J.; Li, D.; Xiao, B.; Miao, Y.; Jiang, Z.; Zhuo, H. Down-regulation of miR-31 expression in gastric cancer tissues and its clinical significance. Med. Oncol. 2010, 27, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussing, C.; Kampmann, M.L.; Mogensen, H.S.; Morling, C.B.N. Comparison of techniques for quantification of next-generation sequencing libraries. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e276–e278. [Google Scholar] [CrossRef]

| OSCC | Control | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variables | N | Mean | ±SEM | Pa | Pb | N | Mean | ±SEM | Pa |
| Age (years) | |||||||||
| <58 | 56 | 43.25 | ±6.31 | 0.385 | 24 | 27.88 | ±5.51 | 0.938 | |
| ≥58 | 65 | 61.24 | ±11.03 | 26 | 20.35 | ±3.88 | |||
| Gender | |||||||||
| Male | 113 | 53.90 | ±7.10 | 0.851 | 43 | 22.64 | ±3.35 | 0.935 | |
| Female | 8 | 41.25 | ±13.71 | 7 | 32.04 | ±12.36 | |||
| Tumor size | |||||||||
| T1–T2 | 48 | 33.24 | ±4.24 | 0.024 * | |||||
| T3–T4 | 73 | 66.09 | ±10.49 | ||||||
| Nodal stage | |||||||||
| N0 | 84 | 43.73 | ±5.59 | 0.009 * | |||||
| N+ | 37 | 74.25 | ±17.52 | ||||||
| Stage | |||||||||
| I–II | 40 | 27.95 | ±4.30 | 0.001 * | |||||
| III–IV | 81 | 65.46 | ±9.49 | ||||||
| Histological grade | |||||||||
| Well | 88 | 50.96 | ±5.32 | 0.268 | |||||
| Moderate | 25 | 69.87 | ±26.53 | ||||||
| Poor | 8 | 23.61 | ±6.30 | ||||||
| Perineural invasion | |||||||||
| No | 84 | 51.72 | ±8.92 | 0.269 | |||||
| Yes | 37 | 56.11 | ±8.48 | ||||||
| Lymphovascular invasion | |||||||||
| No | 101 | 45.08 | ±4.96 | 0.028 * | |||||
| Yes | 20 | 93.39 | ±30.92 | ||||||
| Variables | Cases (%) | Controls (%) | Crude OR (95% CI) | p | Adjusted a OR (95% CI) | p | ||
|---|---|---|---|---|---|---|---|---|
| cfDNA (ng/mL) | ||||||||
| <20.15 | 43 | (35.5%) | 35 | (70.0%) | Reference | Reference | ||
| ≥20.15 | 78 | (64.5%) | 15 | (30.0%) | 4.233 (2.080–8.611) | <0.001 * | 4.153 (2.155–9.204) | <0.001 * |
| Variables | Subgroups | Odd Ratio (95% CI) | p |
|---|---|---|---|
| Univariate analysis | |||
| Age (years) | ≥58 vs. <58 | 0.746 (0.343–1.619) | 0.458 |
| Gender | Female vs. Male | 1.394 (0.315–6.166) | 0.661 |
| Tumor size | T3–T4 vs. T1–T2 | 3.295 (1.350–8.042) | 0.009 * |
| Perineural invasion | Yes vs. No | 2.720 (0.889–6.373) | 0.017 * |
| Lymphovascular invasion | Yes vs. No | 5.958 (2.134–16.636) | 0.001 * |
| cfDNA (ng/mL) | ≥41.94 vs. <41.94 | 3.481 (1.551–7.810) | 0.002 * |
| Multivariate analysis | |||
| Tumor size | T3–T4 vs. T1–T2 | 2.059 (0.783–5.412) | 0.143 |
| Perineural invasion | Yes vs. No | 1.537 (0.581–4.066) | 0.387 |
| Lymphovascular invasion | Yes vs. No | 3.424 (1.065–11.010) | 0.039 * |
| cfDNA (ng/mL) | ≥41.94 vs. <41.94 | 2.533 (1.055–6.084) | 0.038 * |
| Variables | Subgroups | Hazard Ratio (95%CI) | p |
|---|---|---|---|
| Univariate analysis | |||
| Age (years) | ≥58 vs. <58 | 1.438 (0.509–4.060) | 0.493 |
| Gender | Female vs. Male | 1.073 (0.141–8.165) | 0.946 |
| Tumor size | T3–T4 vs. T1–T2 | 4.474 (1.008–19.850) | 0.049 * |
| Nodal stage | N+ vs. N0 | 15.992 (3.601–71.030) | <0.001 * |
| Perineural invasion | Yes vs. No | 4.871 (1.663–14.268) | 0.004 * |
| Lymphovascular invasion | Yes vs. No | 6.690 (2.365–18.926) | <0.001 * |
| cfDNA (ng/mL) | ≥41.94 vs. <41.94 | 6.637 (1.859–23.702) | 0.004 * |
| Multivariate analysis | |||
| Tumor size | T3–T4 vs. T1–T2 | 2.937 (0.640–13.472) | 0.166 |
| Nodal stage | N+ vs. N0 | 9.529 (2.054–44.195) | 0.004 * |
| Perineural invasion | Yes vs. No | 2.747 (0.877–8.602) | 0.083 |
| Lymphovascular invasion | Yes vs. No | 1.886 (0.625–5.685) | 0.260 |
| cfDNA (ng/mL) | ≥41.94 vs. <41.94 | 4.432 (1.214–16.178) | 0.024 * |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, L.-H.; Chang, K.-W.; Kao, S.-Y.; Cheng, H.-W.; Liu, C.-J. Increased Plasma Circulating Cell-Free DNA Could Be a Potential Marker for Oral Cancer. Int. J. Mol. Sci. 2018, 19, 3303. https://doi.org/10.3390/ijms19113303
Lin L-H, Chang K-W, Kao S-Y, Cheng H-W, Liu C-J. Increased Plasma Circulating Cell-Free DNA Could Be a Potential Marker for Oral Cancer. International Journal of Molecular Sciences. 2018; 19(11):3303. https://doi.org/10.3390/ijms19113303
Chicago/Turabian StyleLin, Li-Han, Kuo-Wei Chang, Shou-Yen Kao, Hui-Wen Cheng, and Chung-Ji Liu. 2018. "Increased Plasma Circulating Cell-Free DNA Could Be a Potential Marker for Oral Cancer" International Journal of Molecular Sciences 19, no. 11: 3303. https://doi.org/10.3390/ijms19113303
APA StyleLin, L.-H., Chang, K.-W., Kao, S.-Y., Cheng, H.-W., & Liu, C.-J. (2018). Increased Plasma Circulating Cell-Free DNA Could Be a Potential Marker for Oral Cancer. International Journal of Molecular Sciences, 19(11), 3303. https://doi.org/10.3390/ijms19113303