A Novel Design Combining Isothermal Exponential Amplification and Gold-Nanoparticles Visualization for Rapid Detection of miRNAs
Abstract
:1. Introduction
2. Results and Discussion
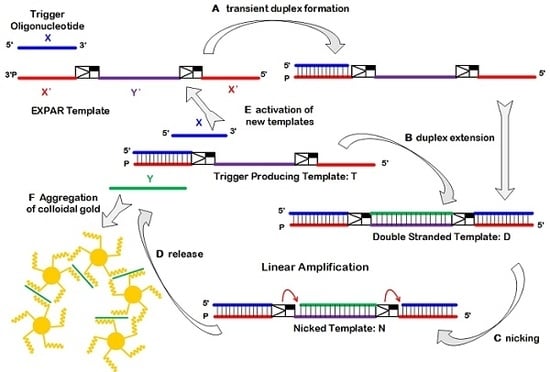
2.1. Principle of EXPAR-AuNP Visual Detection
2.2. Optimization of EXPAR-AuNP Experimental Conditions
2.2.1. Reaction Time
2.2.2. Reaction Volume and Temperature
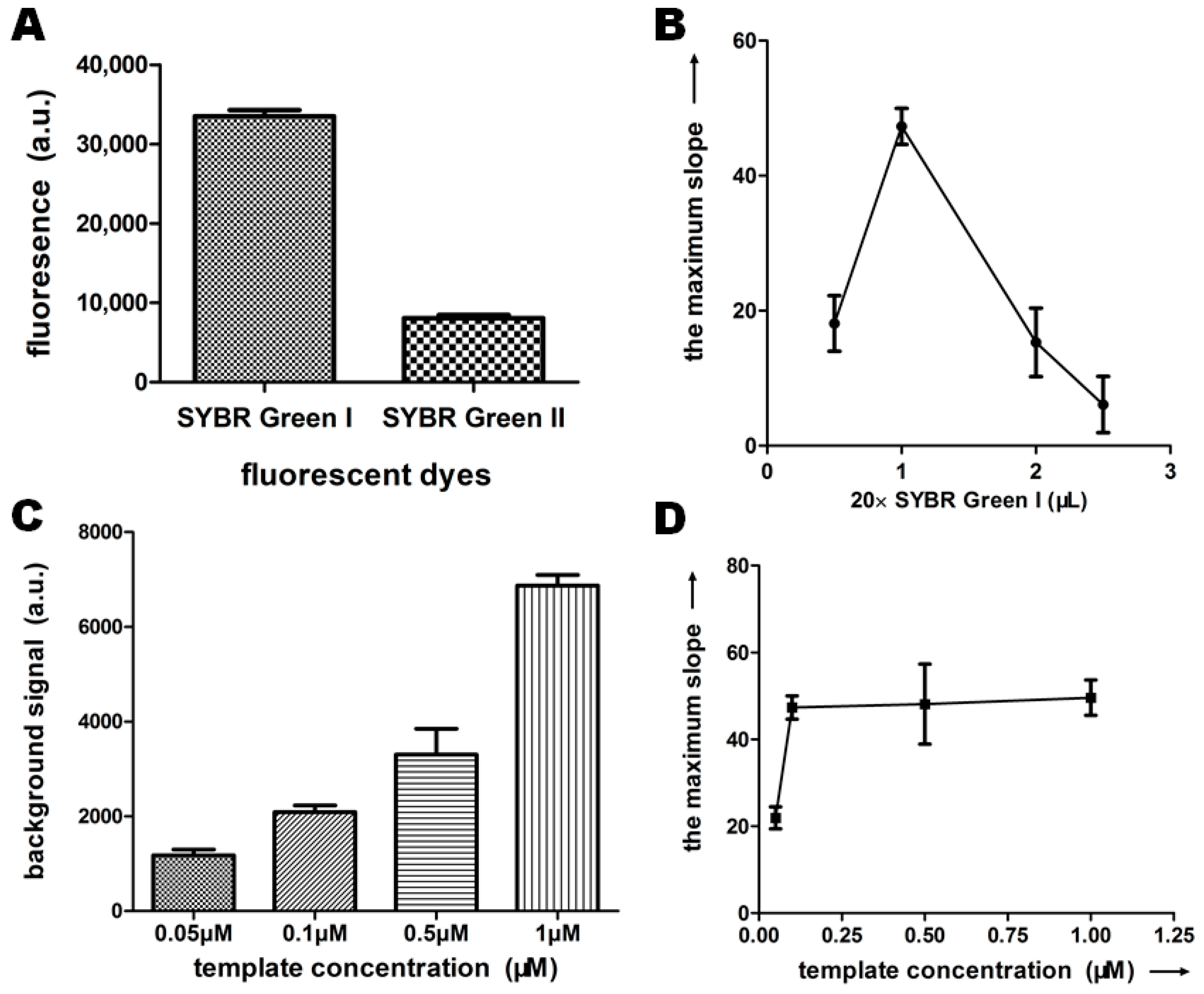
2.2.3. Fluorescent Dyes and Reaction Template
2.2.4. Enzymes
2.3. Evaluation of EXPAR-AuNPs Visual Detection
2.3.1. Sensitivity of the EXPAR-AuNP Assay
2.3.2. Specificity of the EXPAR-AuNP Assay
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Apparatus
3.3. EXPAR Experiments
3.4. Synthesis of Gold Nanoparticles
3.5. Immobilization of DNA Probes on AuNPs
3.6. AuNP-Based Colorimetric Assay
3.7. Non-Denatured Polyacrylamide Gel Electrophoresis (nd-PAGE)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AuNPs | gold nanoparticles |
| EXPAR | exponential amplification reaction |
| LOD | limit of detection |
| POI | point of inflection |
References
- Xu, Y.; Wang, Y.; Liu, S.; Yu, J.; Wang, H.; Guo, Y.; Huang, J. Ultrasensitive and rapid detection of miRNA with three-way junction structure-based trigger-assisted exponential enzymatic amplification. Biosens. Bioelectron. 2016, 81, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Chen, Z.; Shi, L.; Yang, F.; Pan, J.; Zhang, B.; Sun, D. Ultrasensitive electrochemical detection of microRNA based on an arched probe mediated isothermal exponential amplification. Anal. Chem. 2014, 86, 8200–8205. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Cui, Y.; Li, L. Dumbbell probe-mediated cascade isothermal amplification: A novel strategy for label-free detection of microRNAs and its application to real sample assay. Anal. Chim. Acta 2013, 760, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Li, Z.; Liu, C.; Cheng, Y. Ultrasensitive detection of microRNAs by exponential isothermal amplification. Angew. Chem. 2010, 49, 5498–5501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, C.Y. Sensitive detection of microRNA with isothermal amplification and a single-quantum-dot-based nanosensor. Anal. Chem. 2012, 84, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.P.; Yin, B.C.; Wang, P.; Ye, B.C. Highly sensitive detection of microRNAs based on isothermal exponential amplification-assisted generation of catalytic G-quadruplex DNAzyme. Biosens. Bioelectron. 2013, 42, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hou, T.; Wu, M.; Li, F. Label-free fluorescence strategy for sensitive microRNA detection based on isothermal exponential amplification and graphene oxide. Talanta 2016, 148, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, K.; Lv, Z.; Zhu, X.; Zhu, L.; Zhou, F. Ultrasensitive detection of microRNA with isothermal amplification and a time-resolved fluorescence sensor. Biosens. Bioelectron. 2014, 57, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Kiesling, T.; Cox, K.; Davidson, E.A.; Dretchen, K.; Grater, G.; Hibbard, S.; Lasken, R.S.; Leshin, J.; Skowronski, E.; Danielsen, M. Sequence specific detection of DNA using nicking endonuclease signal amplification (NESA). Nucleic Acids Res. 2007, 35, e117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Z.; Zhang, C.Y. Highly sensitive detection of protein with aptamer-based target-triggering two-stage amplification. Anal. Chem. 2012, 84, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, K.; Liu, Y.; Wang, H.; Wu, J.; Zhu, F.; Zou, P. Binding-induced and label-free colorimetric method for protein detection based on autonomous assembly of hemin/G-quadruplex DNAzyme amplification strategy. Biosens. Bioelectron. 2015, 64, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chen, C.C.; Wei, S.C.; Lu, H.H.; Liang, Y.H.; Lin, C.W. Diagnostic devices for isothermal nucleic acid amplification. Sensors 2012, 12, 8319–8337. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhou, J.; Zheng, Y.; Gamson, A.S.; Roembke, B.T.; Nakayama, S.; Sintim, H.O. Isothermal amplified detection of DNA and RNA. Mol. Biosyst. 2014, 10, 970–1003. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Gao, Z. Bioanalytical applications of isothermal nucleic acid amplification techniques. Anal. Chim. Acta 2015, 853, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Zanoli, L.M.; Spoto, G. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 2013, 3, 18–43. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J. Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosens. Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Van Ness, J.; van Ness, L.K.; Galas, D.J. Isothermal reactions for the amplification of oligonucleotides. Proc. Natl. Acad. Sci. USA 2003, 100, 4504–4509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asiello, P.J.; Baeumner, A.J. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip 2011, 11, 1420–1430. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Lv, Y.; Xu, S.; Zhang, Y.; Wang, L.; Li, R.; Yue, Q.; Li, H.; Gu, X.; Zhang, S.; et al. Highly sensitive fluorescence assay of DNA methyltransferase activity by methylation-sensitive cleavage-based primer generation exponential isothermal amplification-induced G-quadruplex formation. Biosens. Bioelectron. 2015, 66, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Neonakis, I.K.; Spandidos, D.A.; Petinaki, E. Use of loop-mediated isothermal amplification of DNA for the rapid detection of Mycobacterium tuberculosis in clinical specimens. Eur. J. Clin. Microbial. Infect. Dis. 2011, 30, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Biswas, G.; Sakai, M. Loop-mediated isothermal amplification (LAMP) assays for detection and identification of aquaculture pathogens: Current state and perspectives. Appl. Microbial. Biotechnol. 2014, 98, 2881–2895. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Yang, K.; Zhang, C.Y. Sensitive detection of methylated DNA using the short linear quencher-fluorophore probe and two-stage isothermal amplification assay. Biosens. Bioelectron. 2013, 49, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Chen, Z.; Jian, W.; Sun, D.; Zhang, B.; Li, X.; Yao, M. Ultrasensitive electrochemical detection of avian influenza A (H7N9) virus DNA based on isothermal exponential amplification coupled with hybridization chain reaction of DNAzyme nanowires. Biosens. Bioelectron. 2015, 64, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Zhang, D.W.; Tie, C.; Zhou, Y.L.; Zhang, X.X. G-quadruplex based two-stage isothermal exponential amplification reaction for label-free DNA colorimetric detection. Biosens. Bioelectron. 2014, 56, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Ferguson, T.M.; Shinde, D.N.; Ramirez-Borrero, A.J.; Hintze, A.; Adami, C.; Niemz, A. Sequence dependence of isothermal DNA amplification via EXPAR. Nucleic Acids Res. 2012, 40, e87. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Niu, C.; Xiao, X.; Zhu, W.; Dai, Z.; Zou, X. Chemical-oxidation cleavage triggered isothermal exponential amplification reaction for attomole gene-specific methylation analysis. Anal. Chem. 2015, 87, 2945–2951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, J.; Zhang, C.Y. Sensitive detection of transcription factors by isothermal exponential amplification-based colorimetric assay. Anal. Chem. 2012, 84, 9544–9549. [Google Scholar] [CrossRef] [PubMed]
- Seetang-Nun, Y.; Jaroenram, W.; Sriurairatana, S.; Suebsing, R.; Kiatpathomchai, W. Visual detection of white spot syndrome virus using DNA-functionalized gold nanoparticles as probes combined with loop-mediated isothermal amplification. Mol. Cell. Probes 2013, 27, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, X.; Ji, X.; Li, X. Formation of G-quadruplex-hemin DNAzyme based on human telomere elongation and its application in telomerase activity detection. Biosens. Bioelectron. 2011, 26, 4095–4098. [Google Scholar] [CrossRef] [PubMed]
- Suebsing, R.; Prombun, P.; Kiatpathomchai, W. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) combined with colorimetric gold nanoparticle (AuNP) probe assay for visual detection of Penaeus vannamei nodavirus (PvNV). Lett. Appl. Microbial. 2013, 56, 428–435. [Google Scholar] [CrossRef] [PubMed]






| Sequences | ||
|---|---|---|
| EXPAR template | 5′-AACTATACAACCTACTACCTCAAACAGACTCCTGCATTGCCGCAGACTGCATTGCCGCAGAAACAGACTCAAACTATACAACCTACTACCTCAA-P-3′ | The nucleotides of the normal font are the target gene binding sequences; the nucleotides labeled in bold are the complementary sequences to the recognition site and cleavage site of the cleavage enzyme; the italicized nucleotide is a sequence that can produce reporter Y |
| let-7a | 5′-UGAGGUAGUAGGUUGUAUAGUU-3′ | The base of the bold marker is a nucleotide that differs from in let-7a |
| let-7b | 5′-UGAGGUAGUAGGUUGUGUGGUU-3′ | |
| let-7c | 5′-UGAGGUAGUAGGUUGUAUGGUU-3′ | |
| let-7d | 5′-AGAGGUAGUAGGUUGCAUAGU-3′ | |
| let-7e | 5′-UGAGGUAGGAGGUUGUAUAGU-3′ | |
| let-7f | 5′-UGAGGUAGUAGAUUGUAUAGUU-3′ | |
| let-7g | 5′-UGAGGUAGUAGUUUGUACAGU-3′ | |
| let-7i | 5′-UGAGGUAGUAGUUUGUGCUGUU-3′ | |
| DNA probes labeled on AuNP | Au-SH-(CH2)6-TTTTTTTGGGGGGAAAAAAACTGCATTGCCGCAGA-3′ | Boldly labeled nucleotides are reporter Y binding sequences |
| Reporter Y | 5′-TCTGCGGCAATGCAGTCTGCGGCAATGCAG-3′ | Sequences that complementarily bind to AuNPs-DNA probe |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Zhang, B.; Zhang, C.; Guan, Y. A Novel Design Combining Isothermal Exponential Amplification and Gold-Nanoparticles Visualization for Rapid Detection of miRNAs. Int. J. Mol. Sci. 2018, 19, 3374. https://doi.org/10.3390/ijms19113374
Jiang J, Zhang B, Zhang C, Guan Y. A Novel Design Combining Isothermal Exponential Amplification and Gold-Nanoparticles Visualization for Rapid Detection of miRNAs. International Journal of Molecular Sciences. 2018; 19(11):3374. https://doi.org/10.3390/ijms19113374
Chicago/Turabian StyleJiang, Jiquan, Bin Zhang, Chi Zhang, and Yifu Guan. 2018. "A Novel Design Combining Isothermal Exponential Amplification and Gold-Nanoparticles Visualization for Rapid Detection of miRNAs" International Journal of Molecular Sciences 19, no. 11: 3374. https://doi.org/10.3390/ijms19113374
APA StyleJiang, J., Zhang, B., Zhang, C., & Guan, Y. (2018). A Novel Design Combining Isothermal Exponential Amplification and Gold-Nanoparticles Visualization for Rapid Detection of miRNAs. International Journal of Molecular Sciences, 19(11), 3374. https://doi.org/10.3390/ijms19113374