Interactions Between Spermine-Derivatized Tentacle Porphyrins and The Human Telomeric DNA G-Quadruplex
Abstract
:1. Introduction
2. Results and Discussion
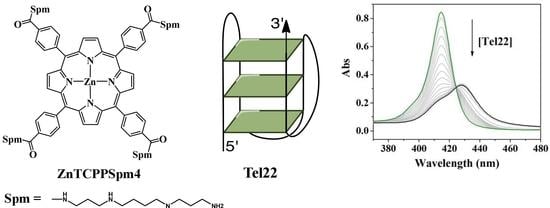
2.1. UV-Vis Spectroscopy Demonstrates that TCPPSpm4 and ZnTCPPSpm4 Bind Tightly to Tel22
2.2. RLS Indicates the Formation of Discrete Stoichiometric Porprhyrin-Tel22 Complexes
2.3. Fluorescence of TCPPSpm4 and ZnTCPPSpm4 Decreases in the Presence of Tel22 Suggesting DNA-Assisted Porphyrin Self-Association
2.4. FRET Studies Indicate that Both Porphyrins Have Exceptional Stabilizing Ability and Modest Selectivity toward Tel22 GQ
2.5. Circular Dichroism (CD) Signal Decreases upon Addition of Porphyrins Signifying Interaction between Porphyrins and Tel22
2.6. The Presence of Induced CD (iCD) Confirms Close Contacts between Porphyrins and Tel22 Aromatic Systems
4. Materials and Methods
4.1. Porphyrins and Oligonucleotides
4.2. UV-Vis Titrations and Job Plot
4.3. Fluorescence Spectroscopy
4.3.1. Resonance Light Scattering (RLS)
4.3.2. Fluorescent Titrations
4.4. Circular Dichroism (CD) Spectroscopy
4.5. Fluorescence Resonance Energy Transfer (FRET) Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GQ | Guanine Quadruplex |
| FRET | Fluorescence Resonance Energy Transfer |
| CD | Circular Dichroism |
| iCD | Induced Circular Dichroism |
| TCPPSpm4 | meso-tetrakis-(4-carboxysperminephenyl)porphyrin |
| ZnTCPPSpm4 | Zn(II) meso-tetrakis-(4-carboxysperminephenyl)porphyrin |
| CT DNA | Calf Thymus DNA |
| F21D | 5′-6-FAM-GGG(TTAGGG)3-Dabcyl-3′ |
References
- Yatsunyk, L.A.; Mendoza, O.; Mergny, J.L. “Nano-oddities”: Unusual nucleic acid assemblies for DNA-based nanostructures and nanodevices. Acc. Chem. Res. 2014, 47, 1836–1844. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability. Met. Ions. Life. Sci. 2016, 16, 203–258. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.H.; Neidle, S. G-quadruplexes and metal ions. Met. Ions. Life Sci. 2012, 10, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedrat, A.; Lacroix, L.; Mergny, J.L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Tseng, T.Y.; Chen, Y.T.; Chang, C.C.; Wang, Z.F.; Wang, C.L.; Hsu, T.N.; Li, P.T.; Chen, C.T.; Lin, J.J.; et al. Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 2015, 43, 10102–10113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, H.; Wang, L.; Liu, Y.; Chen, H.; Li, Q.; Guan, A.; Liu, M.; Tang, Y. Real-time monitoring of DNA G-quadruplexes in living cells with a small-molecule fluorescent probe. Nucleic Acids Res. 2018, 46, 7522–7532. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Carver, M.; Yang, D. Polymorphism of human telomeric quadruplex structures. Biochimie 2008, 90, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2010, 277, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Correia, J.J.; Wang, L.; Trent, J.O.; Chaires, J.B. Not so crystal clear: The structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005, 33, 4649–4659. [Google Scholar] [CrossRef] [PubMed]
- Heddi, B.; Phan, A.T. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011, 133, 9824–9833. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Kan, Z.Y.; Wang, Q.; Yao, Y.; Liu, J.; Hao, Y.H.; Tan, Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007, 129, 11185–11191. [Google Scholar] [CrossRef] [PubMed]
- Nicoludis, J.M.; Barrett, S.P.; Mergny, J.-L.; Yatsunyk, L.A. Interaction of G-quadruplex DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 2012, 40, 5432–5447. [Google Scholar] [CrossRef] [PubMed]
- Nicoludis, J.M.; Miller, S.T.; Jeffrey, P.D.; Barrett, S.P.; Rablen, P.R.; Lawton, T.J.; Yatsunyk, L.A. Optimized end-stacking provides specificity of N-methyl mesoporphyrin IX for human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2012, 134, 20446–20456. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Renciuk, D.; Kejnovska, I.; Skolakova, P.; Bednarova, K.; Motlova, J.; Vorlickova, M. Arrangement of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009, 37, 6625–6634. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef]
- Phan, A.T.; Luu, K.N.; Patel, D.J. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006, 34, 5715–5719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phan, A.T.; Kuryavyi, V.; Luu, K.N.; Patel, D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007, 35, 6517–6525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Noguchi, Y.; Sugiyama, H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006, 14, 5584–5591. [Google Scholar] [CrossRef] [PubMed]
- Luu, K.N.; Phan, A.T.; Kuryavyi, V.; Lacroix, L.; Patel, D.J. Structure of the human telomere in K+ solution: An intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006, 128, 9963–9970. [Google Scholar] [CrossRef] [PubMed]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, K.W.; Amrane, S.; Bouaziz, S.; Xu, W.; Mu, Y.; Patel, D.J.; Luu, K.N.; Phan, A.T. Structure of the human telomere in K+ solution: A stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009, 131, 4301–4309. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Neidle, S. Quadruplex Nucleic Acids as Novel Therapeutic Targets. J. Med. Chem. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ohnmacht, S.A.; Neidle, S. Small-molecule quadruplex-targeted drug discovery. Bioorg. Med. Chem. Lett. 2014, 24, 2602–2612. [Google Scholar] [CrossRef] [PubMed]
- Anantha, N.V.; Azam, M.; Sheardy, R.D. Porphyrin binding to quadrupled T4G4. Biochemistry 1998, 37, 2709–2714. [Google Scholar] [CrossRef] [PubMed]
- Fiel, R.J.; Howard, J.C.; Mark, E.H.; Datta Gupta, N. Interaction of DNA with a porphyrin ligand: Evidence for intercalation. Nucleic Acids Res. 1979, 6, 3093–3118. [Google Scholar] [CrossRef] [PubMed]
- D’Urso, A.; Fragalà, M.E.; Purrello, R. Non-covalent interactions of porphyrinoids with duplex DNA. In Applications of Porphyrinoids; Springer: Berlin/Heidelberg, Germany, 2013; pp. 139–174. [Google Scholar]
- Georgiou, G.N.; Ahmet, M.T.; Houlton, A.; Silver, J.; Cherry, R.J. Measurement of the rate of uptake and subsellular localization of porphyrins in cells using fluorescence digital imaging microscopy. Photochem. Photobiol. 1994, 59, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Benimetskaya, L.; Takle, G.B.; Vilenchik, M.; Lebedeva, I.; Miller, P.; Stein, C.A. Cationic porphyrins: Novel delivery vehicles for antisense oligodeoxynucleotides. Nucleic Acids Res. 1998, 26, 5310–5317. [Google Scholar] [CrossRef] [PubMed]
- Izbicka, E.; Wheelhouse, R.T.; Raymond, E.; Davidson, K.K.; Lawrence, R.A.; Sun, D.; Windle, B.E.; Hurley, L.H.; Von Hoff, D.D. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999, 59, 639–644. [Google Scholar] [PubMed]
- Sabharwal, N.C.; Savikhin, V.; Turek-Herman, J.R.; Nicoludis, J.M.; Szalai, V.A.; Yatsunyk, L.A. N-methylmesoporphyrin IX fluorescence as a reporter of strand orientation in guanine quadruplexes. FEBS J. 2014, 281, 1726–1737. [Google Scholar] [CrossRef] [PubMed]
- Han, F.X.; Wheelhouse, R.T.; Hurley, L.H. Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J. Am. Chem. Soc. 1999, 121, 3561–3570. [Google Scholar] [CrossRef]
- Shi, D.F.; Wheelhouse, R.T.; Sun, D.; Hurley, L.H. Quadruplex-interactive agents as telomerase inhibitors: Synthesis of porphyrins and structure-activity relationship for the inhibition of telomerase. J. Med. Chem. 2001, 44, 4509–4523. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.J.; Ahluwalia, K.; Taylor, S.; Jin, O.; Nicoludis, J.M.; Buscaglia, R.; Chaires, J.B.; Kornfilt, D.J.P.; Marquardt, D.G.S.; Yatsunyk, L.A. Induction of G-quadruplex DNA structure by Zn(II) 5,10,15,20-tetrakis(N-methyl-4-pyridyl)porphyrin. Biochimie 2011, 93, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Sabharwal, N.C.; Mendoza, O.; Nicoludis, J.M.; Ruan, T.; Mergny, J.-L.; Yatsunyk, L.A. Investigation of the interactions between Pt(II) and Pd(II) derivatives of 5,10,15,20-tetrakis (N-methyl-4-pyridyl) porphyrin and G-quadruplex DNA. J. Biol. Inorg. Chem. 2016, 21, 227–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, T.L.; Davis, S.J.; Powell, B.M.; Harbeck, C.P.; Habdas, J.; Habdas, P.; Yatsunyk, L.A. Lowering the overall charge on TMPyP4 improves its selectivity for G-quadruplex DNA. Biochimie 2017, 132, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zhang, S. Interaction between cationic zinc porphyrin and lead ion induced telomeric guanine quadruplexes: Evidence for end-stacking. J. Biol. Inorg. Chem. 2009, 14, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Song, D.; Qin, T.; Yang, C.; Yu, Z.; Li, X.; Liu, K.; Su, H. Interaction between G-Quadruplex and Zinc Cationic Porphyrin: The Role of the Axial Water. Sci. Rep. 2017, 7, 10951. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.; Gaw, H.Y.; Patel, D.J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005, 1, 167. [Google Scholar] [CrossRef] [PubMed]
- Le, V.H.; Nagesh, N.; Lewis, E.A. Bcl-2 promoter sequence G-quadruplex interactions with three planar and non-planar cationic porphyrins: TMPyP4, TMPyP3, and TMPyP2. PLoS ONE 2013, 8, e72462. [Google Scholar] [CrossRef] [PubMed]
- Lubitz, I.; Borovok, N.; Kotlyar, A. Interaction of monomolecular G4-DNA nanowires with TMPyP: Evidence intercalation. Biochemistry 2007, 46, 12925–12929. [Google Scholar] [CrossRef] [PubMed]
- Cavallari, M.; Garbesi, A.; Di Felice, R. Porphyrin intercalation in G4-DNA quadruplexes by molecular dynamics simulations. J. Phys. Chem. B. 2009, 113, 13152–13160. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Wang, L.; Jia, G.; Zhou, J.; Han, G.; Li, C. The binding mode of porphyrins with cation side arms to (TG4T)4 G-quadruplex: Spectroscopic evidence. Biophys. Chem. 2009, 143, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Ghosh, R.; Neidle, S. Structural basis for binding of porphyrin to human telomeres. Biochemistry 2007, 46, 2390–2397. [Google Scholar] [CrossRef] [PubMed]
- McClure, J.E.; Baudouin, L.; Mansuy, D.; Marzilli, L.G. Interactions of DNA with a new electron-deficient tentacle porphyrin: Meso-tetrakis[2,3,5,6-tetrafluoro-4-(2-trimethylammoniumethyl-amine)phenyl]porphyrin. Biopolymers 1997, 42, 203–217. [Google Scholar] [CrossRef]
- Mukundan, N.E.; Petho, G.; Dixon, D.W.; Kim, M.S.; Marzilli, L.G. Interactions of an electron-rich tetracationic tentacle porphyrin with calf thymus DNA. Inorg. Chem. 1994, 33, 4676–4687. [Google Scholar] [CrossRef]
- Mukundan, N.E.; Petho, G.; Dixon, D.W.; Marzilli, L.G. DNA-tentacle porphyrin interactions: AT over GC selectivity exhibited by an outside binding self-stacking porphyrin. Inorg. Chem. 1995, 34, 3677–3687. [Google Scholar] [CrossRef]
- Marzilli, L.G.; Petho, G.; Lin, M.; Kim, M.S.; Dixon, D.W. Tentacle porphyrins: DNA interactions. J. Am. Chem. Soc. 1992, 114, 7575–7577. [Google Scholar] [CrossRef]
- Thomas, T.J.; Tajmir-Riahi, H.A.; Thomas, T. Polyamine-DNA interactions and development of gene delivery vehicles. Amino Acids. 2016, 48, 2423–2431. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.J.; Thomas, T. Collapse of DNA in packaging and cellular transport. Int. J. Biol. Macromol. 2018, 109, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Ouameur, A.A.; Tajmir-Riahi, H.-A. Structural analysis of DNA interactions with biogenic polyamines and cobalt(III)hexamine studied by fourier transform infrared and capillary electrophoresis. J. Biol. Chem. 2004, 279, 42041–42054. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, A.; Hawken, M.; Hall, M.; Sanders, K.J.; Rodger, A. Amine induced Z-DNA in poly (dG-dC)· poly (dG-dC): Circular dichroism and gel electrophoresis study. Phys. Chem. Chem. Phys. 2000, 2, 5469–5478. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L.; Teulade-Fichou, M.-P.; Hounsou, C.; Guittat, L.; Hoarau, M.; Arimondo, P.B.; Vigneron, J.-P.; Lehn, J.-M.; Riou, J.-F.; et al. Telomerase inhibitors based on quadruplex ligands selected by a fluorescence assay. Proc. Natl. Acad. Sci. USA 2001, 98, 3062–3067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Huang, J.; Zhang, M.; Zhou, Y.; Zhang, D.; Wu, Z.; Wang, S.; Weng, X.; Zhou, X.; Yang, G. Bis(benzimidazole)pyridine derivative as a new class of G-quadruplex inducing and stabilizing ligand. Chem. Commun. 2008, 4564–4566. [Google Scholar] [CrossRef] [PubMed]
- Collie, G.W.; Promontorio, R.; Hampel, S.M.; Micco, M.; Neidle, S.; Parkinson, G.N. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 2012, 134, 2723–2731. [Google Scholar] [CrossRef] [PubMed]
- Guyen, B.; Schultes, C.M.; Hazel, P.; Mann, J.; Neidle, S. Synthesis and evaluation of analogues of 10H-indolo[3,2-b]quinoline as G-quadruplex stabilizing ligands and potential inhibitors of the enzyme telomerase. Org. Biomol. Chem. 2004, 2, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Schultes, C.M.; Guyen, B.; Cuesta, J.; Neidle, S. Synthesis, biophysical and biological evaluation of 3,6-bis-amidoacridines with extended 9-anilino substituents as potent G-quadruplex-binding telomerase inhibitors. Bioorg. Med. Chem. Letters 2004, 14, 4347–4351. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Mandal, A.; Johansson, H.E.; Orjalo, A.V.; Park, M.H. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 2169–2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerner, E.W.; Meyskens, F.L. Polyamines and cancer: Old molecules, new understanding. Nat. Rev. Cancer 2004, 4, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E.; Casero, R.A. Current status of the polyamine research field. Methods Mol. Biol. 2011, 720, 3–35. [Google Scholar] [CrossRef] [PubMed]
- Carlisle, D.L.; Devereux, W.L.; Hacker, A.; Woster, P.M.; Casero, R.A. Growth status significantly affects the response of human lung cancer cells to antitumor polyamine-analogue exposure. J. Clin. Cancer Res. 2002, 8, 2684–2689. [Google Scholar]
- Cullis, P.M.; Green, R.E.; Merson-Davies, L.; Travis, N. Probing the mechanism of transport and compartmentalisation of polyamines in mammalian cells. Nat. Chem. Biol. 1999, 717–729. [Google Scholar] [CrossRef]
- Wang, C.; Delcros, J.-G.; Biggerstaff, J.; Phanstiel, O.I. Synthesis and biological evaluation of N1-(anthracen-9-ylmethyl)triamines as molecular recognition elements for the polyamine transporter. J. Med. Chem. 2003, 2663–2671. [Google Scholar] [CrossRef] [PubMed]
- Gangemi, C.M.A.; Randazzo, R.; Fragala, M.E.; Tomaselli, G.A.; Ballistreri, F.P.; Pappalardo, A.; Toscano, R.M.; Sfrazzetto, G.T.; Purrello, R.; D’Urso, A. Hierarchically controlled protonation/aggregation of a porphyrin–spermine derivative. New J. Chem. 2015, 39, 6722–6725. [Google Scholar] [CrossRef]
- D’Urso, A.; Randazzo, R.; Rizzo, V.; Gangemi, C.; Romanucci, V.; Zarrelli, A.; Tomaselli, G.; Milardi, D.; Borbone, N.; Purrello, R. Stabilization vs. destabilization of G-quadruplex superstructures: The role of the porphyrin derivative having spermine arms. Phys. Chem. Chem. Phys. 2017, 19, 17404–17410. [Google Scholar] [CrossRef] [PubMed]
- Pasternack, R.F.; Briganid, R.A.; Abrams, M.J.; Williams, A.P.; Gibbs, E.J. Interactions of porphyrins and metalloporphyrins with single-stranded poly(dA). Inorg. Chem. 1990, 29, 4483–4486. [Google Scholar] [CrossRef]
- Job, P. Formation and Stability of Inorganic Complexes in Solution. Annali di Chimica Applicata 1928, 9, 113–203. [Google Scholar]
- Huang, C.Y. Determination of binding stoichiometry by the continuous variation method: The Job plot. Methods Enzymol. 1982, 87, 509–525. [Google Scholar] [PubMed]
- Boschi, E.; Davis, S.; Taylor, S.; Butterworth, A.; Chirayath, L.A.; Purohit, V.; Siegel, L.K.; Buenaventura, J.; Sheriff, A.H.; Jin, R.; et al. Interaction of a Cationic Porphyrin and Its Metal Derivatives with G-Quadruplex DNA. J. Phys. Chem. B. 2016, 120, 12807–12819. [Google Scholar] [CrossRef] [PubMed]
- Pasternack, R.; Collings, P. Resonance light scattering: A new technique for studying chromophore aggregation. Science 1995, 269, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.M.; Tossi, A.B.; McConnell, D.J.; OhUigin, C. A study of the interactions of some polypyridylruthenium (II) complexes with DNA using fluorescence spectroscopy, topoisomerisation and thermal denaturation. Nucleic Acids Res. 1985, 13, 6017–6034. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.M.; Murphy, M.J.; McConnell, D.J.; OhUigin, C. A comparative study of the interaction of 5,10,15,20-tetrakis (N-methylpyridinium-4-yl)porphyrin and its zinc complex with DNA using fluorescence spectroscopy and topoisomerisation. Nucleic Acids Res. 1985, 13, 167–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Cian, A.; Guittat, L.; Kaiser, M.; Saccà, B.; Amrane, S.; Bourdoncle, A.; Alberti, P.; Teulade-Fichou, M.-P.; Lacroix, L.; Mergny, J.-L. Fluorescence-based melting assays for studying quadruplex ligands. Methods 2007, 42, 183–195. [Google Scholar] [CrossRef] [PubMed]
- DuPont, J.I.; Henderson, K.L.; Metz, A.; Le, V.H.; Emerson, J.P.; Lewis, E.A. Calorimetric and spectroscopic investigations of the binding of metallated porphyrins to G-quadruplex DNA. Biochim. Biophys. Acta. 2016, 1860, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.J.; Wingate, K.L.; Silwal, J.; Leeper, T.C.; Basu, S. The porphyrin TMPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res. 2012, 40, 4137–4145. [Google Scholar] [CrossRef] [PubMed]
- Waller, Z.A.E.; Sewitz, S.A.; Hsu, S.-T.D.; Balasubramanian, S. A small molecule that disrupts G-quadruplex DNA structure and enhances gene expression. J. Am. Chem. Soc. 2009, 131, 12628–12633. [Google Scholar] [CrossRef] [PubMed]
- Kaluzhny, D.; Ilyinsky, N.; Shchekotikhin, A.; Sinkevich, Y.; Tsvetkov, P.O.; Tsvetkov, V.; Veselovsky, A.; Livshits, M.; Borisova, O.; Shtil, A.; et al. Disordering of human telomeric G-quadruplex with novel antiproliferative anthrathiophenedione. PLoS ONE 2011, 6, e27151. [Google Scholar] [CrossRef] [PubMed]
- Marchand, A.; Granzhan, A.; Iida, K.; Tsushima, Y.; Ma, Y.; Nagasawa, K.; Teulade-Fichou, M.-P.; Gabelica, V. Ligand-induced conformational changes with cation ejection upon binding to human telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2015, 137, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Pasternack, R.F. Circular dichroism and the interactions of water soluble porphyrins with DNA—A minireview. Chirality 2003, 15, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Pasternack, R.F.; Gibbs, E.J.; Villafranca, J.J. Interactions of porphyrins with nucleic acids. Biochemistry 1983, 22, 2406–2414. [Google Scholar] [CrossRef] [PubMed]
- Gangemi, C.M.; D’Agostino, B.; Randazzo, R.; Gaeta, M.; Fragalà, M.E.; Purrello, R.; D’Urso, A. Interaction of spermine derivative porphyrin with DNA. J. Porphyr. Phthalocyanines 2018, 2, 1–7. [Google Scholar] [CrossRef]
- Gangemi, C.M.A.; D’Urso, A.; Tomaselli, G.A.; Berova, N.; Purrello, R. A novel porphyrin-based molecular probe ZnTCPPSpm4 with catalytic, stabilizing and chiroptical diagnostic power towards DNA B-Z transition. J. Inorg. Biochem. 2017, 173, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Tataurov, A.V.; You, Y.; Owczarzy, R. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 2008, 133, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Cantor, C.R.; Warshaw, M.M.; Shapiro, H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers 1970, 9, 1059–1077. [Google Scholar] [CrossRef] [PubMed]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabharwal, N.C.; Chen, J.; Lee, J.H.; Gangemi, C.M.A.; D'Urso, A.; Yatsunyk, L.A. Interactions Between Spermine-Derivatized Tentacle Porphyrins and The Human Telomeric DNA G-Quadruplex. Int. J. Mol. Sci. 2018, 19, 3686. https://doi.org/10.3390/ijms19113686
Sabharwal NC, Chen J, Lee JH, Gangemi CMA, D'Urso A, Yatsunyk LA. Interactions Between Spermine-Derivatized Tentacle Porphyrins and The Human Telomeric DNA G-Quadruplex. International Journal of Molecular Sciences. 2018; 19(11):3686. https://doi.org/10.3390/ijms19113686
Chicago/Turabian StyleSabharwal, Navin C., Jessica Chen, Joo Hyun (June) Lee, Chiara M. A. Gangemi, Alessandro D'Urso, and Liliya A. Yatsunyk. 2018. "Interactions Between Spermine-Derivatized Tentacle Porphyrins and The Human Telomeric DNA G-Quadruplex" International Journal of Molecular Sciences 19, no. 11: 3686. https://doi.org/10.3390/ijms19113686
APA StyleSabharwal, N. C., Chen, J., Lee, J. H., Gangemi, C. M. A., D'Urso, A., & Yatsunyk, L. A. (2018). Interactions Between Spermine-Derivatized Tentacle Porphyrins and The Human Telomeric DNA G-Quadruplex. International Journal of Molecular Sciences, 19(11), 3686. https://doi.org/10.3390/ijms19113686