Gramicidin Lateral Distribution in Phospholipid Membranes: Fluorescence Phasor Plots and Statistical Mechanical Model
Abstract
1. Introduction
2. Results
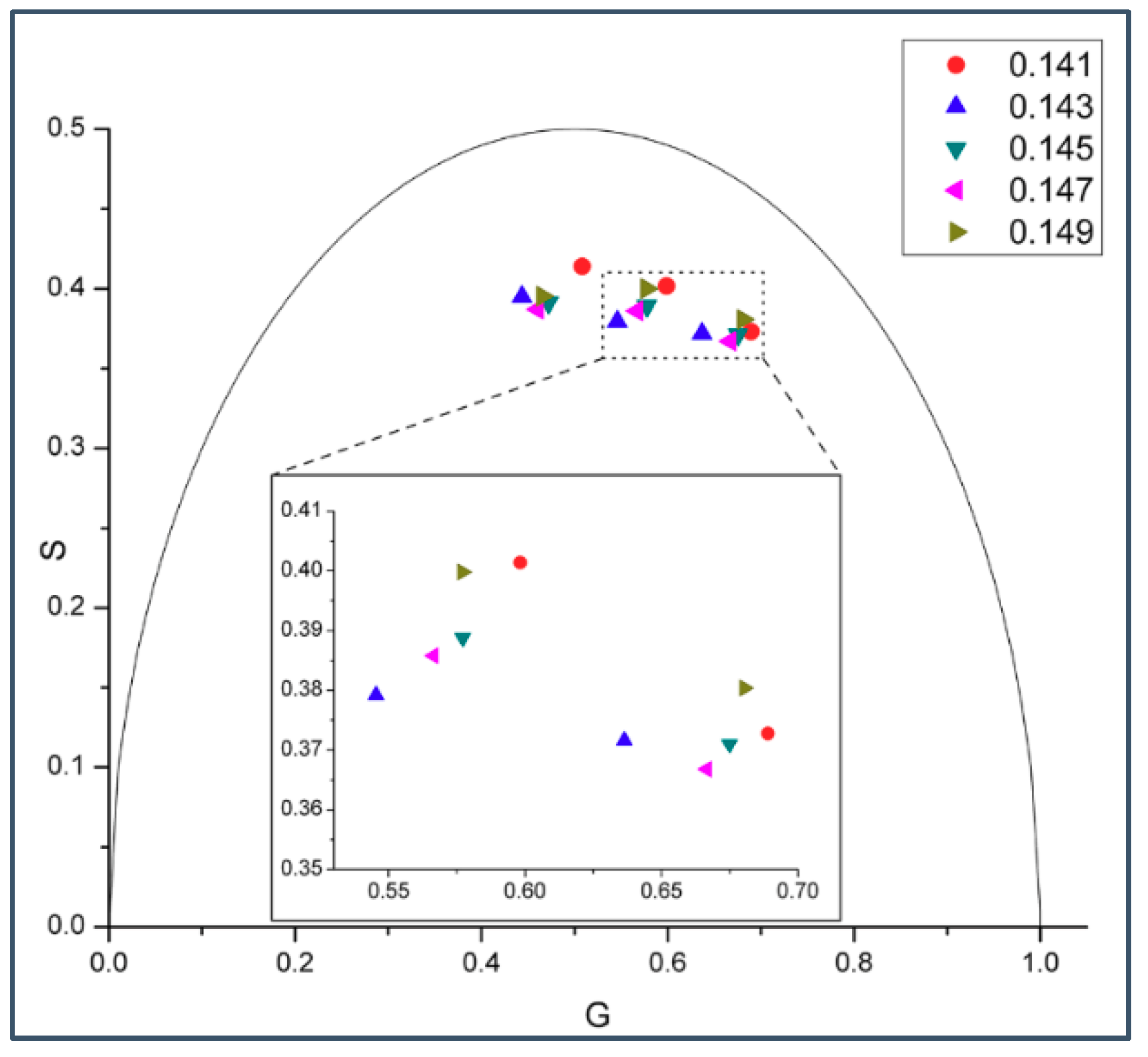
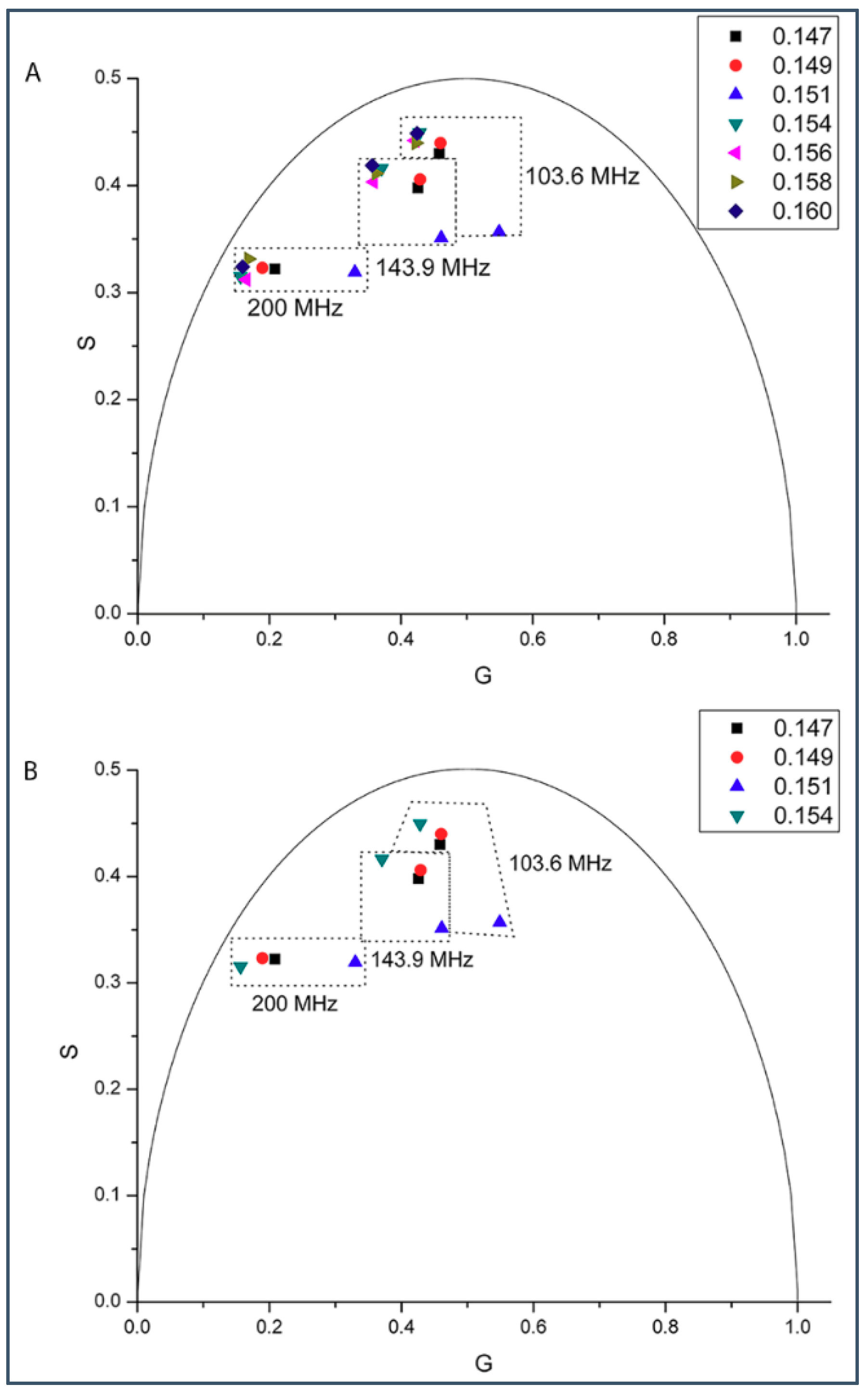
2.1. Phasor Plots of Intrinsic Protein Fluorescence in Gramicidins/DMPC Mixtures
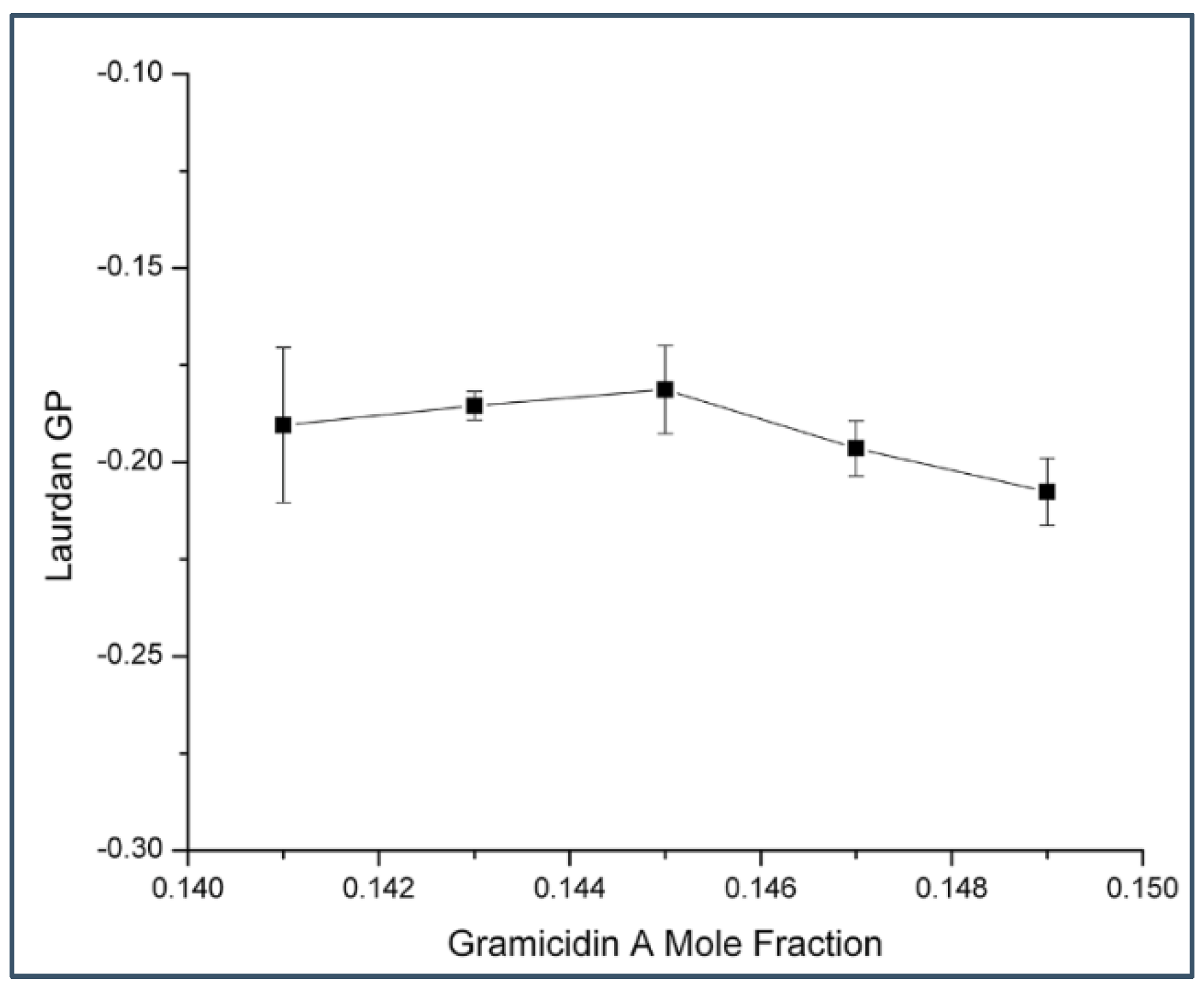
2.2. Generalized Polarization of Laurdan Fluorescence in Gramicidin A/DMPC Mixtures
2.3. Model
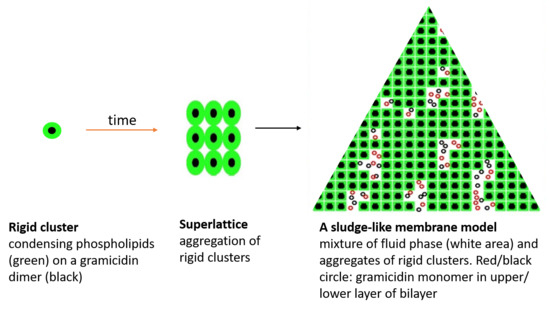
2.3.1. On the Condensing Effect of Gramicidin
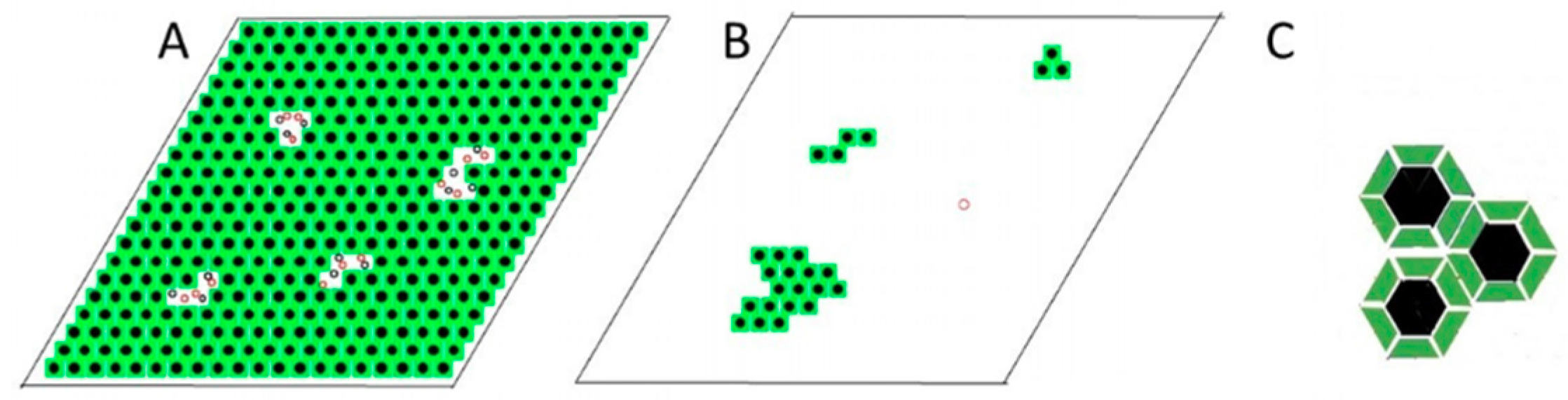
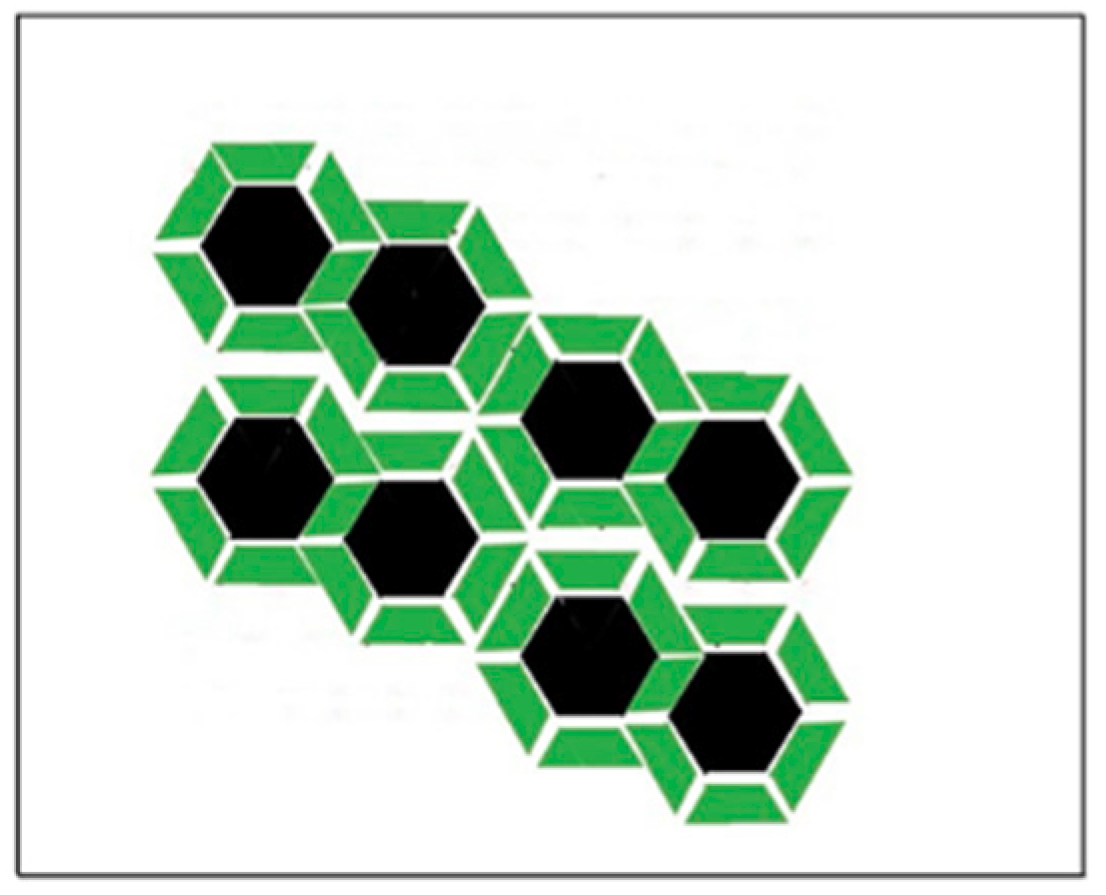
2.3.2. Modeling Gramicidin/Phospholipid Mixture—Qualitative Description
2.3.3. Modeling Gramicidin/Phospholipid Mixture—Statistical Mechanical Description
Calculating
Free Energy of the Lattice
On the Solubility Limit of Gramicidin
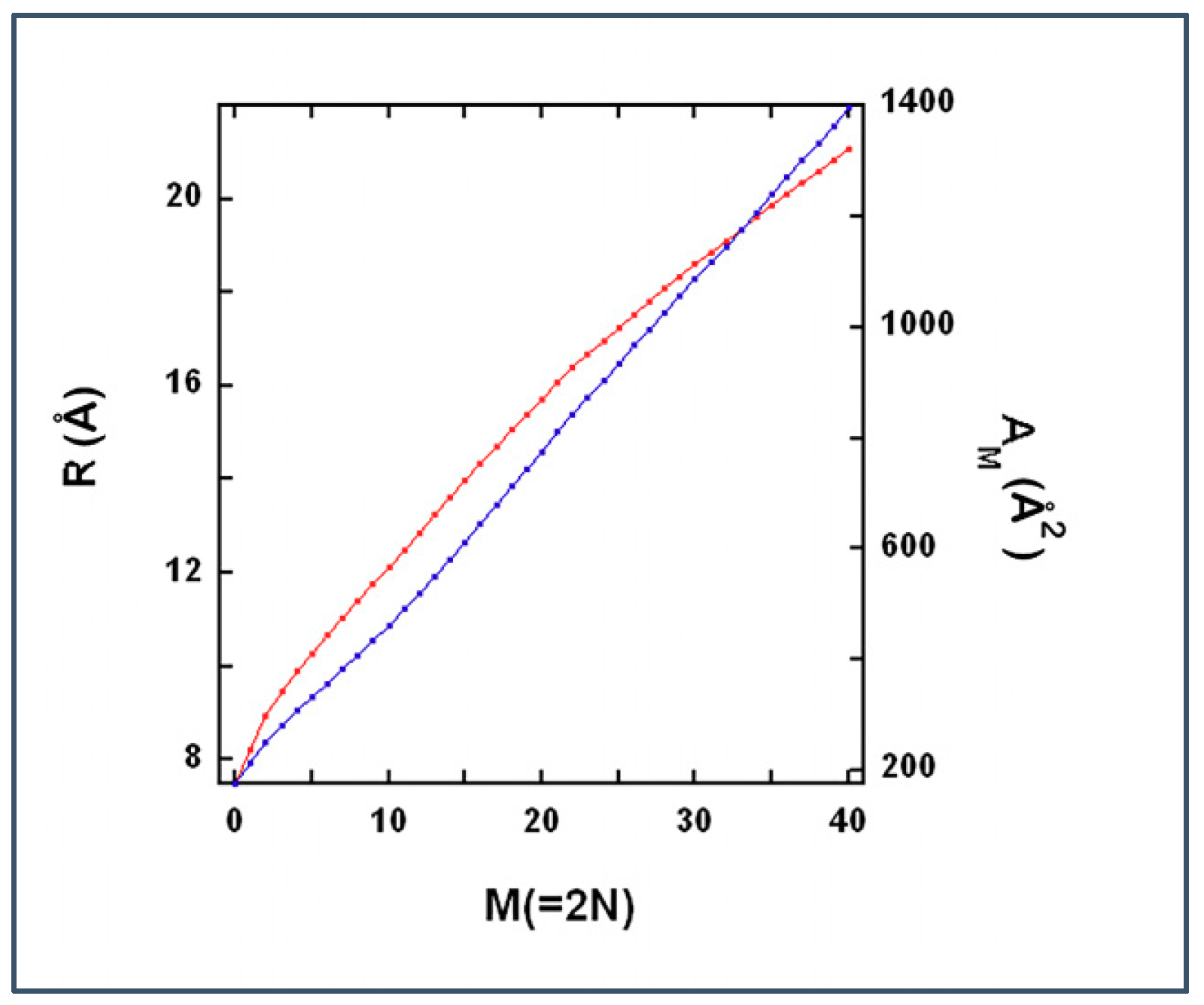
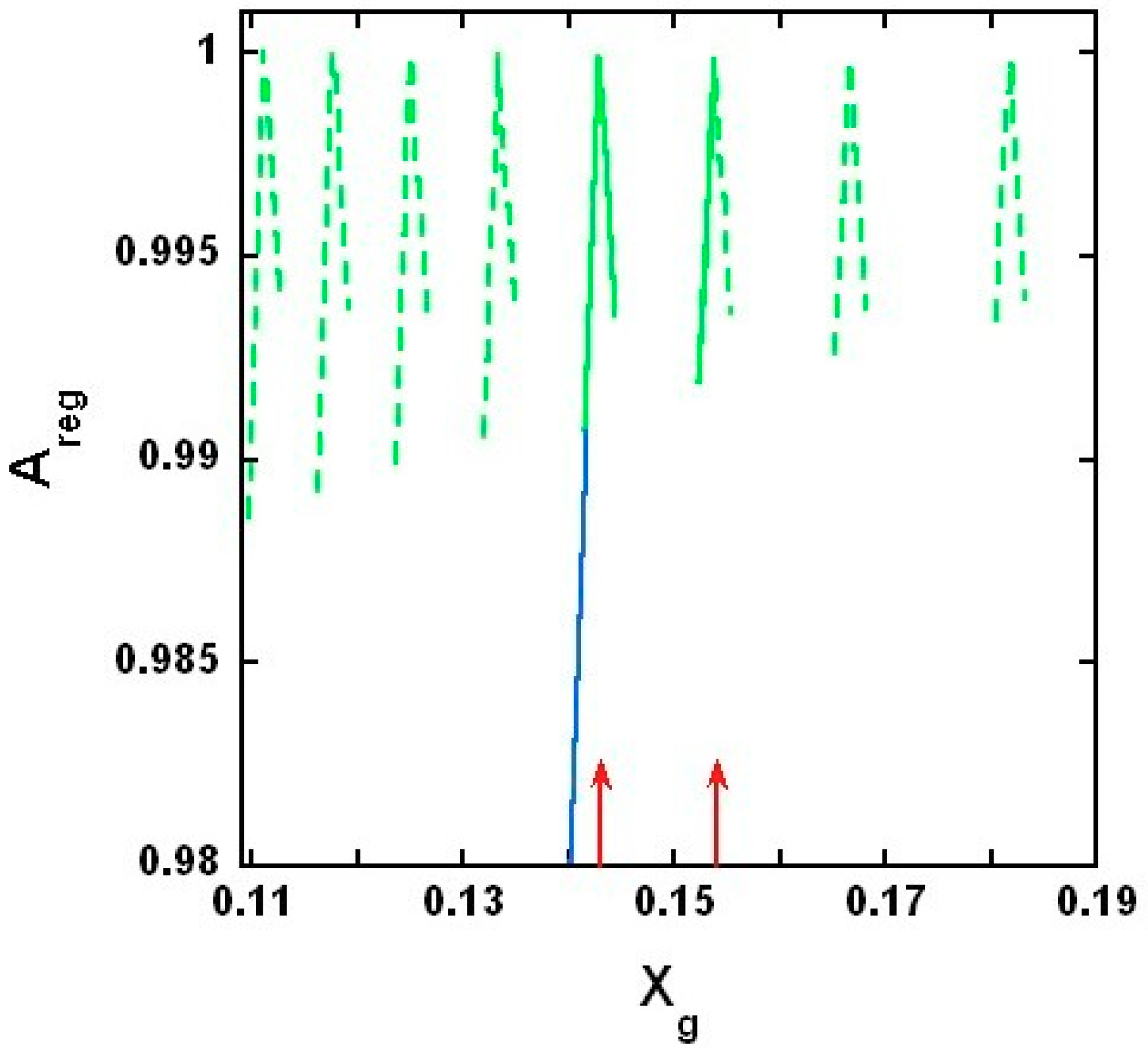
2.3.4. Results of the Theoretical Model
3. Discussion
3.1. On Measured and Predicted Critical Mole Fractions
3.2. On the Upper and Lower Limit of Critical Mole Fractions
3.3. Comparing the Results of the Model with Other Experimental Data
3.4. Similarities and Differences between Gramicidin/DMPC and Cholesterol/DMPC Mixtures
3.5. On Gramicidin/DMPE Mixtures
3.6. Biophysical and Functional Implications
4. Materials and Methods
4.1. Preparation of Gramicidin/DMPC Mixtures
4.2. Fluorescence Lifetime Measurements
4.3. Measurements of Generalized Polarization (GP) of Laurdan Fluorescence
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Andersen, O.S. Gramicidin Channels. Ann. Rev. Physiol. 1984, 46, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.A. Structure of Gramicidine A. Biophys. J. 1986, 49, 295–304. [Google Scholar] [CrossRef]
- Ketchem, R.R.; Hu, W.; Cross, T.A. High-Resolution Conformation of Gramicidin A in a Lipid Bilayer by Solid-State NMR. Science 1993, 261, 1457–1460. [Google Scholar] [CrossRef] [PubMed]
- Ketchem, R.R.; Lee, K.C.; Huo, S.; Cross, T.A. Macromolecular Structural Elucidation with Solid-State NMR-Derived Orientational Constraint. J. Biomol. NMR 1996, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ketchem, R.R.; Roux, B.; Cross, T.A. High-Resolution Polypeptide Structure in a Lamellar Phase Lipid Environment from Solid State NMR Derived Orientational Constraints. Structure 1997, 5, 1655–1669. [Google Scholar] [CrossRef]
- Bouchard, M.; Auger, M. Solvent History Dependence of Gramicidin-Lipid Interactions: A Raman and Infrared Spectroscopic Study. Biophys. J. 1993, 65, 2484–2492. [Google Scholar] [CrossRef]
- Kelkar, D.A.; Chattopadhyay, A. The Gramicidin Ion Channel: A Model Membrane Protein. Biochim. Biophys. Acta 2007, 1768, 2011–2025. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Sampaio, J.L. Membrane Organization and Lipid Rafts. Cold Spring Harb. Perspect. Biol. 2011, 3, a004697. [Google Scholar] [CrossRef] [PubMed]
- Doeven, M.K.; Folgering, J.H.A.; Krasnikov, V.; Geertsma, E.R.; van den Bogaart, G.; Poolman, B. Distribution, Lateral Mobility and Function of Membrane Proteins Incorporated into Giant Unilamellar Vesicles. Biophys. J. 2005, 88, 1134–1142. [Google Scholar] [CrossRef] [PubMed]
- Chapman, D.; Cornell, B.A.; Eliasz, A.W.; Perry, A. Interactions of Helical Polypeptide Segments Which Span the Hydrocarbon Region of Lipid Bilayers. Studies of the Gramicidin A Lipid-Water System. J. Mol. Biol. 1977, 113, 517–538. [Google Scholar] [CrossRef]
- Kota, Z.; Pali, T.; Marsh, D. Orientation and Lipid-Peptide Interactions of Gramicidin A in Lipid Membranes: Polarized Attenuated Total Reflection Infrared Spectroscopy and Spin-Labeled Electron Spin Resonance. Biophys. J. 2004, 86, 1521–1531. [Google Scholar] [CrossRef]
- Oradd, G.; Lindblom, G. NMR Studies of Lipid Lateral Diffusion in the DMPC. Gramicidin D/water System: Peptide Aggregation and Obstruction Effects. Biophys. J. 2004, 87, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Chiu, S.; Subramaniam, S.; Jakobsson, E. Simulation Study of a Gramicidin/lipid Bilayer System in Excess Water and Lipid. I. Structure of the Molecular Complex. Biophys. J. 1999, 76, 1929–1938. [Google Scholar] [CrossRef]
- Kim, T.; Lee, K.I.; Morris, P.; Pastor, R.W.; Andersen, O.S.; Im, W. Influence of Hydrophobic Mismatch on Structures and Dynamics of Gramicidin A and Lipid Bilayers. Biophys. J 2012, 102, 1551–1560. [Google Scholar] [CrossRef] [PubMed]
- Rokitskaya, T.I.; Antonenko, Y.N.; Kotova, E.A. Photodynamic inactivation of Gramicidin Channels: A Flash-Photolysis Study. Biochim. Biophys. Acta 1996, 1275, 116–221. [Google Scholar] [CrossRef]
- Harroun, T.A.; Heller, W.T.; Weiss, T.M.; Yang, L.; Huang, H.W. Experimental Evidence for Hydrophobic Matching and Membrane Mediated Interactions in Lipid Bilayers Containing Gramicidin. Biophys. J. 1999, 76, 937–945. [Google Scholar] [CrossRef]
- Sugar, I.P.; Chong, P.L.-G. A Statistical Mechanical Model of cholesterol/phospholipid Mixtures: Linking Condensed Complexes, Superlattices, and the Phase Diagram. J. Am. Chem. Soc. 2012, 134, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.L.-G. Evidence for Regular Distribution of Sterols in Liquid Crystalline Phosphatidylcholine Bilayers. Proc. Natl. Acad. Sci. USA 1994, 91, 10069–10073. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.L.-G.; Zhu, W.; Venegas, B. On the Lateral Structure of Model Membranes Containing Cholesterol. Biochim. Biophys. Acta 2009, 1788, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Somerharju, P.; Virtanen, J.A.; Cheng, K.H.; Hermansson, M. The Superlattice Model of Lateral Organization of Membranes and its Implications on Membrane Lipid Homeostasis. Biochim. Biophys. Acta 2009, 1788, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Štefla, M.; Jamesb, N.G.; Rossb, J.A.; Jameson, D.M. Applications of Phasors to in Vitro Time-Resolved Fluorescence Measurements. Anal. Biochem. 2011, 410, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.L.-G.; Tang, D.; Sugar, I.P. Exploration of Physical Principles Underlying Lipid Regular Distribution: Effects of Pressure, Temperature, and Radius of Curvature on E/M Dips in Pyrene-Labeled PC/DMPC Binary Mixtures. Biophys. J. 1994, 66, 2029–2038. [Google Scholar] [CrossRef]
- Jameson, D.M.; Gratton, E.; Hall, R. The Measurement and Analysis of Heterogeneous Emissions by Multifrequency Phase and Modulation Fluorometry. Appl. Spectrosc. Rev. 1984, 20, 55–106. [Google Scholar] [CrossRef]
- Venegas, B.; Sugar, I.P.; Chong, P.L.-G. Critical Factors for Detection of Biphasic Changes in Membrane Properties at Specific Sterol Mole Fractions for Maximal Superlattice Formation. J. Phys. Chem. B 2007, 111, 5180–5192. [Google Scholar] [CrossRef] [PubMed]
- Nagle, J.F.; Tristram-Nagle, S. Structure of Lipid Bilayers. Biochim. Biophys. Acta 2000, 1469, 159–195. [Google Scholar] [CrossRef]
- Ipsen, J.H.; Karlstrom, G.; Mouritsen, O.G.; Wennerstrom, H.; Zuckermann, M.J. Phase Equilibria in the Phosphatidylcholine-Cholesterol System. Biochim. Biophys. Acta 1987, 905, 162–172. [Google Scholar] [CrossRef]
- McConnell, H.M.; Radhakrishnan, A. Condensed Complexes of Cholesterol and Phospholipids. Biochim. Biophys. Acta 2003, 1610, 159–173. [Google Scholar] [CrossRef]
- Huang, K. Statistical Mechanics; Wiley, John & Sons: New York, NY, USA, 1990. [Google Scholar]
- Vilfan, I. Lecture Notes in Statistical Mechanics; The Abdus Salam ICTP: Trieste, Italy; The J. Stefan Institute: Ljubljana, Slovenia, 2002; p. 73. [Google Scholar]
- Hill, T.L. Statistical Mechanics: Principles and Selected Applications; Dover: New York, NY, USA, 1987. [Google Scholar]
- Jameson, D.M. Introduction to Fluorescence; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Liu, F.; Sugar, I.P.; Chong, P.L.-G. Cholesterol and Ergosterol Superlattices in Three-Component Liquid Crystalline Lipid Bilayers as Revealed by Dehydroergosterol Fluorescence. Biophys. J. 1997, 72, 2243–2254. [Google Scholar] [CrossRef]
- Parasassi, T.; Di Stefano, M.; Loiero, M.; Ravagnan, G.; Gratton, E. Influence of Cholesterol on Phospholipid Bilayers Phase Domains as Detected by Laurdan Fluorescence. Biophys. J. 1994, 66, 120–132. [Google Scholar] [CrossRef]
- Bagatolli, L.A. LAURDAN Fluorescence Properties in Membranes: A Journey from the Fluorometer to the Microscope; Mely, Y., Duportail, G., Hof, M., Eds.; Fluorescent Methods to study Biological Membranes; Springer: New York, NY, USA, 2013; pp. 3–36. [Google Scholar]
- Berkowitz, M.L. Detailed Molecular Dynamics Simulations of Model Biological Membranes Containing Cholesterol. Biochim. Biophys. Acta—Biomembr. 2009, 1788, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Koeppe, R.E., II; Vogt, T.C.B.; Greathouse, D.V.; Killian, J.A.; De Kruijff, B. Conformation of the Acylation Site of Palmitoylgramicidin in Lipid Bilayers of Dimyristoylphosphatidylcholine. Biochemistry 1996, 35, 3641–3648. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.; Escrive, C.; Laguerre, M.; Pebay-Peyroula, E.; Neri, W.; Pott, T.; Katsaras, J.; Dufourc, E.J. Location of Cholesterol in DMPC Membranes. A Comparative Study by Neutron Diffraction and Molecular Mechanics Simulation. Langmuir 2001, 17, 2019–2030. [Google Scholar] [CrossRef]
- Robinson, A.L.; Richards, W.G.; Thomas, P.J.; Hann, M.M. Behavior of Cholesterol and its Effect on Head Group and Chain Conformations in Lipid Bilayers: A Molecular Dynamics Study. Biophys. J. 1995, 68, 164–170. [Google Scholar] [CrossRef]
- Chaudhuri, A.; Chattopadhyay, A. Transbilayer Organization of Membrane Cholesterol at Low Concentrations: Implications in Health and Disease. Biochim. Biophys. Acta 2011, 1808, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Lazo, N.D.; Hu, W.; Cross, T.A. Low-Temperature Solid-State 15N NMR Characterization of Polypeptide Samples for Characterization of High Definition Dynamics by Solid-State NMR Spectroscopy. J. Magn. Reson. 1995, 107B, 43–50. [Google Scholar] [CrossRef]
- Killian, J.A.; de Kruijff, B. Thermodynamic, Motional, and Structural Aspects of Gramicidin-Induced Hexagonal HII Formation in Phosphatidylethanolamine. Biochemistry 1985, 24, 7881–7890. [Google Scholar] [CrossRef] [PubMed]
- Michonova-Alexova, E.I.; Sugar, I.P. Component and Phase Separation in DMPC/DSPC Lipid Bilayers: A Monte Carlo Simulation study. Biophys. J. 2002, 83, 1820–1833. [Google Scholar] [PubMed]
- Liou, J.; Hung, Y.; Yang, C.; Chen, Y. The Antimicrobial Activity of Gramicidin A is Associated with Hydroxyl Radical Formation. PLoS ONE 2015, 10, e0117065. [Google Scholar] [CrossRef] [PubMed]
- Olsher, M.; Yoon, S.-I.; Chong, P.L.-G. Role of Sterol Superlattice in Free Radical-Induced Sterol Oxidation in Lipid Membranes. Biochemistry 2005, 44, 2080–2087. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chong, P.L.-G. Evidence for a Regulatory Role of Cholesterol Superlattices in the Hydrolytic Activity of Secretory Phospholipase A2 in Lipid Membranes. Biochemistry 1999, 38, 3867–3873. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.M.; Olsher, M.; Sugar, I.P.; Chong, P.L.-G. Cholesterol Superlattice Modulates the Activity of Cholesterol Oxidase in Lipid Membranes. Biochemistry 2004, 43, 2159–2166. [Google Scholar] [CrossRef] [PubMed]
- Gomma, A.I.; Martinent, C.; Hammami, R.; Fliss, I.; Subirade, M. Dual Coating of Liposomes as Encapsulating Matrix of Antimicrobial Peptides: Development and Characterization. Front. Chem. 2017, 5, 103. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, D.A.; Chattopadhyay, A. Modulation of Gramicidin Channel Conformation and Organization by Hydrophobic Mismatch in Saturated Phosphatidylcholine Bilayers. Biochim. Biophys. Acta—Biomembr. 2007, 1768, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, G.R. Phosphorus Assay in Column Chromatography. J. Biol. Chem. 1959, 234, 466–468. [Google Scholar] [PubMed]
- Chong, P.L.-G.; Olsher, M. Fluorometric Assay for Detection of Sterol Oxidation in Liposomal Membranes. Methods Mol. Biol. 2007, 400, 145–158. [Google Scholar] [PubMed]
- Chong, P.L.-G.; Venegas, B.; Olsher, M. Fluorescence Detection of Signs of Sterol Superlattice Formation in Lipid Membranes. Methods Mol. Biol. 2007, 400, 159–170. [Google Scholar] [PubMed]
- James, N.G.; Ross, J.A.; Štefl, M.; Jameson, D.M. Applications of Phasor Plots to in Vitro Protein Studies. Anal. Biochem. 2011, 410, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.L.S.; Araujo, A.P.U.; Jameson, D.M. Investigation of the Conformational Flexibility of DGAT1 Peptides using Tryptophan Fluorescence. Methods Appl. Fluoresc. 2015, 3, 025003. [Google Scholar] [CrossRef] [PubMed]
- Weber, G. Resolution of the Fluorescence Lifetime in a Heterogeneous System by Phase and Modulation Measurements. J. Phys. Chem. 1981, 85, 949–953. [Google Scholar] [CrossRef]
- Bagatolli, L.A.; Gratton, E. Two-Photon Fluorescence Microscopy Observation of Shape Changes at the Phase Transition in Phospholipid Giant Unilamellar Vesicles. Biophys. J. 1999, 77, 2090–2101. [Google Scholar] [CrossRef]
- Parasassi, T.; De Stasio, G.; Ravagnan, G.; Rusch, R.M.; Gratton, E. Quantitation of Lipid Phases in Phospholipid Vesicles by the Generalized Polarization of Laurdan Fluorescence. Biophys. J. 1991, 60, 179–189. [Google Scholar] [CrossRef]
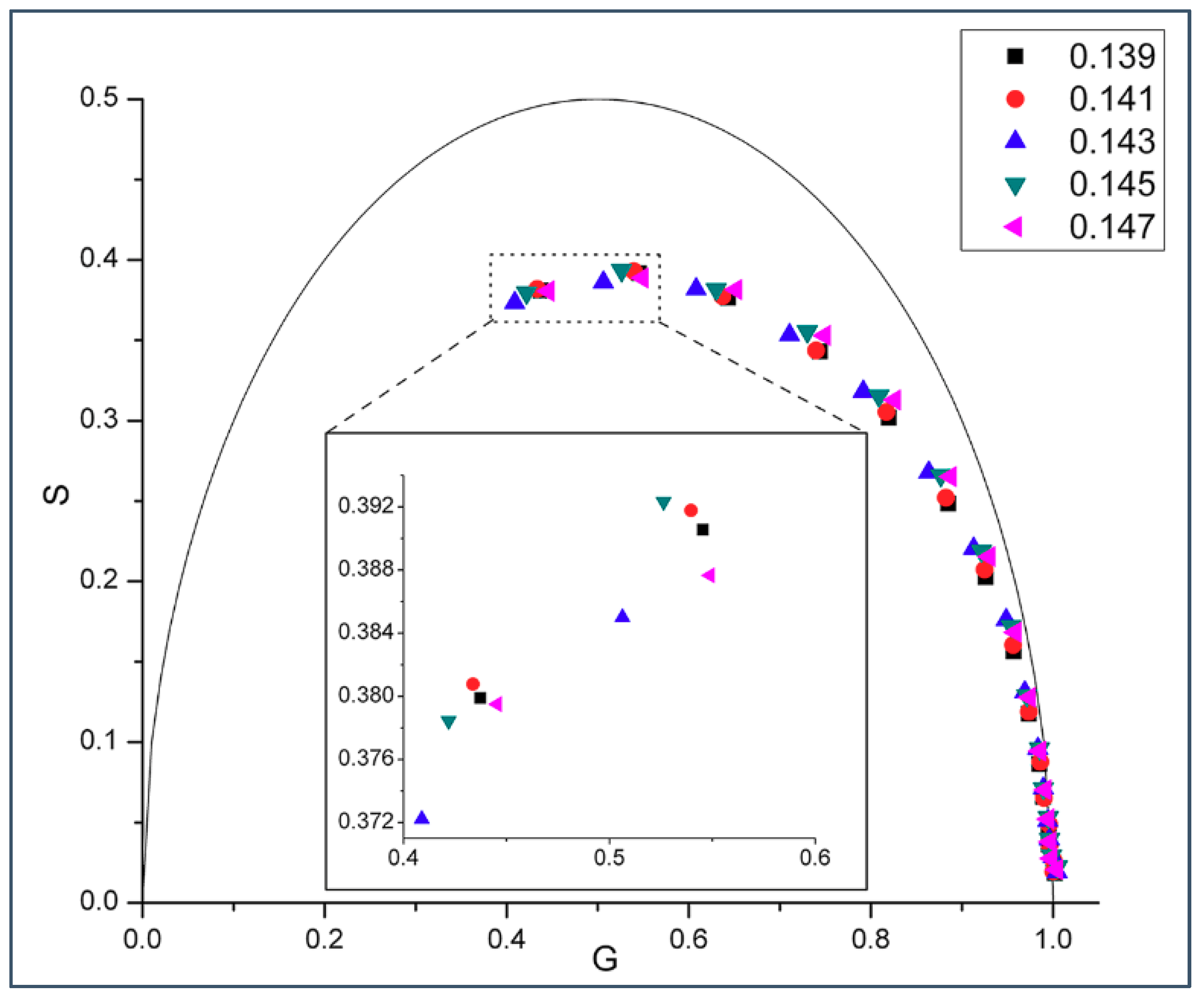
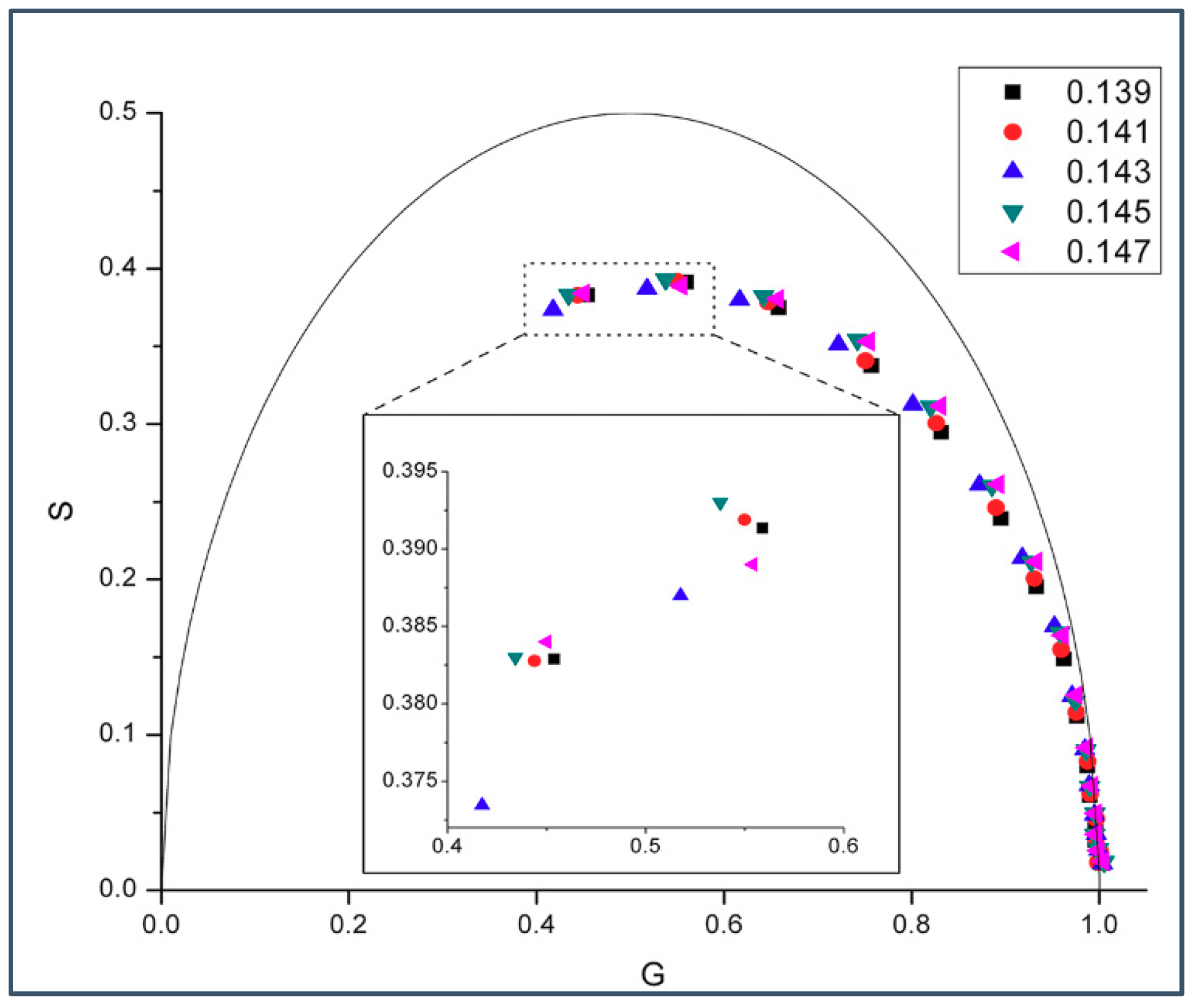
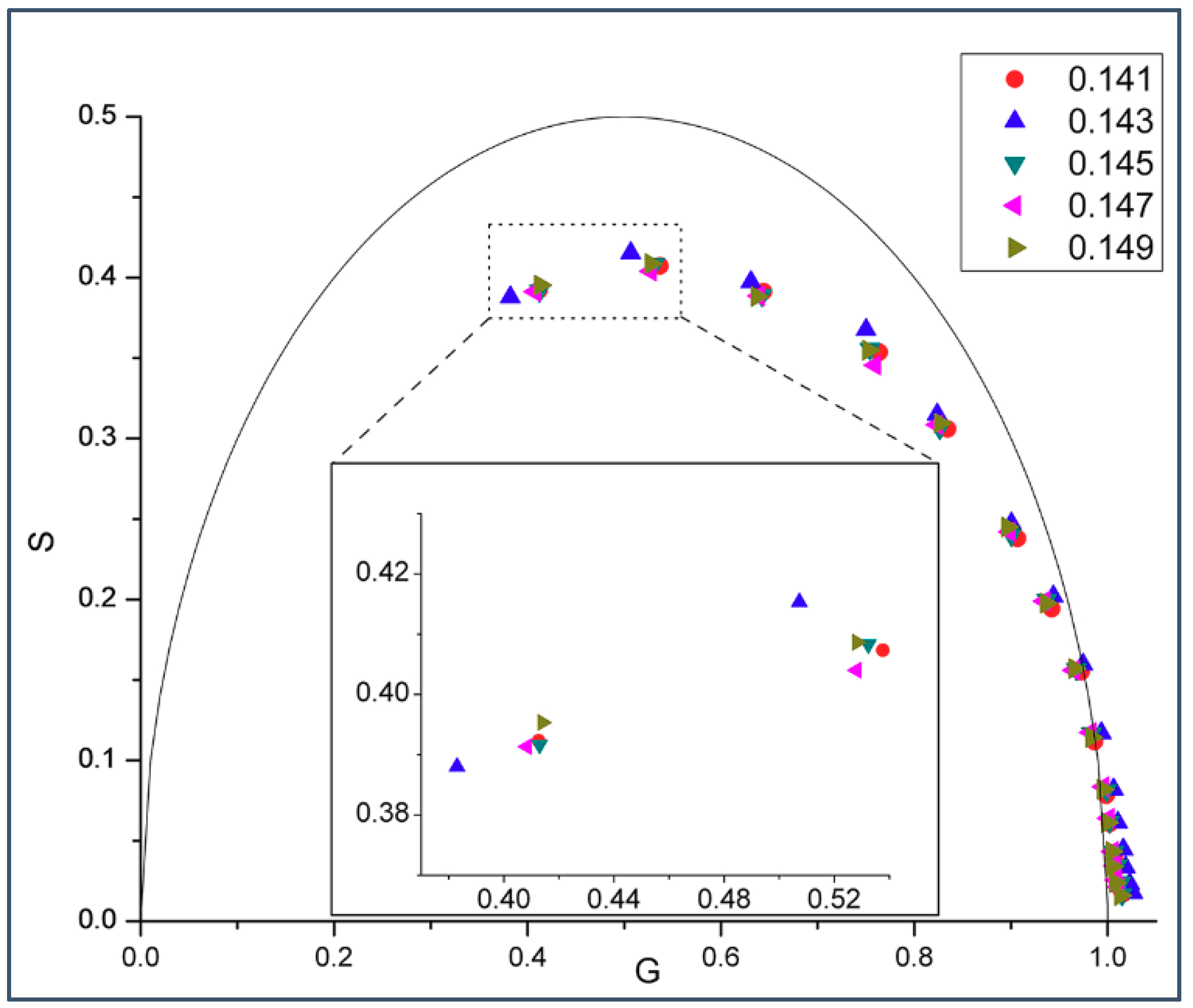
| M | w (cal/mol) | w (cal/mol) | |
|---|---|---|---|
| 9 | 0.182 | 489.2 | 447.0 |
| 10 | 0.166 | 486.8 | 444.1 |
| 11 | 0.154 | 485.2 | 442.3 |
| 12 | 0.143 | 484.1 | 441.5 |
| 13 | 0.133 | 483.7 | 441.4 |
| 14 | 0.125 | 483.8 | 442.0 |
| 15 | 0.118 | 484.3 | 443.2 |
| 16 | 0.111 | 485.3 | 444.9 |
| 17 | 0.105 | 486.6 | 447.0 |
| 18 | 0.100 | 488.3 | 450.0 |
| 19 | 0.0952 | 490.2 | 452.4 |
| 20 | 0.0909 | 492.4 | 455.5 |
| 21 | 0.0870 | 494.8 | 459.0 |
| 22 | 0.0833 | 497.4 | 462.5 |
| 23 | 0.0800 | 500.3 | 466.4 |
| 24 | 0.0769 | 503.3 | 470.4 |
| 25 | 0.0741 | 506.4 | 474.5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugár, I.P.; Bonanno, A.P.; Chong, P.L.-G. Gramicidin Lateral Distribution in Phospholipid Membranes: Fluorescence Phasor Plots and Statistical Mechanical Model. Int. J. Mol. Sci. 2018, 19, 3690. https://doi.org/10.3390/ijms19113690
Sugár IP, Bonanno AP, Chong PL-G. Gramicidin Lateral Distribution in Phospholipid Membranes: Fluorescence Phasor Plots and Statistical Mechanical Model. International Journal of Molecular Sciences. 2018; 19(11):3690. https://doi.org/10.3390/ijms19113690
Chicago/Turabian StyleSugár, István P., Alexander P. Bonanno, and Parkson Lee-Gau Chong. 2018. "Gramicidin Lateral Distribution in Phospholipid Membranes: Fluorescence Phasor Plots and Statistical Mechanical Model" International Journal of Molecular Sciences 19, no. 11: 3690. https://doi.org/10.3390/ijms19113690
APA StyleSugár, I. P., Bonanno, A. P., & Chong, P. L.-G. (2018). Gramicidin Lateral Distribution in Phospholipid Membranes: Fluorescence Phasor Plots and Statistical Mechanical Model. International Journal of Molecular Sciences, 19(11), 3690. https://doi.org/10.3390/ijms19113690