Insect Transcription Factors: A Landscape of Their Structures and Biological Functions in Drosophila and beyond
Abstract
:1. Introduction
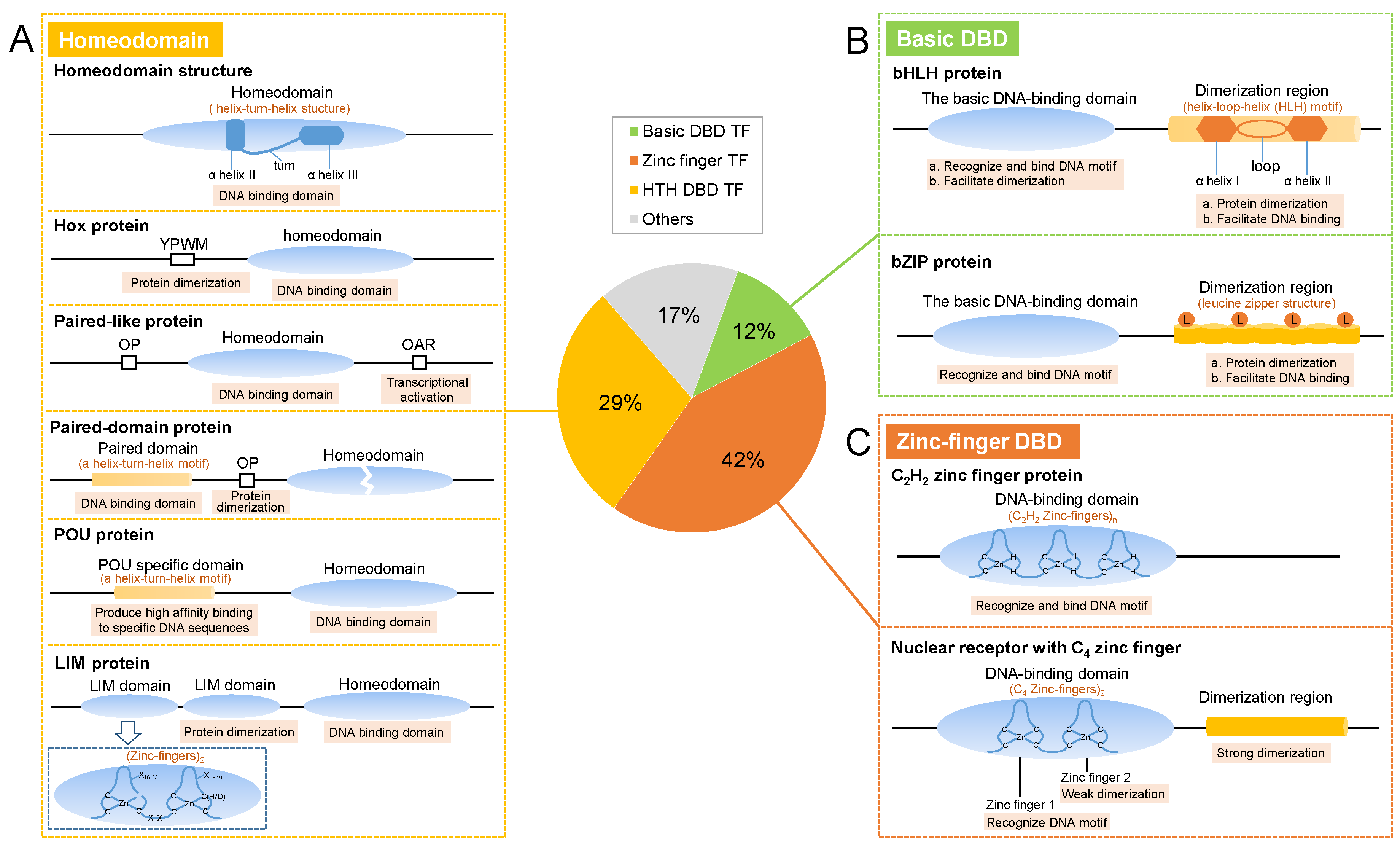
2. Structures
2.1. Homeodomain TFs
2.2. Basic DBD TFs
2.2.1. bHLH Proteins
2.2.2. bZIP proteins
2.3. Zinc-Finger TFs
2.3.1. C2H2 Zinc Finger
2.3.2. Nuclear Receptors
3. Biological Functions
3.1. Internal Responses
3.1.1. Embryonic Axis Establishment
3.1.2. Nervous System Development
3.1.3. Eye Development
3.1.4. Trachea and Gland Formation
3.1.5. Sex Determination
3.1.6. Wing Imaginal Disc Development
3.1.7. Lipid Metabolism
3.1.8. Circadian Clock Adjustment
3.1.9. Diapause Control
3.1.10. Cuticular Protein Synthesis
3.1.11. Cuticle Coloration Dictation
3.1.12. Silk Protein Production
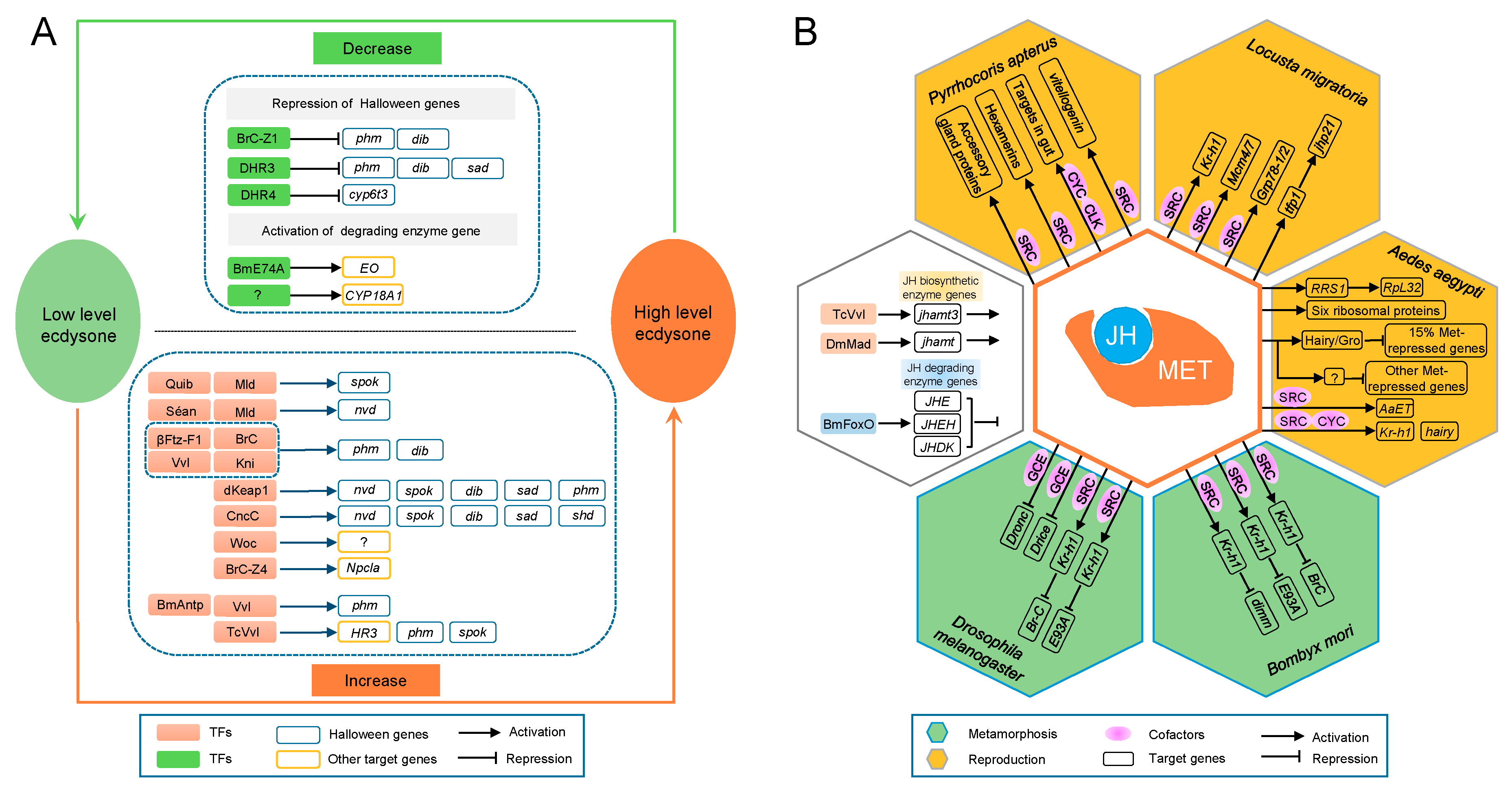
3.1.13. Molting and Metamorphosis Initiation
3.1.14. Reproduction Manipulation
3.2. External Responses
3.2.1. Biotic Factor Responses
3.2.2. Abiotic Factor Responses
4. Research Methods
4.1. TF-Binding Site (TFBS) Prediction
4.2. DNA–Protein Interaction Detection
4.2.1. Dual-Luciferase Reporter Assay System
4.2.2. Electrophoretic Mobility Shift Assay (EMSA)
4.2.3. Yeast One-Hybrid System (Y1H)
4.2.4. Chromatin Immunoprecipitation (ChIP)
4.2.5. CRISPR Affinity Purification In Situ of Regulatory Elements (CAPTURE)
4.3. TF Function Verification
4.3.1. CRISPR/Cas9 system
4.3.2. Yeast Two-Hybrid Assay (Y2H)
4.3.3. Expression Read Depth GWAS (eRD-GWAS)
5. Discussion and Prospects
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Harshman, L.G.; James, A.A. Differential gene expression in insects: Transcriptional control. Annu. Rev. Entomol. 1998, 43, 671–700. [Google Scholar] [CrossRef] [PubMed]
- Papantonis, A.; Swevers, L.; Iatrou, K. Chorion genes: A landscape of their evolution, structure, and regulation. Annu. Rev. Entomol. 2015, 60, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Ratcliffe, O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000, 3, 423–434. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Wingender, E.; Schoeps, T.; Haubrock, M.; Krull, M.; Dönitz, J. TFClass: Expanding the classification of human transcription factors to their mammalian orthologs. Nucleic Acids Res. 2018, 46, D343–D347. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Furlong, E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Reiter, F.; Wienerroither, S.; Stark, A. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 2017, 43, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, A.S.; Li, C.H.; Zamudio, A.V.; Sigova, A.A.; Hannett, N.M.; Day, D.S.; Abraham, B.J.; Cohen, M.A.; Nabet, B.; Buckley, D.L.; et al. YY1 is a structural regulator of enhancer-promoter loops. Cell 2017, 171, 1573–1588. [Google Scholar] [CrossRef] [PubMed]
- King-Jones, K.; Thummel, C.S. Nuclear receptors–a perspective from Drosophila. Nat. Rev. Genet. 2005, 6, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Valanne, S.; Wang, J.H.; Rämet, M. The Drosophila Toll signaling pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Niwa, Y.S.; Niwa, R. Transcriptional regulation of insect steroid hormone biosynthesis and its role in controlling timing of molting and metamorphosis. Dev. Growth Differ. 2016, 58, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Mussabekova, A.; Daeffler, L.; Imler, J.L. Innate and intrinsic antiviral immunity in Drosophila. Cell Mol. Life Sci. 2017, 74, 2039–2054. [Google Scholar] [CrossRef] [PubMed]
- Wingender, E.; Schoeps, T.; Dönitz, J. TFClass: An expandable hierarchical classification of human transcription factors. Nucleic Acids Res. 2013, 41, D165–D170. [Google Scholar] [CrossRef] [PubMed]
- Wingender, E.; Schoeps, T.; Haubrock, M.; Dönitz, J. TFClass: A classification of human transcription factors and their rodent orthologs. Nucleic Acids Res. 2015, 43, D97–D102. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.J.; Qian, Y.Q.; Billeter, M.; Furukubo-Tokunaga, K.; Schier, A.F.; Resendez-Perez, D.; Affolter, M.; Otting, G.; Wuthrich, K. Homeodomain-DNA recognition. Cell 1994, 78, 211–223. [Google Scholar] [CrossRef]
- Gehring, W.J.; Affolter, M.; Bürglin, T. Homeodomain proteins. Annu. Rev. Biochem. 1994, 63, 487–526. [Google Scholar] [CrossRef] [PubMed]
- Latchman, D.S. Families of DNA binding transcription factors. In Eukaryotic Transcription Factors; Latchman, D.S., Ed.; Elsevier: Boston, MA, USA, 2008; Volume 4, pp. 96–160. [Google Scholar]
- Papadopoulos, D.K.; Skouloudaki, K.; Adachi, Y.; Samakovlis, C.; Gehring, W.J. Dimer formation via the homeodomain is required for function and specificity of Sex combs reduced in Drosophila. Dev. Biol. 2012, 367, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.K.; Jaffe, L.; Capovilla, M.; Botas, J.; Mann, R.S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 1994, 78, 603–615. [Google Scholar] [CrossRef]
- Prince, F.; Katsuyama, T.; Oshima, Y.; Plaza, S.; Resendez-Perez, D.; Berry, M.; Kurata, S.; Gehring, W.J. The YPWM motif links Antennapedia to the basal transcriptional machinery. Development 2008, 135, 1669–1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bürglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef] [PubMed]
- Hammonds, A.S.; Bristow, C.A.; Fisher, W.W.; Weiszmann, R.; Wu, S.; Hartenstein, V.; Kellis, M.; Yu, B.; Frise, E.; Celniker, S.E. Spatial expression of transcription factors in Drosophila embryonic organ development. Genome Biol. 2013, 14, R140. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.C.; Rould, M.A.; Weintraub, H.; Pabo, C.O. Crystal structure of MyoD bHLH domain-DNA complex: Perspectives on DNA recognition and implications for transcriptional activation. Cell 1994, 77, 451–459. [Google Scholar] [CrossRef]
- Ledent, V.; Paquet, O.; Vervoort, M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002, 3, research0030.0031. [Google Scholar] [CrossRef] [PubMed]
- Katzenberg, D.; Young, T.; Lin, L.; Finn, L.; Mignot, E. A human period gene (HPER1) polymorphism is not associated with diurnal preference in normal adults. Psychiatr. Genet. 1999, 9, 107–109. [Google Scholar] [CrossRef] [PubMed]
- Ellis, H.M. Embryonic expression and function of the Drosophila helix-loop-helix gene, extramacrochaetae. Mech. Dev. 1994, 47, 65–72. [Google Scholar] [CrossRef]
- Van Doren, M.; Ellis, H.M.; Posakony, J.W. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development 1991, 113, 245–255. [Google Scholar] [PubMed]
- Lavista-Llanos, S.; Centanin, L.; Irisarri, M.; Russo, D.M.; Gleadle, J.M.; Bocca, S.N.; Muzzopappa, M.; Ratcliffe, P.J.; Wappner, P. Control of the hypoxic response in Drosophila melanogaster by the basic helix-loop-helix PAS protein similar. Mol. Cell Biol. 2002, 22, 6842–6853. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.R.; Turner, D.L.; Weintraub, H.; Parkhurst, S.M. Specificity for the Hairy/Enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol. Cell. Biol. 1995, 15, 6923–6931. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 1999, 287, 1023–1040. [Google Scholar] [CrossRef] [PubMed]
- Crozatier, M.; Valle, D.; Dubois, L.; Ibnsouda, S.; Vincent, A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr. Biol. 1996, 6, 707–718. [Google Scholar] [CrossRef]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.; McLachlan, A.D.; Klug, A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985, 4, 1609–1614. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Gippert, G.P.; Soman, K.V.; Case, D.A.; Wright, P.E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science 1989, 245, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Klug, A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Proc. Japan Acad. Ser. 2010, B, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Pabo, C.O.; Peisach, E.; Grant, R.A. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001, 70, 313–340. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, S.A.; Nekludova, L.; Pabo, C.O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 183–212. [Google Scholar] [CrossRef] [PubMed]
- Persikov, A.V.; Osada, R.; Singh, M. Predicting DNA recognition by Cys2His2 zinc finger proteins. Bioinformatics 2009, 25, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Nasiadka, A.; Dietrich, B.H.; Krause, H.M. Anterior-posterior patterning in the Drosophila embryo. In Advances in Developmental Biology and Biochemistry; DePamphilis, M.L., Ed.; Elsevier: Amsterdam, The Netherlands, 2002; Volume 12, pp. 155–204. [Google Scholar]
- Gilbert, S.F. The genetics of axis specification in Drosophila. In Developmental Biology (7th ed.); Gilbert, S.F., Ed.; Sinauer Associates: Sunderland, MA, USA, 2003; pp. 263–304. [Google Scholar]
- Gilbert, S.F. The genetics of axis specification in Drosophila. In Developmental Biology (9th ed.); Gilbert, S.F., Ed.; Sinauer Associates: Sunderland, MA, USA, 2010; pp. 203–239. [Google Scholar]
- Hang, S.; Gergen, J.P. Different modes of enhancer-specific regulation by Runt and Even-skipped during Drosophila segmentation. Mol. Biol. Cell 2017, 28, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Levine, M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 1993, 72, 741–752. [Google Scholar] [CrossRef]
- Shirokawa, J.M.; Courey, A.J. A direct contact between the Dorsal rel homology domain and Twist may mediate transcriptional synergy. Mol. Cell. Biol. 1997, 17, 3345–3355. [Google Scholar] [CrossRef] [PubMed]
- Stathopoulos, A.; Van Drenth, M.; Erives, A.; Markstein, M.; Levine, M. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell 2002, 111, 687–701. [Google Scholar] [CrossRef]
- Ratnaparkhi, G.S.; Jia, S.; Courey, A.J. Uncoupling dorsal-mediated activation from dorsal-mediated repression in the Drosophila embryo. Development 2006, 133, 4409–4414. [Google Scholar] [CrossRef] [PubMed]
- Zeitlinger, J.; Zinzen, R.P.; Stark, A.; Kellis, M.; Zhang, H.; Young, R.A.; Levine, M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007, 21, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Dubnicoff, T.; Valentine, S.A.; Chen, G.; Shi, T.; Lengyel, J.A.; Paroush, Z.; Courey, A.J. Conversion of Dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev. 1997, 11, 2952–2957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Saaib, R.D.; Jia, S.; Courey, A.J. Activation and repression by the C-terminal domain of Dorsal. Development 2001, 128, 1869–1879. [Google Scholar] [PubMed]
- Thor, S. Neuroscience: Stem cells in multiple time zones. Nature 2013, 498, 441–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashraf, S.I.; Hu, X.; Roote, J.; Ip, Y.T. The mesoderm determinant Snail collaborates with related zinc-finger proteins to control Drosophila neurogenesis. EMBO J. 1999, 18, 6426–6438. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.I.; Ip, Y.T. The Snail protein family regulates neuroblast expression of inscuteable and string, genes involved in asymmetry and cell division in Drosophila. Development 2001, 128, 4757–4767. [Google Scholar] [PubMed]
- Lai, S.L.; Miller, M.R.; Robinson, K.J.; Doe, C.Q. The Snail family member Worniu is continuously required in neuroblasts to prevent Elav-induced premature differentiation. Dev. Cell 2012, 23, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Erclik, T.; Bertet, C.; Chen, Z.; Voutev, R.; Venkatesh, S.; Morante, J.; Celik, A.; Desplan, C. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 2013, 498, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Bello, B.C.; Izergina, N.; Caussinus, E.; Reichert, H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xie, Y.; Zhu, S. Notch maintains Drosophila type II neuroblasts by suppressing expression of the Fez transcription factor Earmuff. Development 2016, 143, 2511–2521. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Barshow, S.; Wildonger, J.; Jan, L.Y.; Jan, Y.N. Ets transcription factor Pointed promotes the generation of intermediate neural progenitors in Drosophila larval brains. Proc. Natl. Acad. Sci. USA 2011, 108, 20615–20620. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Li, X.; Deng, X.; Hou, Y.; O’Hara, K.; Urso, A.; Peng, Y.; Chen, L.; Zhu, S. The Ets protein Pointed prevents both premature differentiation and dedifferentiation of Drosophila intermediate neural progenitors. Development 2016, 143, 3109–3118. [Google Scholar] [CrossRef] [PubMed]
- Janssens, D.H.; Hamm, D.C.; Anhezini, L.; Xiao, Q.; Siller, K.H.; Siegrist, S.E.; Harrison, M.M.; Lee, C.Y. An Hdac1/Rpd3-poised circuit balances continual self-renewal and rapid restriction of developmental potential during asymmetric stem cell division. Dev. Cell 2017, 40, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Koe, C.T.; Li, S.; Rossi, F.; Wong, J.J.; Wang, Y.; Zhang, Z.; Chen, K.; Aw, S.S.; Richardson, H.E.; Robson, P.; et al. The Brm-HDAC3-Erm repressor complex suppresses dedifferentiation in Drosophila type II neuroblast lineages. eLife 2014, 3, e01906. [Google Scholar] [CrossRef] [PubMed]
- Pappu, K.S.; Mardon, G. Genetic control of retinal specification and determination in Drosophila. Int. J. Dev. Biol. 2004, 48, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Fichelson, P.; Brigui, A.; Pichaud, F. Orthodenticle and Kruppel homolog 1 regulate Drosophila photoreceptor maturation. Proc. Natl. Acad. Sci. USA 2012, 109, 7893–7898. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Bargmann, B.O.R.; Tsachaki, M.; Fritsch, C.; Sprecher, S.G. Functional genomics identifies regulators of the phototransduction machinery in the Drosophila larval eye and adult ocelli. Dev. Biol. 2016, 410, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Higueras, C.; Sotillos, S.; Castelli-Gair Hombria, J. Common origin of insect trachea and endocrine organs from a segmentally repeated precursor. Curr. Biol. 2014, 24, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Fox, R.M.; Vaishnavi, A.; Maruyama, R.; Andrew, D.J. Organ-specific gene expression: The bHLH protein Sage provides tissue specificity to Drosophila FoxA. Development 2013, 140, 2160–2171. [Google Scholar] [CrossRef] [PubMed]
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R. Sex determination: Why so many ways of doing it? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, A.N.; Lu, H.; Erickson, J.W. A shared enhancer controls a temporal switch between promoters during Drosophila primary sex determination. Proc. Natl. Acad. Sci. USA 2008, 105, 18436–18441. [Google Scholar] [CrossRef] [PubMed]
- Verhulst, E.C.; van de Zande, L.; Beukeboom, L.W. Insect sex determination: It all evolves around transformer. Curr. Opin. Genet. Dev. 2010, 20, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Kozhina, E.; Mahadevaraju, S.; Yang, D.; Avila, F.W.; Erickson, J.W. Maternal Groucho and bHLH repressors amplify the dose-sensitive X chromosome signal in Drosophila sex determination. Dev. Biol. 2008, 323, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Salz, H.K.; Erickson, J.W. Sex determination in Drosophila: The view from the top. Fly 2010, 4, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Verhulst, E.C.; van de Zande, L. Double nexus–Doublesex is the connecting element in sex determination. Brief. Funct. Genomics 2015, 14, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Guo, L.; Jiao, X.; Yang, N.; Yang, X.; Wu, Q.; Wang, S.; Zhou, X.; Zhang, Y. Transcriptomic dissection of sexual differences in Bemisia tabaci, an invasive agricultural pest worldwide. Sci. Rep. 2014, 4, 4088. [Google Scholar] [CrossRef] [PubMed]
- Zinna, R.A.; Gotoh, H.; Kojima, T.; Niimi, T. Recent advances in understanding the mechanisms of sexually dimorphic plasticity: Insights from beetle weapons and future directions. Curr. Opin. Insect Sci. 2018, 25, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Hayashi, S. Specification of the embryonic limb primordium by graded activity of Decapentaplegic. Development 1997, 124, 125–132. [Google Scholar] [PubMed]
- Kubota, K.; Goto, S.; Eto, K.; Hayashi, S. EGF receptor attenuates Dpp signaling and helps to distinguish the wing and leg cell fates in Drosophila. Development 2000, 127, 3769–3776. [Google Scholar] [PubMed]
- Dahmann, C.; Basler, K. Opposing transcriptional outputs of Hedgehog signaling and engrailed control compartmental cell sorting at the Drosophila A/P boundary. Cell 2000, 100, 411–422. [Google Scholar] [CrossRef] [Green Version]
- Milán, M.; Cohen, S.M. A re-evaluation of the contributions of Apterous and Notch to the dorsoventral lineage restriction boundary in the Drosophila wing. Development 2003, 130, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Nienhaus, U.; Aegerter-Wilmsen, T.; Aegerter, C.M. In-vivo imaging of the Drosophila wing imaginal disc over time: Novel insights on growth and boundary formation. PLoS ONE 2012, 7, e47594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatini, V.; Kula-Eversole, E.; Nusinow, D.; Del Signore, S.J. Essential roles for stat92E in expanding and patterning the proximodistal axis of the Drosophila wing imaginal disc. Dev. Biol. 2013, 378, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Recasens-Alvarez, C.; Ferreira, A.; Milán, M. JAK/STAT controls organ size and fate specification by regulating morphogen production and signalling. Nat. Commun. 2017, 8, 13815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terriente, J.; Perea, D.; Suzanne, M.; Díaz-Benjumea, F.J. The Drosophila gene zfh2 is required to establish proximal-distal domains in the wing disc. Dev. Biol. 2008, 320, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Perea, D.; Molohon, K.; Edwards, K.; Díaz-Benjumea, F.J. Multiple roles of the gene zinc finger homeodomain-2 in the development of the Drosophila wing. Mech. Dev. 2013, 130, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Azpiazu, N.; Morata, G. Function and regulation of homothorax in the wing imaginal disc of Drosophila. Development 2000, 127, 2685–2693. [Google Scholar] [PubMed]
- Casares, F.; Mann, R.S. A dual role for homothorax in inhibiting wing blade development and specifying proximal wing identities in Drosophila. Development 2000, 127, 1499–1508. [Google Scholar] [PubMed]
- Shukla, V.; Habib, F.; Kulkarni, A.; Ratnaparkhi, G.S. Gene duplication, lineage-specific expansion, and subfunctionalization in the MADF-BESS family patterns the Drosophila wing hinge. Genetics 2014, 196, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Vihervaara, T.; Puig, O. dFOXO regulates transcription of a Drosophila acid lipase. J. Mol. Biol. 2008, 376, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Liu, Y.; Zhou, S.; Li, K.; Tian, L.; Li, S. 20-Hydroxyecdysone-induced transcriptional activity of FoxO upregulates brummer and acid lipase-1 and promotes lipolysis in Bombyx fat body. Insect Biochem. Mol. Biol. 2013, 43, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Baumann, A.A.; Benoit, J.B.; Michalkova, V.; Mireji, P.; Attardo, G.M.; Moulton, J.K.; Wilson, T.G.; Aksoy, S. Juvenile hormone and insulin suppress lipolysis between periods of lactation during tsetse fly pregnancy. Mol. Cell. Endocrinol. 2013, 372, 30–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, P.; Chang, K.; Liu, Y.; Bouska, M.; Birnbaum, A.; Karashchuk, G.; Thakore, R.; Zheng, W.; Post, S.; Brent, C.S.; et al. Drosophila Kruppel homolog 1 represses lipolysis through interaction with dFOXO. Sci. Rep. 2017, 7, 16369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, Y.X.; Liu, J.; Yang, C.; Feng, Q.L.; Xu, W.H. A regulatory pathway, ecdysone-transcription factor Relish-cathepsin L, is involved in insect fat body dissociation. PLoS Genet. 2013, 9, e1003273. [Google Scholar] [CrossRef] [PubMed]
- Okamura, T.; Shimizu, H.; Nagao, T.; Ueda, R.; Ishii, S. ATF-2 regulates fat metabolism in Drosophila. Mol. Biol. Cell 2007, 18, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Bond, N.D.; Nelliot, A.; Bernardo, M.K.; Ayerh, M.A.; Gorski, K.A.; Hoshizaki, D.K.; Woodard, C.T. ßFTZ-F1 and Matrix metalloproteinase 2 are required for fat-body remodeling in Drosophila. Dev. Biol. 2011, 360, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Vyazunova, I.; Lan, Q. Yellow fever mosquito sterol carrier protein-2 gene structure and transcriptional regulation. Insect Mol. Biol. 2010, 19, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Fu, Q.; Hong, H.; Schwaegler, T.; Lan, Q. THAP and ATF-2 regulated sterol carrier protein-2 promoter activities in the larval midgut of the yellow fever mosquito, Aedes aegypti. PLoS ONE 2012, 7, e46948. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.N.; Zhang, L.L.; Zeng, B.J.; Zheng, S.C.; Feng, Q.L. Transcription factor CAAT/enhancer-binding protein is involved in regulation of expression of sterol carrier protein x in Spodoptera litura. Insect Mol. Biol. 2015, 24, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Glossop, N.R.; Houl, J.H.; Zheng, H.; Ng, F.S.; Dudek, S.M.; Hardin, P.E. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 2003, 37, 249–261. [Google Scholar] [CrossRef]
- Tataroglu, O.; Emery, P. The molecular ticks of the Drosophila circadian clock. Curr. Opin. Insect Sci. 2015, 7, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Gunawardhana, K.L.; Hardin, P.E. VRILLE controls PDF neuropeptide accumulation and arborization rhythms in small ventrolateral neurons to drive rhythmic behavior in Drosophila. Curr. Biol. 2017, 27, 3442–3453. [Google Scholar] [CrossRef] [PubMed]
- Cyran, S.A.; Buchsbaum, A.M.; Reddy, K.L.; Lin, M.C.; Glossop, N.R.; Hardin, P.E.; Young, M.W.; Storti, R.V.; Blau, J. Vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 2003, 112, 329–341. [Google Scholar] [CrossRef]
- Matsumoto, A.; Ukai-Tadenuma, M.; Yamada, R.G.; Houl, J.; Uno, K.D.; Kasukawa, T.; Dauwalder, B.; Itoh, T.Q.; Takahashi, K.; Ueda, R.; et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007, 21, 1687–1700. [Google Scholar] [CrossRef] [PubMed]
- Sivachenko, A.; Li, Y.; Abruzzi, K.C.; Rosbash, M. The transcription factor Mef2 links the Drosophila core Clock to Fas2, neuronal morphology, and circadian behavior. Neuron 2013, 79, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Jaumouillé, E.; Machado Almeida, P.; Stähli, P.; Koch, R.; Nagoshi, E. Transcriptional regulation via nuclear receptor crosstalk required for the Drosophila circadian clock. Curr. Biol. 2015, 25, 1502–1508. [Google Scholar] [CrossRef] [PubMed]
- Kamae, Y.; Uryu, O.; Miki, T.; Tomioka, K. The nuclear receptor genes HR3 and E75 are required for the circadian rhythm in a primitive insect. PLoS ONE 2014, 9, e114899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, B.; Zhang, Z.F.; Tang, S.M.; Yi, Y.Z.; Zhang, T.Y.; Xu, W.H. Protein-DNA interactions in the promoter region of the gene encoding diapause hormone and pheromone biosynthesis activating neuropeptide of the cotton bollworm, Helicoverpa armigera. Biochim. Biophys. Acta 2006, 1759, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.Y.; Xu, W.H. Identification and characterization of a POU transcription factor in the cotton bollworm, Helicoverpa armigera. BMC Mol. Biol. 2009, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.H.; Hong, B.; Xu, W.H. Identification of an E-box DNA binding protein, activated protein 4, and its function in regulating the expression of the gene encoding diapause hormone and pheromone biosynthesis-activating neuropeptide in Helicoverpa armigera. Insect Mol. Biol. 2010, 19, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Hong, B.; Feng, Q.L.; Xu, W.H. Transcription factor fork head regulates the promoter of diapause hormone gene in the cotton bollworm, Helicoverpa armigera, and the modification of SUMOylation. Insect Biochem. Mol. Biol. 2011, 41, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.Y.; Kang, L.; Zhang, Z.F.; Xu, W.H. Identification of a POU factor involved in regulating the neuron-specific expression of the gene encoding diapause hormone and pheromone biosynthesis-activating neuropeptide in Bombyx mori. Biochem. J. 2004, 380, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Zhang, X.; Deng, H.; Wu, F.; Ren, Y.; Xiang, H.; Zheng, S.; Liu, L.; Huang, L.; Zeng, B.; et al. BmILF and i-motif structure are involved in transcriptional regulation of BmPOUM2 in Bombyx mori. Nucleic Acids Res. 2018, 46, 1710–1723. [Google Scholar] [CrossRef] [PubMed]
- Bajgar, A.; Jindra, M.; Dolezel, D. Autonomous regulation of the insect gut by circadian genes acting downstream of juvenile hormone signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 4416–4421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murata, T.; Kageyama, Y.; Hirose, S.; Ueda, H. Regulation of the EDG84A gene by FTZ-F1 during metamorphosis in Drosophila melanogaster. Mol. Cell. Biol. 1996, 16, 6509–6515. [Google Scholar] [CrossRef] [PubMed]
- Rebers, J.E.; Niu, J.; Riddiford, L.M. Structure and spatial expression of the Manduca sexta MSCP14.6 cuticle gene. Insect Biochem. Mol. Biol. 1997, 27, 229–240. [Google Scholar] [CrossRef]
- Deng, H.; Zhang, J.; Li, Y.; Zheng, S.; Liu, L.; Huang, L.; Xu, W.H.; Palli, S.R.; Feng, Q. Homeodomain POU and Abd-A proteins regulate the transcription of pupal genes during metamorphosis of the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. USA 2012, 109, 12598–12603. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Deng, H.; Hu, Q.; Zhu, Z.; Liu, L.; Zheng, S.; Song, Q.; Feng, Q. Identification of the binding domains and key amino acids for the interaction of the transcription factors BmPOUM2 and BmAbd-A in Bombyx mori. Insect Biochem. Mol. Biol. 2017, 81, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Wittkopp, P.J.; Beldade, P. Development and evolution of insect pigmentation: Genetic mechanisms and the potential consequences of pleiotropy. Semin Cell Dev. Biol. 2009, 20, 65–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kronforst, M.R.; Barsh, G.S.; Kopp, A.; Mallet, J.; Monteiro, A.; Mullen, S.P.; Protas, M.; Rosenblum, E.B.; Schneider, C.J.; Hoekstra, H.E. Unraveling the thread of nature’s tapestry: The genetics of diversity and convergence in animal pigmentation. Pigment. Cell Melanoma Res. 2012, 25, 411–433. [Google Scholar] [CrossRef] [PubMed]
- Kunte, K.; Zhang, W.; Tenger-Trolander, A.; Palmer, D.H.; Martin, A.; Reed, R.D.; Mullen, S.P.; Kronforst, M.R. Doublesex is a mimicry supergene. Nature 2014, 507, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Yoda, S.; Yamaguchi, J.; Mita, K.; Yamamoto, K.; Banno, Y.; Ando, T.; Daimon, T.; Fujiwara, H. The transcription factor Apontic-like controls diverse colouration pattern in caterpillars. Nat. Commun. 2014, 5, 4936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.M.; Liu, C.; Jiang, L.J.; Li, Q.Y.; Zhou, M.T.; Cheng, T.C.; Mita, K.; Xia, Q.Y. A juvenile hormone transcription factor Bmdimm-fibroin H chain pathway is involved in the synthesis of silk protein in silkworm, Bombyx mori. J. Biol. Chem. 2015, 290, 972–986. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zha, X.; Shi, P.; Wei, S.; Wang, H.; Zheng, R.; Xia, Q. Multiprotein bridging factor 2 regulates the expression of the fibroin heavy chain gene by interacting with Bmdimmed in the silkworm Bombyx mori. Insect Mol. Biol. 2016, 25, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, C.; Li, Q.; Hu, W.; Zhou, M.; Nie, H.; Zhang, Y.; Peng, Z.; Zhao, P.; Xia, Q. Basic helix-loop-helix transcription factor Bmsage is involved in regulation of fibroin H-chain gene via interaction with SGF1 in Bombyx mori. PLoS ONE 2014, 9, e94091. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.C.; Matsuno, K.; Suzuki, Y. Fibroin gene promoter contains a cluster of homeodomain binding sites that interact with three silk gland factors. J. Mol. Biol. 1990, 213, 651–670. [Google Scholar] [CrossRef]
- Kimoto, M.; Tsubota, T.; Uchino, K.; Sezutsu, H.; Takiya, S. LIM-homeodomain transcription factor Awh is a key component activating all three fibroin genes, fibH, fibL and fhx, in the silk gland of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2015, 56, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Horard, B.; Julien, E.; Nony, P.; Garel, A.; Couble, P. Differential binding of the Bombyx silk gland-specific factor SGFB to its target DNA sequence drives posterior-cell-restricted expression. Mol. Cell. Biol. 1997, 17, 1572–1579. [Google Scholar] [CrossRef] [PubMed]
- Grzelak, K. Control of expression of silk protein genes. Comp. Biochem. Physiol. 1995, 110B, 671–681. [Google Scholar] [CrossRef]
- Julien, E.; Bordeaux, M.C.; Garel, A.; Couble, P. Fork head alternative binding drives stage-specific gene expression in the silk gland of Bombyx mori. Insect Biochem. Mol. Biol. 2002, 32, 377–387. [Google Scholar] [CrossRef]
- Mach, V.; Takiya, S.; Ohno, K.; Handa, H.; Imai, T.; Suzuki, Y. Silk gland factor-1 involved in the regulation of Bombyx sericin-1 gene contains fork head motif. J. Biol. Chem. 1995, 270, 9340–9346. [Google Scholar] [CrossRef] [PubMed]
- Fukuta, M.; Matsuno, K.; Hui, C.C.; Nagata, T.; Takiya, S.; Xu, P.X.; Ueno, K.; Suzuki, Y. Molecular cloning of a POU domain-containing factor involved in the regulation of the Bombyx sericin-1 gene. J. Biol. Chem. 1993, 268, 19471–19475. [Google Scholar] [PubMed]
- Kimoto, M.; Tsubota, T.; Uchino, K.; Sezutsu, H.; Takiya, S. Hox transcription factor Antp regulates sericin-1 gene expression in the terminal differentiated silk gland of Bombyx mori. Dev. Biol. 2014, 386, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Kitagawa, T.; Kobayashi, I.; Nakata, T.; Kuroiwa, A.; Takiya, S. Inhibition of the binding of MSG-intermolt-specific complex, MIC, to the sericin-1 gene promoter and sericin-1 gene expression by POU-M1/SGF-3. Dev. Genes Evol. 2012, 222, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Kayukawa, T.; Jouraku, A.; Ito, Y.; Shinoda, T. Molecular mechanism underlying juvenile hormone-mediated repression of precocious larval-adult metamorphosis. Proc. Natl. Acad. Sci. USA 2017, 114, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, N.; Rewitz, K.F.; O’Connor, M.B. Ecdysone control of developmental transitions: Lessons from Drosophila research. Annu. Rev. Entomol. 2013, 58, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Uryu, O.; Ou, Q.; Komura-Kawa, T.; Kamiyama, T.; Iga, M.; Syrzycka, M.; Hirota, K.; Kataoka, H.; Honda, B.M.; King-Jones, K.; et al. Cooperative control of ecdysone biosynthesis in Drosophila by transcription factors Séance, Ouija board, and Molting defective. Genetics 2017, 208, 605–622. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Tian, L.; Peng, C.; Abdou, M.; Wen, D.; Wang, Y.; Li, S.; Wang, J. DPP-mediated TGFβ signaling regulates juvenile hormone biosynthesis by activating the expression of juvenile hormone acid methyltransferase. Development 2011, 138, 2283–2291. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Ko, A.; Chaieb, L.; Koyama, T.; Sarwar, P.; Mirth, C.K.; Smith, W.A.; Suzuki, Y. The POU factor Ventral veins lacking/Drifter directs the timing of metamorphosis through ecdysteroid and juvenile hormone signaling. PLoS Genet. 2014, 10, e1004425. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Huang, Y.; Xu, J.; Shiotsuki, T.; Bai, H.; Palli, S.R.; Huang, Y.; Tan, A. The FOXO transcription factor controls insect growth and development by regulating juvenile hormone degradation in the silkworm, Bombyx mori. J. Biol. Chem. 2017, 292, 11659–11669. [Google Scholar] [CrossRef] [PubMed]
- Ashok, M.; Turner, C.; Wilson, T.G. Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc. Natl. Acad. Sci. USA 1998, 95, 2761–2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Mead, E.A.; Zhu, J. Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc. Natl. Acad. Sci. USA 2011, 108, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, J.; Sheng, Z.; Sui, Y.; Palli, S.R. Steroid receptor co-activator is required for juvenile hormone signal transduction through a bHLH-PAS transcription factor, methoprene tolerant. J. Biol. Chem. 2011, 286, 8437–8447. [Google Scholar] [CrossRef] [PubMed]
- Charles, J.P.; Iwema, T.; Epa, V.C.; Takaki, K.; Rynes, J.; Jindra, M. Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc. Natl. Acad. Sci. USA 2011, 108, 21128–21133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.W.; Zou, Z.; Saha, T.T.; Raikhel, A.S. bHLH-PAS heterodimer of Methoprene-tolerant and Cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc. Natl. Acad. Sci. USA 2012, 109, 16576–16581. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Sui, Y.; Xu, J.; Zhu, F.; Palli, S.R. Juvenile hormone regulates Aedes aegypti Krüppel homolog 1 through a conserved E box motif. Insect Biochem. Mol. Biol. 2014, 52, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Kayukawa, T.; Shinoda, T. Functional characterization of two paralogous JH receptors, methoprene-tolerant 1 and 2, in the silkworm, Bombyx mori (Lepidoptera: Bombycidae). Appl. Entomol. Zool. 2015, 50, 383–391. [Google Scholar] [CrossRef]
- Godlewski, J.; Wang, S.; Wilson, T.G. Interaction of bHLH-PAS proteins involved in juvenile hormone reception in Drosophila. Biochem. Biophys. Res. Commun. 2006, 342, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sheng, Z.; Liu, H.; Wen, D.; He, Q.; Wang, S.; Shao, W.; Jiang, R.J.; An, S.; Sun, Y.; et al. Juvenile hormone counteracts the bHLH-PAS transcription factors MET and GCE to prevent caspase-dependent programmed cell death in Drosophila. Development 2009, 136, 2015–2025. [Google Scholar] [CrossRef] [PubMed]
- Kayukawa, T.; Minakuchi, C.; Namiki, T.; Togawa, T.; Yoshiyama, M.; Kamimura, M.; Mita, K.; Imanishi, S.; Kiuchi, M.; Ishikawa, Y.; et al. Transcriptional regulation of juvenile hormone-mediated induction of Krüppel homolog 1, a repressor of insect metamorphosis. Proc. Natl. Acad. Sci. USA 2012, 109, 11729–11734. [Google Scholar] [CrossRef] [PubMed]
- Kayukawa, T.; Tateishi, K.; Shinoda, T. Establishment of a versatile cell line for juvenile hormone signaling analysis in Tribolium castaneum. Sci. Rep. 2013, 3, 1570. [Google Scholar] [CrossRef] [PubMed]
- Smykal, V.; Bajgar, A.; Provaznik, J.; Fexova, S.; Buricova, M.; Takaki, K.; Hodkova, M.; Jindra, M.; Dolezel, D. Juvenile hormone signaling during reproduction and development of the linden bug, Pyrrhocoris apterus. Insect Biochem. Mol. Biol. 2014, 45, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Konopova, B.; Jindra, M. Broad-Complex acts downstream of Met in juvenile hormone signaling to coordinate primitive holometabolan metamorphosis. Development 2008, 135, 559–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belles, X.; Santos, C.G. The MEKRE93 (Methoprene tolerant-Krüppel homolog 1-E93) pathway in the regulation of insect metamorphosis, and the homology of the pupal stage. Insect Biochem. Mol. Biol. 2014, 52, 60–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urñna, E.; Chafino, S.; Manjón, C.; Franch-Marro, X.; Martín, D. The occurrence of the holometabolous pupal stage requires the interaction between E93, Krüppel-Homolog 1 and Broad-Complex. PLoS Genet. 2016, 12, e1006020. [Google Scholar]
- Roy, S.; Saha, T.T.; Zou, Z.; Raikhel, A.S. Regulatory pathways controlling female insect reproduction. Annu. Rev. Entomol. 2018, 63, 489–511. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Saha, T.T.; Roy, S.; Shin, S.W.; Backman, T.W.; Girke, T.; White, K.P.; Raikhel, A.S. Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proc. Natl. Acad. Sci. USA 2013, 110, E2173–E2181. [Google Scholar] [CrossRef] [PubMed]
- Saha, T.T.; Shin, S.W.; Dou, W.; Roy, S.; Zhao, B.; Hou, Y.; Wang, X.L.; Zou, Z.; Girke, T.; Raikhel, A.S. Hairy and Groucho mediate the action of juvenile hormone receptor Methoprene-tolerant in gene repression. Proc. Natl. Acad. Sci. USA 2016, 113, E735–E743. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hou, Y.; Saha, T.T.; Pei, G.; Raikhel, A.S.; Zou, Z. Hormone and receptor interplay in the regulation of mosquito lipid metabolism. Proc. Natl. Acad. Sci. USA 2017, 114, E2709–E2718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.; Wu, Z.; Song, J.; Jiang, F.; Wang, Z.; Deng, S.; Walker, V.K.; Zhou, S. Juvenile hormone-receptor complex acts on Mcm4 and Mcm7 to promote polyploidy and vitellogenesis in the migratory locust. PLoS Genet. 2014, 10, e1004702. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Guo, W.; Xie, Y.; Zhou, S. Juvenile hormone activates the transcription of cell-division-cycle 6 (Cdc6) for polyploidy-dependent insect vitellogenesis and oogenesis. J. Biol. Chem. 2016, 291, 5418–5427. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Li, D.; Wang, Z.; Guo, W.; Kang, L.; Zhou, S. Juvenile hormone differentially regulates two Grp78 genes encoding protein chaperones required for insect fat body cell homeostasis and vitellogenesis. J. Biol. Chem. 2017, 292, 8823–8834. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wu, Z.; Wang, Z.; Deng, S.; Zhou, S. Krüppel-homolog 1 mediates juvenile hormone action to promote vitellogenesis and oocyte maturation in the migratory locust. Insect Biochem. Mol. Biol. 2014, 52, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Gujar, H.; Palli, S.R. Juvenile hormone regulation of female reproduction in the common bed bug, Cimex lectularius. Sci. Rep. 2016, 6, 35546. [Google Scholar] [CrossRef] [PubMed]
- Hejnikova, M.; Paroulek, M.; Hodkova, M. Decrease in Methoprene tolerant and Taiman expression reduces juvenile hormone effects and enhances the levels of juvenile hormone circulating in males of the linden bug Pyrrhocoris apterus. J. Insect Physiol. 2016, 93–94, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Uryu, O.; Ameku, T.; Niwa, R. Recent progress in understanding the role of ecdysteroids in adult insects: Germline development and circadian clock in the fruit fly Drosophila melanogaster. Zoological Lett. 2015, 1, 32. [Google Scholar] [CrossRef] [PubMed]
- Sourmeli, S.; Papantonis, A.; Lecanidou, R. A novel role for the Bombyx Slbo homologue, BmC/EBP, in insect choriogenesis. Biochem. Biophys. Res. Commun. 2005, 337, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Sourmeli, S.; Papantonis, A.; Lecanidou, R. BmCbZ, an insect-specific factor featuring a composite DNA-binding domain, interacts with BmC/EBPγ. Biochem. Biophys. Res. Commun. 2005, 338, 1957–1965. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.X.; Pan, P.L.; Xu, J.Y.; Shen, Z.F.; Kang, D.; Lu, J.B.; Hu, Q.L.; Huang, H.J.; Lou, Y.H.; Zhou, N.M.; et al. Forkhead box transcription factor L2 activates Fcp3C to regulate insect chorion formation. Open Biol. 2017, 7, 170061. [Google Scholar] [CrossRef] [PubMed]
- Hetru, C.; Hoffmann, J.A. NF-κB in the immune response of Drosophila. Cold Spring Harb. Perspect. Biol. 2009, 1, a000232. [Google Scholar] [CrossRef] [PubMed]
- Stöven, S.; Ando, I.; Kadalayil, L.; Engström, Y.; Hultmark, D. Activation of the Drosophila NF-κB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000, 1, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Reichhart, J.M.; Hoffmann, J.A. Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 1997, 94, 14614–14619. [Google Scholar] [CrossRef] [PubMed]
- Michel, T.; Reichhart, J.M.; Hoffmann, J.A.; Royet, J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 2001, 414, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Zambon, R.A.; Nandakumar, M.; Vakharia, V.N.; Wu, L.P. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 7257–7262. [Google Scholar] [CrossRef] [PubMed]
- Kleino, A.; Silverman, N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 2014, 42, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Tanji, T.; Yun, E.Y.; Ip, Y.T. Heterodimers of NF-κB transcription factors DIF and Relish regulate antimicrobial peptide genes in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 14715–14720. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Rao, X.J.; Yi, H.Y.; Lin, X.Y.; Huang, X.H.; Yu, X.Q. Co-expression of Dorsal and Rel2 negatively regulates antimicrobial peptide expression in the tobacco hornworm Manduca sexta. Sci. Rep. 2016, 6, 20654. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, V.; Courey, A.J. The MADF-BESS domain factor Dip3 potentiates synergistic activation by Dorsal and Twist. Gene 2002, 299, 173–184. [Google Scholar] [CrossRef]
- Ratnaparkhi, G.S.; Duong, H.A.; Courey, A.J. Dorsal interacting protein 3 potentiates activation by Drosophila Rel homology domain proteins. Dev. Comp. Immunol. 2008, 32, 1290–1300. [Google Scholar] [CrossRef] [PubMed]
- Junell, A.; Uvell, H.; Pick, L.; Engström, Y. Isolation of regulators of Drosophila immune defense genes by a double interaction screen in yeast. Insect Biochem. Mol. Biol. 2007, 37, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Barillas-Mury, C.; Charlesworth, A.; Gross, I.; Richman, A.; Hoffmann, J.A.; Kafatos, F.C. Immune factor Gambif1, a new rel family member from the human malaria vector, Anopheles gambiae. EMBO J. 1996, 15, 4691–4701. [Google Scholar] [CrossRef] [PubMed]
- Osta, M.A.; Christophides, G.K.; Vlachou, D.; Kafatos, F.C. Innate immunity in the malaria vector Anopheles gambiae: Comparative and functional genomics. J. Exp. Biol. 2004, 207, 2551–2563. [Google Scholar] [CrossRef] [PubMed]
- Meister, S.; Kanzok, S.M.; Zheng, X.L.; Luna, C.; Li, T.R.; Hoa, N.T.; Clayton, J.R.; White, K.P.; Kafatos, F.C.; Christophides, G.K.; et al. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2005, 102, 11420–11425. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.W.; Kokoza, V.; Bian, G.; Cheon, H.M.; Kim, Y.J.; Raikhel, A.S. REL1, a homologue of Drosophila Dorsal, regulates Toll antifungal immune pathway in the female mosquito Aedes aegypti. J. Biol. Chem. 2005, 280, 16499–16507. [Google Scholar] [CrossRef] [PubMed]
- Frolet, C.; Thoma, M.; Blandin, S.; Hoffmann, J.A.; Levashina, E.A. Boosting NF-κB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity 2006, 25, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008, 4, e1000098. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.W.; Kokoza, V.; Ahmed, A.; Raikhel, A.S. Characterization of three alternatively spliced isoforms of the Rel/NF-kappa B transcription factor Relish from the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2002, 99, 9978–9983. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.W.; Kokoza, V.; Lobkov, I.; Raikhel, A.S. Relish-mediated immune deficiency in the transgenic mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2003, 100, 2616–2621. [Google Scholar] [CrossRef] [PubMed]
- Antonova, Y.; Alvarez, K.S.; Kim, Y.J.; Kokoza, V.; Raikhel, A.S. The role of NF-κB factor REL2 in the Aedes aegypti immune response. Insect Biochem. Mol. Biol. 2009, 39, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Paradkar, P.N.; Duchemin, J.-B.; Voysey, R.; Walker, P.J. Dicer-2-dependent activation of Culex Vago occurs via the TRAF-Rel2 signaling pathway. PLoS Negl. Trop. Dis. 2014, 8, e2823. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Yamamoto, M.; Moriyama, Y.; Yamao, M.; Furukawa, S.; Sagisaka, A.; Nakazawa, H.; Mori, H.; Yamakawa, M. A novel Rel protein and shortened isoform that differentially regulate antibacterial peptide genes in the silkworm Bombyx mori. Biochim. Biophys. Acta 2005, 1730, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Matsuki, H.; Furukawa, S.; Sagisaka, A.; Kotani, E.; Mori, H.; Yamakawa, M. Identification and functional analysis of Relish homologs in the silkworm, Bombyx mori. Biochim. Biophys. Acta 2007, 1769, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.H.; Nam, K.B.; Oh, C.T.; Nam, H.J.; Kim, S.H.; Yoon, J.H.; Seong, J.K.; Yoo, M.A.; Jang, I.H.; Brey, P.T.; et al. The homeobox gene Caudal regulates constitutive local expression of antimicrobial peptide genes in Drosophila epithelia. Mol. Cell. Biol. 2004, 24, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.H.; Kim, S.H.; Lee, H.Y.; Bai, J.Y.; Nam, Y.D.; Bae, J.W.; Lee, D.G.; Shin, S.C.; Ha, E.M.; Lee, W.J. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 2008, 319, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Junell, A.; Uvell, H.; Davis, M.M.; Edlundh-Rose, E.; Antonsson, A.; Pick, L.; Engström, Y. The POU transcription factor Drifter/Ventral veinless regulates expression of Drosophila immune defense genes. Mol. Cell Biol. 2010, 30, 3672–3684. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Chowdhury, M.; Li, C.F.; Yu, X.Q. Transcription factor forkhead regulates expression of antimicrobial peptides in the tobacco hornworm, Manduca sexta. Sci. Rep. 2017, 7, 2688. [Google Scholar] [CrossRef] [PubMed]
- Mai, T.; Chen, S.; Lin, X.; Zhang, X.; Zou, X.; Feng, Q.; Zheng, S. 20-hydroxyecdysone positively regulates the transcription of the antimicrobial peptide, lebocin, via BmEts and BmBR-C Z4 in the midgut of Bombyx mori during metamorphosis. Dev. Comp. Immunol. 2017, 74, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Gerardo, N.M.; Altincicek, B.; Anselme, C.; Atamian, H.; Barribeau, S.M.; de Vos, M.; Duncan, E.J.; Evans, J.D.; Gabaldon, T.; Ghanim, M.; et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010, 11, R21. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Pan, Y.; Gao, X.; Xi, J.; Zhang, L.; Yang, C.; Bi, R.; Yang, S.; Xin, X.; Shang, Q. Cytochrome P450 CYP6DA2 regulated by cap ‘n’collar isoform C (CncC) is associated with gossypol tolerance in Aphis gossypii Glover. Insect Mol. Biol. 2016, 25, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, M.; Palli, S.R. Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Insect Biochem. Mol. Biol. 2017, 83, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.P.; McDonnell, C.M.; Berenbaum, M.R.; Schuler, M.A. Regulation of an insect cytochrome P450 monooxygenase gene (CYP6B1) by aryl hydrocarbon and xanthotoxin response cascades. Gene 2005, 358, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Chen, X.; Pan, Y.; Zheng, Z.; Wei, X.; Xi, J.; Zhang, J.; Gao, X.; Shang, Q. Transcription factor aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator is involved in regulation of the xenobiotic tolerance-related cytochrome P450 CYP6DA2 in Aphis gossypii Glover. Insect Mol. Biol. 2017, 26, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, X.N.; Li, F.; Zhuang, S.Z.; Huang, L.N.; Ma, J.; Gao, X.W. Yeast one-hybrid screening the potential regulator of CYP6B6 overexpression of Helicoverpa armigera under 2-tridecanone stress. Bull. Entomol. Res. 2016, 106, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Wilding, C.S. Regulating resistance: CncC:Maf, antioxidant response elements and the overexpression of detoxification genes in insecticide resistance. Curr. Opin. Insect Sci. 2018, 27, 89–96. [Google Scholar] [CrossRef] [PubMed]
- King-Jones, K.; Horner, M.A.; Lam, G.; Thummel, C.S. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006, 4, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Ma, D.; Huang, Y.; He, W.; Li, Y.; Vasseur, L.; You, M. Genome-wide investigation of transcription factors provides insights into transcriptional regulation in Plutella xylostella. Mol. Genet. Genomics 2018, 293, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.N.; Li, M.; Gong, Y.H.; Liu, F.; Li, T. Cytochrome P450s-their expression, regulation, and role in insecticide resistance. Pestic. Biochem. Physiol. 2015, 120, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, N. Regulation of P450-mediated permethrin resistance in Culex quinquefasciatus by the GPCR/Galphas/AC/cAMP/PKA signaling cascade. Biochem. Biophys. Rep. 2017, 12, 12–19. [Google Scholar] [PubMed]
- Adang, M.J.; Crickmore, N.; Jurat-Fuentes, J.L. Diversity of Bacillus thuringiensis crystal toxins and mechanism of action. Adv. Insect Physiol. 2014, 47, 39–87. [Google Scholar]
- Guo, Z.J.; Kang, S.; Zhu, X.; Xia, J.X.; Wu, Q.J.; Wang, S.L.; Xie, W.; Zhang, Y.J. Down-regulation of a novel ABC transporter gene (Pxwhite) is associated with Cry1Ac resistance in the diamondback moth, Plutella xylostella (L.). Insect Biochem. Mol. Biol. 2015, 59, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, Y.; Yuan, W.; Xiao, Y.; Liu, C.; Wang, J.; Peng, J.; Peng, R.; Soberón, M.; Bravo, A.; et al. FOXA transcriptional factor modulates insect susceptibility to Bacillus thuringiensis Cry1Ac toxin by regulating the expression of toxin-receptor ABCC2 and ABCC3 genes. Insect Biochem. Mol. Biol. 2017, 88, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cotto, J.J.; Morimoto, R.I. Stress-induced activation of the heat-shock response: Cell and molecular biology of heat-shock factors. Biochem. Soc. Symp. 1999, 64, 105–118. [Google Scholar] [PubMed]
- Guertin, M.J.; Petesch, S.J.; Zobeck, K.L.; Min, I.M.; Lis, J.T. Drosophila heat shock system as a general model to investigate transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jedlicka, P.; Mortin, M.A.; Wu, C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997, 16, 2452–2462. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, T.; Kim, W.S.; Mandal, L.; Banerjee, U. Interaction between Notch and Hif-α in development and survival of Drosophila blood cells. Science 2011, 332, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
- Charoensawan, V.; Wilson, D.; Teichmann, S.A. Genomic repertoires of DNA-binding transcription factors across the tree of life. Nucleic. Acids. Res. 2010, 38, 7364–7377. [Google Scholar] [CrossRef] [PubMed]
- Mathelier, A.; Fornes, O.; Arenillas, D.J.; Chen, C.Y.; Denay, G.; Lee, J.; Shi, W.; Shyr, C.; Tan, G.; Worsley-Hunt, R.; et al. JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids. Res. 2016, 44, D110–D115. [Google Scholar] [CrossRef] [PubMed]
- Hume, M.A.; Barrera, L.A.; Gisselbrecht, S.S.; Bulyk, M.L. UniPROBE, update 2015: New tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids. Res. 2015, 43, D117–D122. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids. Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Huang, C.H. LASAGNA-Search: An integrated web tool for transcription factor binding site search and visualization. BioTechniques 2013, 54, 1417–1453. [Google Scholar] [CrossRef] [PubMed]
- Messeguer, X.; Escudero, R.; Farré, D.; Núñez, O.; Martı́nez, J.; Albà, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef] [PubMed]
- Parra, R.G.; Rohr, C.O.; Koile, D.; Perez-Castro, C.; Yankilevich, P. INSECT 2.0: A web-server for genome-wide cis-regulatory modules prediction. Bioinformatics 2016, 32, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Suzuki-Ogoh, C.; Ohmiya, Y. Dual-reporter assay using two secreted luciferase genes. BioTechniques 2007, 42, 290–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, C.S.; Wight, P.A.; Dobretsova, A.; Bronstein, I. Dual luminescence-based reporter gene assay for luciferase and β-galactosidase. BioTechniques 1996, 21, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Paguio, A.; Stecha, P.; Wood, K.V.; Fan, F. Improved dual-luciferase reporter assays for nuclear receptors. Curr. Chem. Genomics 2010, 4, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Wider, D.; Picard, D. Secreted dual reporter assay with Gaussia luciferase and the red fluorescent protein mCherry. PLoS ONE 2017, 12, e0189403. [Google Scholar] [CrossRef] [PubMed]
- Garner, M.M.; Revzin, A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: Application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981, 9, 3047–3060. [Google Scholar] [CrossRef] [PubMed]
- Fried, M.; Crothers, D.M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981, 9, 6505–6525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wang, C.F.; Zhang, Z. Transcription factor E74A affects the ecdysone titer by regulating the expression of the EO gene in the silkworm, Bomby mori. Biochim. Biophys. Acta 2017, 1861, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; Humphries, S.E. Characterization of DNA-binding proteins using multiplexed competitor EMSA. J. Mol. Biol. 2009, 385, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Karns, K.; Herr, A.E. Microfluidic electrophoretic mobility shift assays for quantitative biochemical analysis. Electrophoresis 2014, 35, 2078–2090. [Google Scholar] [CrossRef] [PubMed]
- Reece-Hoyes, J.S.; Diallo, A.; Lajoie, B.; Kent, A.; Shrestha, S.; Kadreppa, S.; Pesyna, C.; Dekker, J.; Myers, C.L.; Walhout, A.J. Enhanced yeast one-hybrid (eY1H) assays for high-throughput gene-centered regulatory network mapping. Nat. Methods 2011, 8, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Reece-Hoyes, J.S.; Marian Walhout, A.J. Yeast one-hybrid assays: A historical and technical perspective. Methods 2012, 57, 441–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mundade, R.; Ozer, H.G.; Wei, H.; Prabhu, L.; Lu, T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle 2014, 13, 2847–2852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, B.R.; Tan, M. Intra-ChIP: Studying gene regulation in an intracellular pathogen. Curr. Genet. 2016, 62, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Robert, F.; Wyrick, J.J.; Aparicio, O.; Jennings, E.G.; Simon, I.; Zeitlinger, J.; Schreiber, J.; Hannett, N.; Kanin, E.; et al. Genome-wide location and function of DNA binding proteins. Science 2000, 290, 2306–2309. [Google Scholar] [CrossRef] [PubMed]
- Nakato, R.; Shirahige, K. Recent advances in ChIP-seq analysis: From quality management to whole-genome annotation. Brief. Bioinform. 2017, 18, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Rusk, N. Reverse ChIP. Nat. Methods 2009, 6, 187. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Chen, Y.; Li, M.; Zhou, F.; Li, K.; Cao, H.; Ni, M.; Liu, Y.; Gu, Z.; et al. In situ capture of chromatin interactions by biotinylated dCas9. Cell 2017, 170, 1028–1043. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Guo, Z.; Liu, Y.; Zhang, Y. Progress and prospects of CRISPR/Cas systems in insects and other Arthropods. Front. Physiol. 2017, 8, 608. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, Y.; Zeng, B.; Wang, Y.; James, A.A.; Gurr, G.M.; Yang, G.; Lin, X.; Huang, Y.; You, M. CRISPR/Cas9 mediated knockout of the abdominal-A homeotic gene in the global pest, diamondback moth (Plutella xylostella). Insect Biochem. Mol. Biol. 2016, 75, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.S.; Gantz, V.M.; Siomava, N.; Bier, E. CRISPR/Cas9 and active genetics-based trans-species replacement of the endogenous Drosophila kni-L2 CRM reveals unexpected complexity. eLife 2017, 6, e30281. [Google Scholar] [CrossRef] [PubMed]
- Mohamad Ishak, N.S.; Nong, Q.D.; Matsuura, T.; Kato, Y.; Watanabe, H. Co-option of the bZIP transcription factor Vrille as the activator of Doublesex1 in environmental sex determination of the crustacean Daphnia magna. PLoS Genet. 2017, 13, e1006953. [Google Scholar] [CrossRef] [PubMed]
- Fields, S.; Song, O. A novel genetic system to detect protein-protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef] [PubMed]
- Rain, J.C.; Selig, L.; De Reuse, H.; Battaglia, V.; Reverdy, C.; Simon, S.; Lenzen, G.; Petel, F.; Wojcik, J.; Schächter, V.; et al. The protein-protein interaction map of Helicobacter pylori. Nature 2001, 409, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Simonis, N.; Rual, J.F.; Carvunis, A.R.; Tasan, M.; Lemmens, I.; Hirozane-Kishikawa, T.; Hao, T.; Sahalie, J.M.; Venkatesan, K.; Gebreab, F.; et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat. Methods 2009, 6, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Rolland, T.; Tasan, M.; Charloteaux, B.; Pevzner, S.J.; Zhong, Q.; Sahni, N.; Yi, S.; Lemmens, I.; Fontanillo, C.; Mosca, R.; et al. A proteome-scale map of the human interactome network. Cell 2014, 159, 1212–1226. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.V.; Freitas, M.J.; Felgueiras, J.; Fardilha, M. The power of the yeast two-hybrid system in the identification of novel drug targets: Building and modulating PPP1 interactomes. Expert Rev. Proteomics 2015, 12, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Alfatah, M.; Sin, M.F.; Sim, H.M.; Verma, C.S.; Lane, D.P.; Arumugam, P. A yeast two-hybrid system for the screening and characterization of small-molecule inhibitors of protein-protein interactions identifies a novel putative Mdm2-binding site in p53. BMC Biol. 2017, 15, 108. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Liu, Q.; Li, X.; Yang, J.; Liu, S.; Huang, Y.; Scanlon, M.J.; Nettleton, D.; Schnable, P.S. Substantial contribution of genetic variation in the expression of transcription factors to phenotypic variation revealed by eRD-GWAS. Genome Biol. 2017, 18, 192. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The human transcription factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kang, S.; Zhu, X.; Xia, J.; Wu, Q.; Wang, S.; Xie, W.; Zhang, Y. The novel ABC transporter ABCH1 is a potential target for RNAi-based insect pest control and resistance management. Sci. Rep. 2015, 5, 13728. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Names | Organisms | Websites | Descriptions | References |
|---|---|---|---|---|
| TRANSFAC | Eukaryotes | http://gene-regulation.com/ | Partially commercial. License required to access some restricted areas. | [212] |
| JASPAR | Eukaryotes | http://jaspardev.genereg.net/ | Contains a curated, non-redundant set of profiles, derived from published collections of experimentally defined eukaryotic TFBS. | [213] |
| DBD | Cellular organisms | http://www.transcriptionfactor.org/ | Contains TF predictions of more than 1000 cellular organisms. | [212] |
| UniPROBE | Cellular organisms | http://the_brain.bwh.harvard.edu/uniprobe/ | Contains DNA binding data for 638 non-redundant proteins and complexes from a diverse collection of organisms. | [214] |
| PlantTFDB | Plants | http://planttfdb.cbi.pku.edu.cn/ | Contains 320,370 TFs from 165 plant species; enables regulation prediction and functional enrichment analyses. | [215] |
| LASAGNA-Search | Organisms | http://biogrid-lasagna.engr.uconn.edu/lasagna_search/ | An integrated web tool for TFBS search and visualization. | [216] |
| PROMO | Organisms | http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3 | A virtual laboratory for the identification of putative TFBS in DNA sequences from a species or groups of species of interest. | [217] |
| MatInspector | Organisms | http://www.genomatix.de/matinspector.html | A software tool that utilizes a large library of matrix descriptions for TFBS to locate matches in DNA sequences. | [218] |
| INSECT 2.0 | Insects | http://bioinformatics.ibioba-mpsp-conicet.gov.ar/INSECT2/ | A web-server for genome-wide cis-regulatory module prediction. | [219] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Z.; Qin, J.; Zhou, X.; Zhang, Y. Insect Transcription Factors: A Landscape of Their Structures and Biological Functions in Drosophila and beyond. Int. J. Mol. Sci. 2018, 19, 3691. https://doi.org/10.3390/ijms19113691
Guo Z, Qin J, Zhou X, Zhang Y. Insect Transcription Factors: A Landscape of Their Structures and Biological Functions in Drosophila and beyond. International Journal of Molecular Sciences. 2018; 19(11):3691. https://doi.org/10.3390/ijms19113691
Chicago/Turabian StyleGuo, Zhaojiang, Jianying Qin, Xiaomao Zhou, and Youjun Zhang. 2018. "Insect Transcription Factors: A Landscape of Their Structures and Biological Functions in Drosophila and beyond" International Journal of Molecular Sciences 19, no. 11: 3691. https://doi.org/10.3390/ijms19113691
APA StyleGuo, Z., Qin, J., Zhou, X., & Zhang, Y. (2018). Insect Transcription Factors: A Landscape of Their Structures and Biological Functions in Drosophila and beyond. International Journal of Molecular Sciences, 19(11), 3691. https://doi.org/10.3390/ijms19113691