Proteomic Characterization of Bradyrhizobium diazoefficiens Bacteroids Reveals a Post-Symbiotic, Hemibiotrophic-Like Lifestyle of the Bacteria within Senescing Soybean Nodules
Abstract
:1. Introduction
2. Results
2.1. Nodule Mass and Leghemoglobin Content
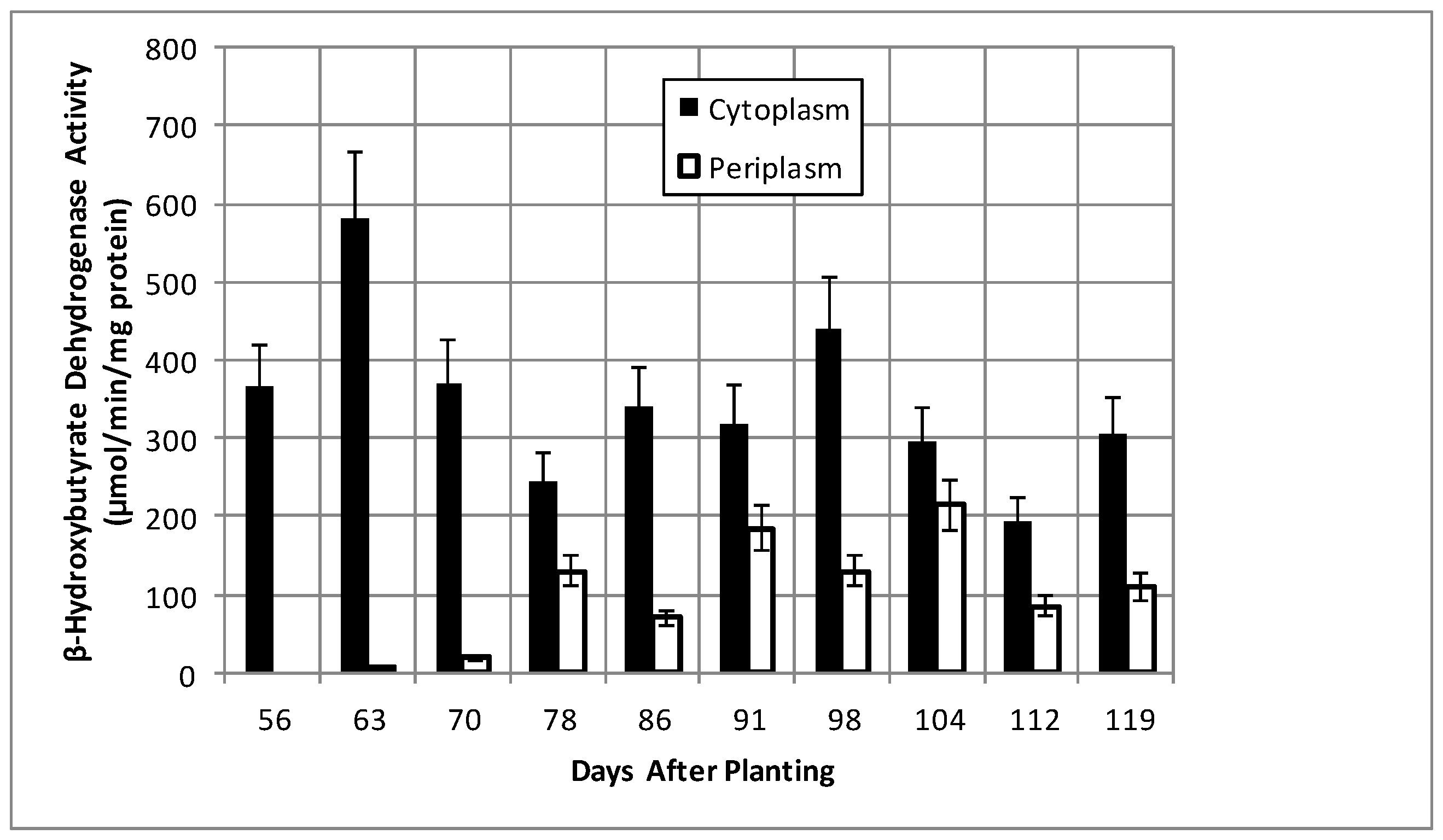
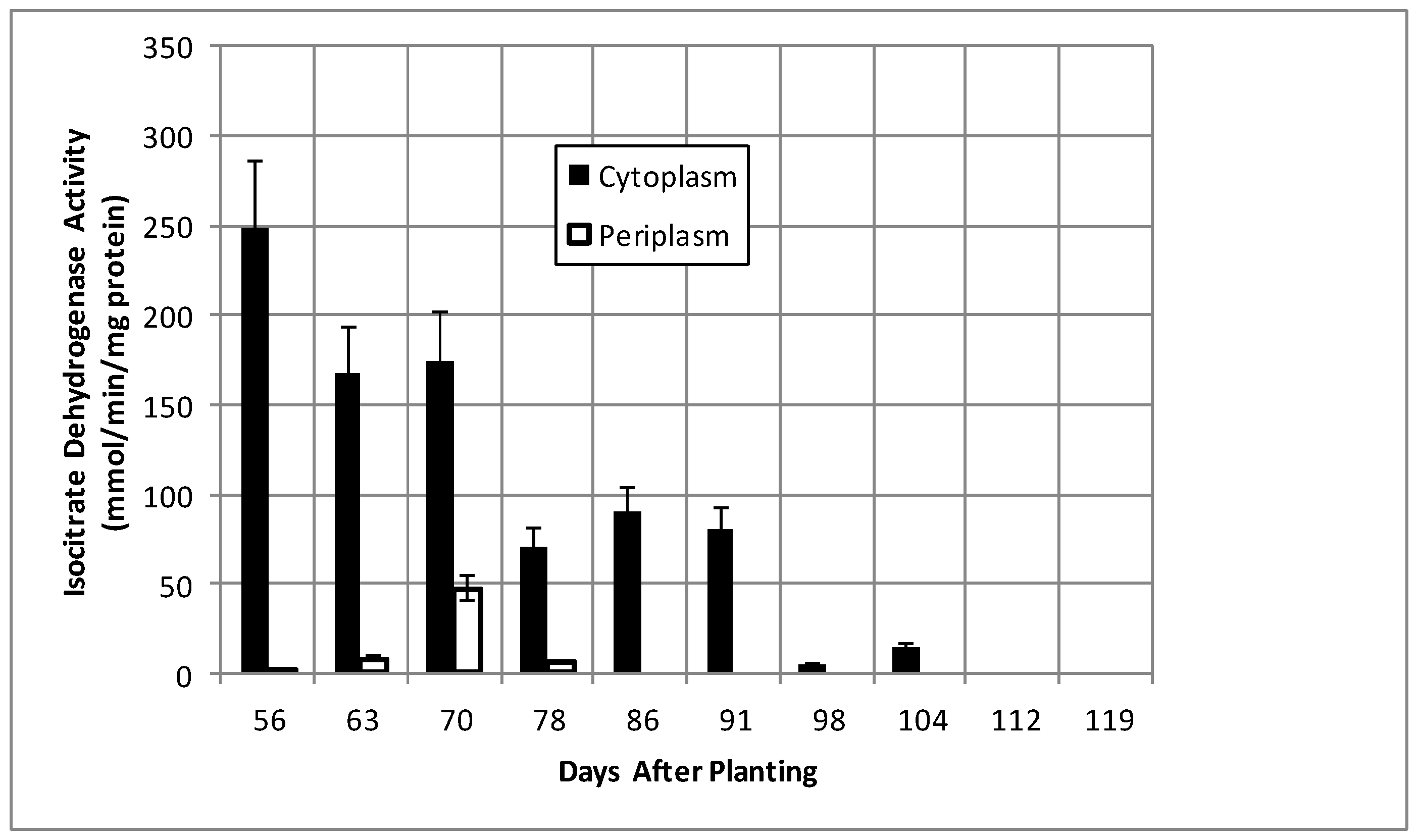
2.2. Bacteroid Protein and Poly-β-hydroxybutyrate (PHB) Content and Enzymes Activities in the Post-Symbiotic Period
2.3. Proteomics Time Course
2.4. Proteins That Declined Following Symbiosis
2.5. Proteins That Increased Following Symbiosis
2.6. Constitutive Proteins
3. Discussion
4. Materials and Methods
4.1. Source of Nodules and Bacteroid Preparations
4.2. Enzymatic, Leghemoglobin, and Poly-β-hydroxybutyrate (PHB) Analysis
4.3. Protein Isolation and Identification
4.4. Mass Spectrometry Analysis
4.5. Database Searching and Spectral Analysis
4.6. Peptide Match Filtering
4.7. Protein Expression Trends
Author Contributions
Funding
Acknowledgements
Conflicts of Interest
References
- Douglas, A. The Symbiotic Habit; Princeton University Press: Princeton, NJ, USA, 2010; p. 202. ISBN 0-19-854294-1. [Google Scholar] [CrossRef]
- Hellriegel, H.; Wilfarth, H. Untersuchungen uber die Stickstoffnahrung der Gramineon und Leguminosen; Beilageheft zu der Ztschr. Ver. Rü benzucker-Industrie Deutschen Reichs Germany; Food and Agriculture Organization of the United Nations: Roma, Italy, 1888; p. 234, ISBN 10: 1110628293, ISBN 13: 9781110628292. [Google Scholar]
- Cermola, M.; Fedorova, E.; Tate, F.R.; Patriarca, E.J. Nodule invasion and symbiosome differentiation during Rhizobium etli-Phaseolus vulgaris symbiosis. Mol. Plant Microbe Interact. 2000, 13, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Groten, K.; Dutilleul, C.; van Heerden, P.D.; Vanacker, H.; Bernard, S.; Finkemeier, I.; Dietz, K.J.; Fover, C.H. Redox regulation of peroxiredoxin and proteinases by ascorbate and thiols during pea root nodule senescence. FEBS Lett. 2006, 580, 1269–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, J.; Wiemken, A.; Boller, T. Redifferentiation of bacteria isolated from Lotus japonicus root nodules colonized by Rhizobium sp. NGR234. J. Exp. Bot. 2001, 52, 2181–2186. [Google Scholar] [CrossRef] [PubMed]
- Puppo, A.; Groten, K.; Bastain, F.; Carzaniga, R.; Soussi, M.; Lucas, M.M.; de Felipe, M.R.; Harrison, J.; Vanacker, H.; Fover, C.H. Legume nodule senescence: Roles for redox and hormone signalling in the orchestration of the natural aging process. New Phytol. 2005, 165, 683–701. [Google Scholar] [CrossRef]
- Vasse, J.; De Billy, F.; Camut, S.; Truchet, G. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol. 1990, 172, 4295–4306. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Rose, J.K.C. Mediation of the transition from biotrophy to necrotrophy in hemibriotrophic plant pathogens by secreted effector proteins. Plant Signal. Behav. 2010, 5, 769–772. [Google Scholar] [CrossRef]
- Spanu, P.D.; Panstruga, R. Editorial: Biotrophic Plant-Microbe Interactions. Front. Plant Sci. 2017, 8, 192. [Google Scholar] [CrossRef]
- Hiltner, L. Über neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie unter bessonderer Berücksichtigung der Gründung und Brache. Arb. Dtsch. Landwirtsch. Ges. Berl. 1904, 98, 59–78. [Google Scholar]
- Marschner, H. Mineral Nutrition of Higher Plants; Academic Press: London, UK, 1995; 889p. [Google Scholar] [CrossRef]
- Sarma, A.D.; Emerich, D.W. Global protein expression pattern of Bradyrhizobium japonicum bacteroids: A prelude to functional proteomics. Proteomics 2005, 5, 4170–4184. [Google Scholar] [CrossRef]
- Delmotte, N.; Ahrens, C.J.; Knief, C.; Qeli, E.; Koch, M.; Fischer, H.M.; Vorholt, J.A.; Hennecke, H. An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean nodules. Proteomics 2010, 10, 1391–1400. [Google Scholar] [CrossRef]
- Sarath, G.; Preiffer, N.E.; Sodhi, C.S.; Wagner, F.W. Bacteroids are stable during dark induced senescence of soybean root nodules. Plant Physiol. 1986, 82, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Pladys, D.; Vance, C.P. Proteolysis during development and senescence of effective and plant gene-controlled ineffective alfalfa nodules. Plant Physiol. 1993, 103, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.J.; Gallesi, D.; Mathieu, C.; Hernandez, M.J.; de Felipe, M.; Halliwell, B.; Puppo, A. Oxidative stress occurs during soybean nodule senescence. Planta 1999, 208, 73–78. [Google Scholar] [CrossRef]
- Karr, D.B.; Waters, J.K.; Suzuki, F.; Emerich, D.W. Enzymes of the β-hydroxybutyrate and citric acid cycles of Rhizobium japonicum bacteroids. Plant Physiol. 1984, 75, 1158–1162. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Emerich, D.W. Isocitrate Dehydrogenase of Bradyrhizobium japonicum is not required for symbiotic nitrogen fixation with soybean. J. Bacteriol. 2006, 188, 7600–7608. [Google Scholar] [CrossRef] [PubMed]
- Karr, D.B.; Emerich, D.W. Protein synthesis by Bradyrhizobium japonicum bacteroids declines as a function of nodule age. Appl. Environ. Microbiol. 1996, 62, 3757–3761. [Google Scholar] [PubMed]
- Scotti, M.A.; Chanfon, A.; Hennecke, H.; Fischer, H.-M. Disparate oxygen responsiveness of two regulatory cascades that control expression of symbiotic gene in Bradyrhizobium japonicum. J. Bacteriol. 2003, 185, 5639–5642. [Google Scholar] [CrossRef]
- Lindemann, A.; Moser, A.; Pessi, G.; Hauser, F.; Friberg, M.; Hennecke, H.; Fischer, H.M. New target genes controlled by the Bradyrhizobium japonicum two-component regulatory system RegSR. J. Bacteriol. 2007, 189, 8928–8943. [Google Scholar] [CrossRef]
- Green, L.S.; Li, Y.; Emerich, D.W.; Bergersen, F.J.; Day, D.A. Catabolism of α-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: Evidence for an alternative tricarboxylic acid cycle. J. Bacteriol. 2000, 182, 2838–2844. [Google Scholar] [CrossRef]
- Fischer, H.M.; Babst, M.; Kaspar, T.; Acuna, G.; Arigoni, F.; Hennecke, H. One member of a gro-ESL-like chaperonin multigene family in Bradyrhziobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO 1993, 12, 2901–2912. [Google Scholar] [CrossRef]
- Hauser, F.; Pessi, G.; Friberg, M.; Weber, C.; Rusca, N.; Lindemann, A.; Fischer, H.M.; Hennecke, H. Dissection of the Bradyrhizobium japonicum NifA+sigma54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol. Genet. Genom. 2007, 278, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Cytryn, E.J.; Sangurdekar, D.P.; Streeter, J.G.; Franck, W.L.; Chang, W.-S.; Stacey, G.; Emerich, D.W.; Trupti, J.; Xu, D.; Sadowsky, M.J. Transcriptional and physiological responses of Bradyrhizobium japonicum to dessication-induced stress. J. Bacteriol. 2007, 189, 6751–6762. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Yokoyama, T.; Minamisawa, K.; Mitsui, H.; Itakura, M.; Kaneko, T.; Tabata, S.; Saeki, K.; Omori, H.; Tajima, S.; et al. Soybean seed extracts preferentially express genomic loci of Bradyrhizobium japonicum in the initial interaction with soybean, Glycine max (L). Merr. DNA Res. 2008, 15, 201–214. [Google Scholar] [CrossRef]
- Parke, D.; Ornston, L.N. Enzymes of the beta-ketoadipate pathway are inducible in Rhizobium and Agrobacterium spp. and constitutive in Bradyrhizobium spp. J. Bacteriol. 1986, 165, 288–292. [Google Scholar] [CrossRef]
- Hillmer, P.; Gest, H. H2 metabolism in the photosynthetic bacterium Rhodopseudomonas capsulate: Production and utilization of H2 by resting cells. J. Bacteriol. 1977, 129, 732–739. [Google Scholar]
- Richardson, D.J.; King, G.F.; Kelly, D.J.; McEwan, A.G.; Ferguson, S.J.; Jackson, J.B. The role of auxiliary oxidant in maintaining redox balance during phototrophic growth of Rhodobacter capsulatus on propionate or butyrate. Arch. Microbiol. 1988, 150, 131–137. [Google Scholar] [CrossRef]
- Falcone, D.L.; Tabita, F.R. Expression of endogenous and foreign ribulose 1,5-bisphosphate carboxylase-oxygenase (Rubisco) genes in a Rubisco delection mutant of Rhodobacter sphaeroides. J. Bacteriol. 1991, 173, 2099–2108. [Google Scholar] [CrossRef] [PubMed]
- McKinlay, J.B.; Harwood, C.S. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 11669–11675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gourion, B.; Delmotte, N.; Bonaldi, K.; Nouwen, N.; Vorholt, J.A.; Girau, E. Bacterial RubisCO is required for efficient Bradyrhizobium/Aeschynomene symbiosis. PLoS ONE 2011, 6, e21900. [Google Scholar] [CrossRef] [PubMed]
- Franck, S.; Strodtman, K.N.; Qiu, J.; Emerich, D.W. Transcriptomic analysis of Bradyrhizobium diazoefficiens bacteroids reveals a post-symbiotic, hemibiotrophic lifestyle of the bacteria within senescing soybean nodules. Internat. J. Mol. Sci. 2018, 19, 3918. [Google Scholar] [CrossRef]
- McDermott, T.R.; Graham, P.H.; Brandwein, D.H. Viability of Bradyrhizobium japonicum bacteroids. Arch. Microbiol. 1987, 148, 100–106. [Google Scholar] [CrossRef]
- Tsien, H.C.; Cain, P.S.; Schmidt, E.L. Viability of rhizobium bacteroids. Appl. Environ. Microbiol. 1977, 34, 854–856. [Google Scholar] [PubMed]
- Wong, P.P.; Evans, H.J. Poly-β-hydroxybutyrate utilization by soybean (Glycine max Merr) nodules and assessment of its role in maintenance of nitrogenase activity. Plant. Physiol. 1971, 47, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Luiz, J.; Rodigues, M.; Melotto, M.; Gomes-Silveria, J.A.; Silveria, D. Nodulin activities, hydrogenase expression and nitrogen fixation in cowpea inoculated with Hup+ and Hup− symbionts. Rev. Bras. Fisiol. Veg. 1998, 10, 85–90. [Google Scholar]
- Pfeiffer, N.E.; Torres, C.M.; Wagner, F.M. Proteolytic activity in soybean root nodules. Activity in host cell cytosol and bacteroids throughout physiological development and senescence. Plant Physiol. 1983, 71, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Winzer, T.; Bairl, A.; Linder, M.; Werner, D.; Muller, P. A novel 53-kDa nodulin of the symbiosome membrane of soybean nodules controlled by Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 1999, 12, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Manen, J.F.; Simon, P.; Van Slooten, J.C.; Osteras, M.; Frutiger, S.; Hughes, G.J. A nodulin specifically expressed in senescent nodules of winged bean is a protease inhibitor. Plant Cell 1991, 3, 259–270. [Google Scholar] [CrossRef]
- Kereszt, A.; Mergaert, P.; Kondorosi, E. Bacteroid development in legume nodules: Evolution of mutual benefit or of sacrificial victims? Mol. Plant Microbe Interact. 2011, 24, 1300–1309. [Google Scholar] [CrossRef]
- Strodtman, K.N.; Stevenson, S.E.; Waters, J.K.; Mawhinney, T.P.; Thelen, J.J.; Polacco, J.C.; Emerich, D.W. The bacteroid periplasm in soybean nodules is an interkingdom symbiotic space. Mol. Microbe Plant Interact. 2017, 30, 997–1008. [Google Scholar] [CrossRef]
- Streeter, J.G.; Le Rudlier, D. Release of periplasmic enzymes from Rhizobium leguminosarum bv phaseoli bacteroids by lysozyme is enhanced by pretreatment of cells at low pH. Curr. Microbiol. 1990, 21, 169–173. [Google Scholar] [CrossRef]
- Rice, E.W. Rapid determination of total hemoglobin as hemoglobin cyanide in total blood containing carboxyhemoglobin. Clin. Chim. Acta 1967, 18, 89–91. [Google Scholar] [CrossRef]
- Karr, D.B.; Waters, J.K.; Emerich, D.W. Analysis of Poly-3-Hydroxybutyrate in Rhizobium japonicum bacteroids by ion exclusion HPLC and UV detection. Appl. Environ. Microbiol. 1983, 46, 1339–1343. [Google Scholar] [PubMed]
- Carvalho, P.C.; Fisher, J.S.; Chen, E.I.; Yates, J.R., III; Barbosa, V.C. PatternLab for proteomics: A tool for differential shotgun proteomics. BMC Bioinform. 2008, 9, 316. [Google Scholar] [CrossRef] [PubMed]







| Function | Rhizobase | Accession Number | Location |
|---|---|---|---|
| 67 kDa Myosin-cross-reactive streptococcal antigen homolog | bll0025 | NP_766665 | Cytoplasm |
| Dihydrolipoamide dehydrogenase | bll0449 | NP_767089 | Cytoplasm |
| Succinyl-CoA synthetase beta chain | bll0455 | NP_767095 | Cytoplasm |
| Malate dehydrogenase | bll0456 | NP_767096 | Cytoplasm |
| 3-isopropylmalate dehydrogenase | bll0504 | NP_767143 | Cytoplasm |
| Protein-export protein | bll0641 | NP_767281 | Cytoplasm |
| ABC transporter glycerol-3-phosphate-binding protein | bll0733 | NP_767373 | Cytoplasm |
| Polyribonucleotide nucleotidyltransferase | bll0779 | NP_767419 | Cytoplasm |
| ABC transporter substrate-binding protein | bll0887 | NP_767527 | Cytoplasm |
| Fructose bisphosphate aldolase | bll1520 | NP_768160 | Cytoplasm |
| Hypothetical protein | bll1875 | NP_768515 | Cytoplasm |
| GroEL3 chaperonin | bll2059 | NP_768699 | Both |
| GroES3 chaperonin | bll2060 | NP_768700 | Cytoplasm |
| phenolhydroxylase homolog | bll2063 | NP_768703 | Both |
| MoxR family protein | bll2465 | NP_769105 | Cytoplasm |
| Cystathionine beta lyase | bll4445 | NP_771085 | Cytoplasm |
| Hypothetical protein | bll4781 | NP_771421 | Cytoplasm |
| Pyruvate dehydrogenase beta subunit | bll4782 | NP_771422 | Cytoplasm |
| Enolase | bll4794 | NP_771434 | Cytoplasm |
| Translation elongation factor Ts | bll4860 | NP_771500 | Periplasm |
| Prolyl-tRNA synthetase | bll4877 | NP_771517 | Cytoplasm |
| ATP-dependent protease LA | bll4942 | NP_771582 | Cytoplasm |
| Trigger factor | bll4945 | NP_771585 | Cytoplasm |
| 50S ribosomal protein L14 | bll5390 | NP_772030 | Cytoplasm |
| Elongation factor TU | bll5402 | NP_772042 | Cytoplasm |
| Translation elongation factor G | bll5403 | NP_772043 | Cytoplasm |
| 30S ribosomal protein S7 | bll5404 | NP_772044 | Cytoplasm |
| DNA-directed RNA polymerase beta chain | bll5409 | NP_772049 | Cytoplasm |
| DNA-directed RNA polymerase beta chain | bll5410 | NP_772050 | Cytoplasm |
| Carbon monoxide dehydrogenase large chain | bll5914 | NP_772554 | Cytoplasm |
| Putative hydrolase serine protease transmembrane protein | bll6508 | NP_773148 | Cytoplasm |
| 3-oxoadipate CoA-transferase subunit A | bll7093 | NP_773733 | Cytoplasm |
| DNA-binding stress response protein, Dps family | bll7374 | NP_774014 | Cytoplasm |
| Phosphoenolpyruvate carboxykinase | bll8141 | NP_774781 | Cytoplasm |
| Unknown protein | bll8309 | NP_774949 | Periplasm |
| Hypothetical protein | blr0028 | NP_766668 | Cytoplasm |
| 3-isopropylmalate dehydratase large subunit | blr0488 | NP_767128 | Cytoplasm |
| Succinate dehydrogenase flavoprotein subunit | blr0514 | NP_767154 | Cytoplasm |
| 30S ribosomal protein S1 | blr0740 | NP_767380 | Cytoplasm |
| Catalase | blr0778 | NP_767418. | Cytoplasm |
| Succinate-semialdehyde dehydrogenase | blr0807 | NP_767447 | Cytoplasm |
| Putative aminotransferase protein | blr1686 | NP_768326 | Both |
| Nitrogenase molybdenum-iron protein alpha chain (NifD) | blr1743 | NP_768383 | Both |
| Nitrogenase molybdenum-iron protein beta chain (NifK) | blr1744 | NP_768384 | Both |
| Flavoprotein (FixC) | blr1774 | NP_768414 | Both |
| Pantoate--beta-alanine ligase | blr1852 | NP_768492 | Cytoplasm |
| 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase | blr2068 | NP_768708 | Cytoplasm |
| Unknown protein | blr2372 | NP_769012 | Cytoplasm |
| Glutaryl-CoA dehydrogenase | blr2616 | NP_769256 | Cytoplasm |
| Aldehyde dehydrogenase | blr2816 | NP_769456 | Cytoplasm |
| Nucleoside diphosphate kinase | blr4119 | NP_770759 | Cytoplasm |
| Aminotransferase | blr4134 | NP_770774 | Cytoplasm |
| NAD-dependent malic enzyme | blr4145 | NP_770785 | Cytoplasm |
| Citrate synthase | blr4839 | NP_771479 | Both |
| Glutamine synthetase I | blr4949 | NP_771589 | Cytoplasm |
| Anti-oxidant protein | blr5308 | NP_771948. | Cytoplasm |
| 60 KDA chaperonin | blr5626 | NP_772266 | Both |
| Adenylosuccinate lyase | blr5690 | NP_772330 | Cytoplasm |
| 30S ribosomal protein S4 | blr5706 | NP_772346 | Cytoplasm |
| Hypothetical protein | blr6718 | NP_773358 | Cytoplasm |
| Hypothetical protein | blr7070 | NP_773710 | Cytoplasm |
| Hypothetical zinc protease | blr7485 | NP_774125 | Cytoplasm |
| Glutamate synthase large subunit | blr7743 | NP_774383 | Cytoplasm |
| ABC transporter substrate-binding protein | blr8117 | NP_774757 | Cytoplasm |
| Two-component response regulator | blr8144 | NP_774784 | Cytoplasm |
| 30S ribosomal protein S18 | bsl4078 | NP_770718 | Cytoplasm |
| Function | Rhizobase | Accession | Location |
|---|---|---|---|
| Hypothetical protein | bll2012 | NP_768652 | Cytoplasm |
| Unknown protein | bll4278 | NP_770918 | Cytoplasm |
| Peptidyl prolyl cis-trans isomerase | bll4690 | NP_771330 | Cytoplasm |
| Hypothetical protein | bll5155 | NP_771795 | Cytoplasm |
| Hypothetical protein | bll7395 | NP_774035 | Cytoplasm |
| CheY, two-component response regulator | bll7795 | NP_774435. | Periplasm |
| Probable fatty oxidation complex, alpha subunit | bll7821 | NP_774461 | Periplasm |
| Acetyl-CoA carboxylase carboxyl transferase α-subunit | blr0191 | NP_766831 | Cytoplasm |
| Carboxy-terminal protease | blr0434 | NP_767074 | Periplasm |
| Acyl-CoA thiolase | blr1159 | NP_767799 | Periplasm |
| Enoyl-CoA hydratase | blr1160 | NP_767800 | Periplasm |
| Unknown protein | blr1830 | NP_768470 | Cytoplasm |
| Hypothetical protein | blr3804 | NP_770444 | Cytoplasm |
| ABC transporter substrate-binding protein | blr4039 | NP_770679 | Periplasm |
| Hypothetical protein | blr5502 | NP_772142 | Cytoplasm |
| Cold shock protein | bsl3986 | NP_770626 | Cytoplasm |
| Function | Rhizobase | Accession Number | Fraction |
|---|---|---|---|
| N-utilization substance protein A | bll0785 | NP_767425 | Periplasm |
| Transketolase | bll1524 | NP_768164 | Pperiplasm |
| Alkyl hydroperoxide reductase | bll1777 | NP_768417 | Cytoplasm |
| Carbonic anhydrase | bll2065 | NP_768705 | Cytoplasm |
| Nodulate formation efficiency C protein | bll2067 | NP_768707 | Periplasm |
| Probable amino acid binding protein | bll2909 | NP_769549 | Both |
| Glycine hydroxymethyltransferase | bll5033 | NP_771673 | Periplasm |
| ABC transporter substrate-binding protein | bll5596 | NP_772236 | Cytoplasm |
| RecA protein | bll5755 | NP_772395 | Periplasm |
| Hypothetical protein | bll6069 | NP_772709 | Periplasm |
| PQQ-dependent alcohol dehydrogenase | bll6220 | NP_772860 | Cytoplasm |
| Peptidyl-dipeptidase | bll7756 | NP_774396 | Periplasm |
| Superoxide dismutase | bll7774 | NP_774414 | Cytoplasm |
| ABC transporter substrate-binding protein (Putative oligopeptide binding protein) | bll7921 | NP_774561 | Both |
| Hypothetical protein | bll8229 | NP_774869 | Periplasm |
| Heat shock protein 70 | blr0678 | NP_767319 | Periplasm |
| Beta-ketoadipyl CoA thiolase | blr0925 | NP_767565 | Periplasm |
| Peptidoglycan acetylation protein | blr0936 | NP_767576 | Periplasm |
| ATP-dependent protease, ATP-binding subunit | blr1404 | NP_768044 | Periplasm |
| Dehydrogenase | blr2146 | NP_768786 | Periplasm |
| Methylmalonate-semialdehyde dehydrogenase | blr3954 | NP_770594 | Periplasm |
| Probable transaldolase | blr6758 | NP_773398 | Periplasm |
| Histone H4 | - | Glyma02g38920 | Periplasm |
| Histone H3 | - | Glyma06g32880 | Periplasm |
| Glu/leu/phe/val dehydrogenase | - | Glyma16g04560 | Periplasm |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strodtman, K.N.; Frank, S.; Stevenson, S.; Thelen, J.J.; Emerich, D.W. Proteomic Characterization of Bradyrhizobium diazoefficiens Bacteroids Reveals a Post-Symbiotic, Hemibiotrophic-Like Lifestyle of the Bacteria within Senescing Soybean Nodules. Int. J. Mol. Sci. 2018, 19, 3947. https://doi.org/10.3390/ijms19123947
Strodtman KN, Frank S, Stevenson S, Thelen JJ, Emerich DW. Proteomic Characterization of Bradyrhizobium diazoefficiens Bacteroids Reveals a Post-Symbiotic, Hemibiotrophic-Like Lifestyle of the Bacteria within Senescing Soybean Nodules. International Journal of Molecular Sciences. 2018; 19(12):3947. https://doi.org/10.3390/ijms19123947
Chicago/Turabian StyleStrodtman, Kent N., Sooyoung Frank, Severin Stevenson, Jay J. Thelen, and David W. Emerich. 2018. "Proteomic Characterization of Bradyrhizobium diazoefficiens Bacteroids Reveals a Post-Symbiotic, Hemibiotrophic-Like Lifestyle of the Bacteria within Senescing Soybean Nodules" International Journal of Molecular Sciences 19, no. 12: 3947. https://doi.org/10.3390/ijms19123947
APA StyleStrodtman, K. N., Frank, S., Stevenson, S., Thelen, J. J., & Emerich, D. W. (2018). Proteomic Characterization of Bradyrhizobium diazoefficiens Bacteroids Reveals a Post-Symbiotic, Hemibiotrophic-Like Lifestyle of the Bacteria within Senescing Soybean Nodules. International Journal of Molecular Sciences, 19(12), 3947. https://doi.org/10.3390/ijms19123947