Oleuropein Induces AMPK-Dependent Autophagy in NAFLD Mice, Regardless of the Gender
Abstract
1. Introduction
2. Results
2.1. ND and HFD Liver Histology before and after Ole Treatment
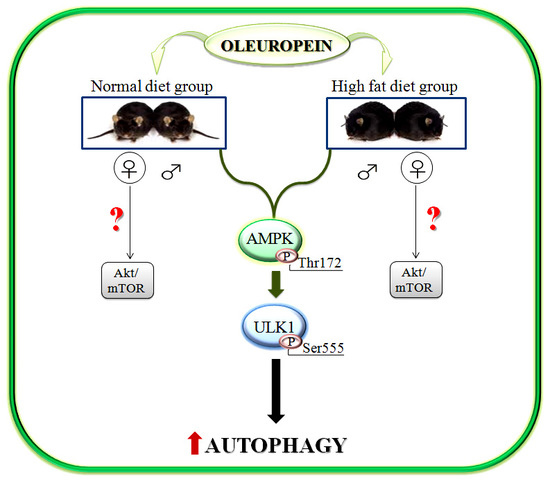
2.2. Gender Specific Activation of Akt/mTOR Pathway by Ole in ND and HFD Mice
2.3. Activation of AMPK/ULK1 Pathway by Ole in ND and HFD Mice
2.4. Ole is Able to Induce Early Autophagic Machinery
2.5. Ole Does Not Affect the Expression of Caspase 3 and Bcl-2 Apoptotic Proteins in HFD Mice
3. Discussion
4. Materials and Methods
4.1. Mice Experimental Protocol
4.2. Histological Analysis
4.3. Immunohistochemistry for Caspase 3 and Bcl2
4.4. RNA Extraction and cDNA Synthesis
4.5. Real-Time Quantitative Polymerase Chain Reaction PCR (Q-PCR) Analysis
4.6. Western Blot Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ND | Normal diet |
| HFD | High-fat diet |
| Ole | Oleuropein |
| NAFLD | Non-alcoholic fatty liver disease |
References
- Tarantino, G.; Saldalamacchia, G.; Conca, P.; Arena, A. Non-alcoholic fatty liver disease: Further expression of the metabolic syndrome. J. Gastroenterol. Hepatol. 2007, 22, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Araújo, A.R.; Rosso, N. Global epidemiology of non-alcoholic fatty liver disease/non-alcoholic steatohepatitis: What we need in the future. Liver Int. 2018, 38, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Visioli, F.; Bernardini, E. Extra virgin olive oil’s polyphenols: Biological activities. Curr. Pharm. Des. 2011, 17, 786–804. [Google Scholar] [CrossRef] [PubMed]
- Barbaro, B.; Toietta, G.; Maggio, R.; Arciello, M.; Tarocchi, M.; Galli, A.; Balsano, C. Effects of the olive-derived polyphenol oleuropein on human health. Int. J. Mol. Sci. 2014, 15, 18508–18524. [Google Scholar] [CrossRef]
- Lavallard, V.J.; Gual, P. Autophagy and non-alcoholic fatty liver disease. Biomed. Res. Int. 2014, 2014, 120179. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Yu, F.; Wang, J.; Guo, C.; Fan, X. Autophagy: A new target for nonalcoholic fatty liver disease therapy. Hepat. Med. 2016, 8, 27–37. [Google Scholar] [CrossRef]
- Buytaert, E.; Dewaele, M.; Agostinis, P. Molecular effectors of multiple cell death pathways initiated by photodynamic therapy. Biochim. Biopys. Acta 2007, 1776, 86–107. [Google Scholar] [CrossRef]
- Lefranc, F.; Facchini, V.; Kiss, R. Proautophagic drugs: A novel means to combat apoptosis-resistant cancer, with a special emphasis on glioblastomas. Oncologist 2007, 12, 1395–1403. [Google Scholar] [CrossRef]
- Fazi, B.; Bursch, W.; Fimia, G.M.; Nardacci, R.; Piacentini, M.; Di Sano, F.; Piredda, L. Fenretinide induces autophagic cell death in caspase-defective breast cancer cells. Autophagy 2008, 4, 435–441. [Google Scholar] [CrossRef]
- Hu, X.; Xuan, Y. Bypassing cancer drug resistance by activating multiple death pathways-a proposal from the study of circumventing cancer drug resistance by induction of necroptosis. Cancer Lett. 2008, 259, 127–137. [Google Scholar] [CrossRef]
- Shen, S.; Kepp, O.; Michaud, M.; Martins, I.; Minoux, H.; Métivier, D.; Maiuri, M.C.; Kroemer, R.T.; Kroemer, G. Association and dissociation of autophagy, apoptosis and necrosis by systematic chemical study. Oncogene 2011, 30, 4544–4556. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Debnath, J. Autophagyat the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015, 16, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Bursch, W.; Karwan, A.; Mayer, M.; Dornetshuber, J.; Fröhwein, U.; Schulte-hermann, R.; Fazi, B.; Di Sano, F.; Piredda, L.; Piacentini, M.; et al. Cell death and autophagy: Cytokines, drugs, nutritional factors. Toxicology 2008, 254, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Scherz-Shouval, R.; Elazar, Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007, 17, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Rubinsztein, D.C.; Gestwicki, J.E.; Murphy, L.O.; Klionsky, D.J. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007, 6, 304–312. [Google Scholar] [CrossRef]
- Kopelovich, L.; Fay, J.R.; Sigmann, C.C.; Crowell, J.A. The mammalian target of rapamycin pathway as a potential target of cancer chemoprevention. Cancer Epidemiol. Biomarkers Prev. 2007, 1687, 1330–1340. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell. 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Schläfli, A.M.; Adams, O.; Galván, J.A.; Gugger, M.; Savic, S.; Bubendorf, L.; Schmid, R.A.; Becker, K.F.; Tschan, M.P.; Langer, R.; et al. Prognostic value of the autophagy markers LC3 and p62/SQSTM1 in early-stage non-small cell lung cancer. Oncotarget 2016, 7, 39544–39555. [Google Scholar] [CrossRef]
- Marino, M.L.; Fais, S.; Djavaheri-Mergny, M.; Villa, A.; Meschini, S.; Lozupone, F.; Venturi, G.; Della Mina, P.; Pattingre, S.; Rivoltini, L.; et al. Proton pump inhibition induces autophagy as a survival mechanism following oxidative stress in human melanoma cells. Cell Death Dis. 2010, 1e87. [Google Scholar] [CrossRef]
- Omar, S.H. Oleuropein in olive and its pharmacological effects. Sci. Pharm. 2010, 78, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Choi, Y.; Um, S.J.; Yoon, S.K.; Park, T. Oleuropein attenuates hepatic steatosis induced by high-fat diet in mice. J. Hepatol. 2011, 54, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Choi, Y.; Park, T. Hepatoprotective effect of oleuropein in mice: Mechanisms uncovered by gene expression profiling. Biotechnol. J. 2010, 5, 950–960. [Google Scholar] [CrossRef]
- Chen, C.P.; Lai, T.C.; Chern, S.R.; Li, S.H.; Chou, H.C.; Chen, Y.W.; Lin, S.T.; Lu, Y.C.; Wu, C.L.; Li, J.M.; et al. Proteome differences between male and female fetal cells in amniotic fluid. OMICS. 2013, 17, 16–26. [Google Scholar] [CrossRef] [PubMed]
- De Bock, M.; Thorstensen, E.B.; Derraik, J.G.; Henderson, H.V.; Hofman, P.L.; Cutfield, W.S. Human absorption and metabolism of oleuropein and hydroxytyrosol ingested as olive (Olea europaea L.) leaf extract. Mol. Nutr. Food. Res. 2013, 57, 2079–2085. [Google Scholar] [CrossRef]
- Surwit, R.S.; Kuhn, C.M.; Cochrane, C.; McCubbin, J.A.; Feinglos, M.N. Diet-induced type II diabetes in C57BL/6J mice. Diabetes 1988, 37, 1163–1167. [Google Scholar] [CrossRef]
- Wang, L.; Yu, S.; Chan, A.W.H. Pathology of Non-Alcoholic Fatty Liver Disease. Int. J. Dig. Dis. 2016, 2, 71–83. [Google Scholar] [CrossRef]
- Tandra, S.; Yeh, M.M.; Brunt, E.M.; Vuppalanchi, R.; Cummings, O.W.; Ünalp-Arida, A.; Wilson, L.A.; Chalasani, N. Presence and significance of microvesicular steatosis in nonalcoholic fatty liver disease. J. Hepatol. 2011, 55, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Noda, N.N.; Inagaki, F. Mechanisms of Autophagy. Annu Rev Biophys. 2015, 44, 101–122. [Google Scholar] [CrossRef]
- Lee, S.; Kim, J.S. Mitophagy: Therapeutic potentials for liver disease and beyond. Toxicol Res. 2014, 30, 243–250. [Google Scholar] [CrossRef]
- Vakifahmetoglu-Norberg, H.; Xia, H.G.; Yuan, J. Pharmacologic agents targeting autophagy. J. Clin. Invest. 2015, 125, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Kimball, S.R.; Ravi, S.; Gordon, B.S.; Dennis, M.D.; Jefferson, L.S. Amino Acid-Induced Activation of mTORC1 in Rat Liver Is Attenuated by Short-Term Consumption of a High-Fat Diet. J. Nutr. 2015, 145, 2496–2502. [Google Scholar] [CrossRef]
- Lim, H.; Lim, Y.M.; Kim, K.H.; Jeon, Y.E.; Park, K.; Kim, J.; Hwang, H.Y.; Lee, D.J.; Pagire, H.; Kwon, H.J.; et al. A novel autophagy enhancer as therapeutic agent against metabolic syndrome and diabetes. Nat. Commun 2018, 9, 1438. [Google Scholar] [CrossRef] [PubMed]
- Wirawan, E.; Vande-Walle, L.; Kersse, K.; Cornelis, S.; Claerhout, S.; Vanoverberghe, I.; Roelandt, R.; de-Rycke, R.; Verspurten, J.; Declercq, W.; et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010, 1, e18. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Gao, P.; Zhang, J. Crosstalk between Autophagy and Apoptosis: Potential and Emerging Therapeutic Targets for Cardiac Diseases. Int. J. Mol. Sci. 2016, 3, 332. [Google Scholar] [CrossRef]
- Galluzzi, L.; Bravo-San Pedro, J.M.; Levine, B.; Green, D.R.; Kroemer, G. Pharmacological modulation of autophagy: Therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2017, 16, 487–511. [Google Scholar] [CrossRef]
- Martini, M.; Cenci, T.; D’Alessandris, G.Q.; Cesarini, V.; Cocomazzi, A.; Ricci-Vitiani, L.; De Maria, R.; Pallini, R.; Larocca, L.M. Epigenetic silencing of Id4 identifies a glioblastoma subgroup with a better prognosis as a consequence of an inhibition of angiogenesis. Cancer 2013, 119, 1004–1012. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porcu, C.; Sideri, S.; Martini, M.; Cocomazzi, A.; Galli, A.; Tarantino, G.; Balsano, C. Oleuropein Induces AMPK-Dependent Autophagy in NAFLD Mice, Regardless of the Gender. Int. J. Mol. Sci. 2018, 19, 3948. https://doi.org/10.3390/ijms19123948
Porcu C, Sideri S, Martini M, Cocomazzi A, Galli A, Tarantino G, Balsano C. Oleuropein Induces AMPK-Dependent Autophagy in NAFLD Mice, Regardless of the Gender. International Journal of Molecular Sciences. 2018; 19(12):3948. https://doi.org/10.3390/ijms19123948
Chicago/Turabian StylePorcu, Cristiana, Silvia Sideri, Maurizio Martini, Alessandra Cocomazzi, Andrea Galli, Giovanni Tarantino, and Clara Balsano. 2018. "Oleuropein Induces AMPK-Dependent Autophagy in NAFLD Mice, Regardless of the Gender" International Journal of Molecular Sciences 19, no. 12: 3948. https://doi.org/10.3390/ijms19123948
APA StylePorcu, C., Sideri, S., Martini, M., Cocomazzi, A., Galli, A., Tarantino, G., & Balsano, C. (2018). Oleuropein Induces AMPK-Dependent Autophagy in NAFLD Mice, Regardless of the Gender. International Journal of Molecular Sciences, 19(12), 3948. https://doi.org/10.3390/ijms19123948