Messenger RNA Life-Cycle in Cancer Cells: Emerging Role of Conventional and Non-Conventional RNA-Binding Proteins?
Abstract
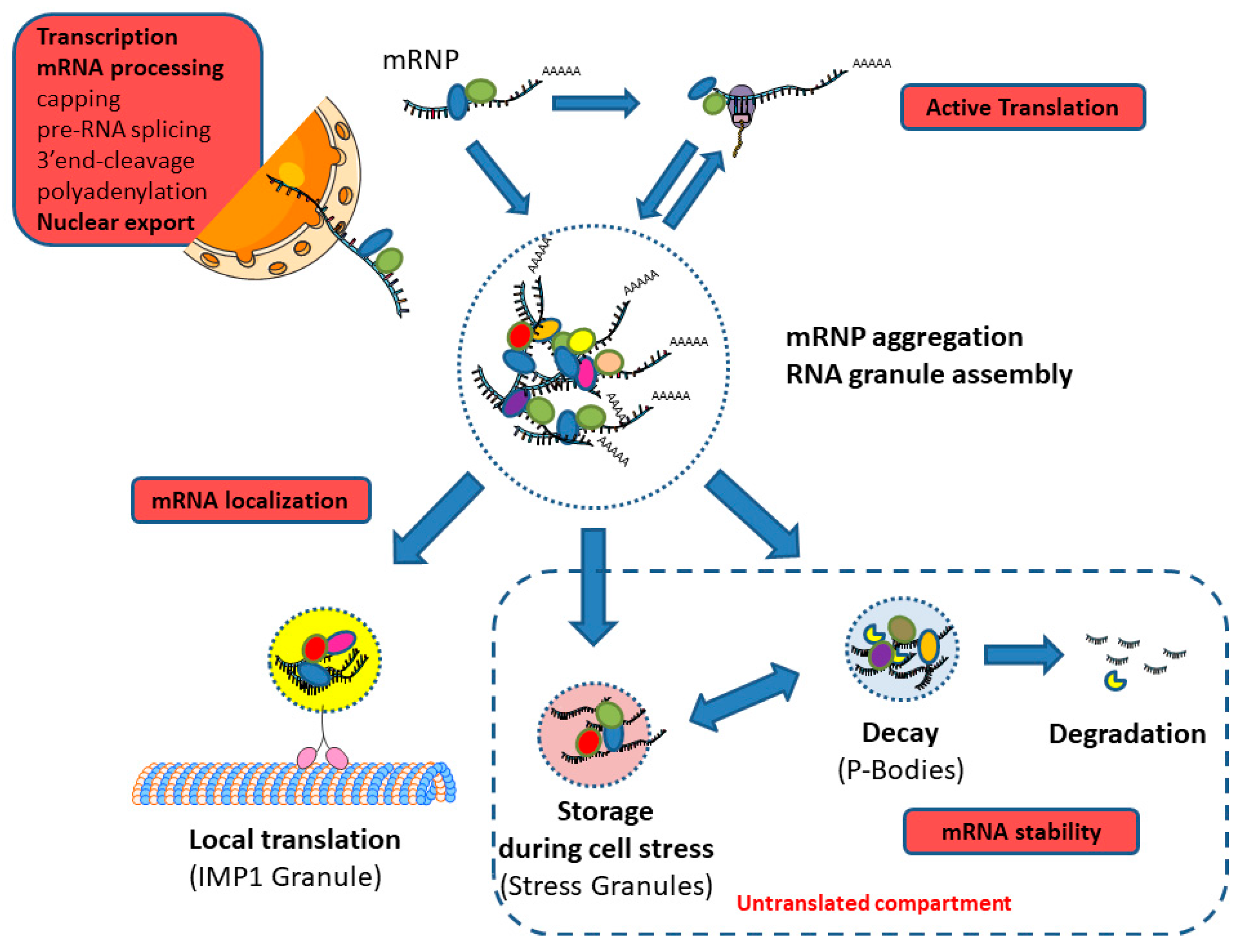
1. Introduction to the Messenger RNA (mRNA) Life-Cycle
2. Major Actors of the mRNA Life-Cycle: The RNA-Binding Proteins (RBPs)
- -
- One type of mRNA may be associated with many mRBPs and each mRBP may have hundreds or thousands of different RNA targets [36].
- -
- For each transcript, the composition of the proteins within the mRNPs can vary as it is influenced by different pathophysiological parameters.
- -
- Use of alternative gene promoters during transcription or alternative polyadenylation sites in mRNA maturation may also constitute a source of gain or loss of cis-binding sequences for RBPs in the mRNA [1].
3. Co-Transcriptional Processes in the mRNA Life-Cycle
3.1. Pre-mRNA Splicing
3.2. Cap and Poly(A) Tail Addition
4. Post-Transcriptional Processes in the mRNA Life-Cycle
4.1. Nuclear Export
4.2. mRNA Cytoplasmic Degradation
- -
- be degraded in the 3′ to 5′ direction by the exosome, a multiprotein complex that recruits RNAses and cofactors, the activity of which is regulated by the SKI (Sloan-Kettering Institute) complex; or
- -
- be decapped by decapping protein 2 (DCP2) and its co-activator, DCP1. This step is followed by an exonucleolytic degradation in the 5′ to 3′ direction performed by the exoribonuclease, XRN1.
4.3. Factors Modulating mRNA Stability
4.3.1. Poly(A) Tail and PABP
4.3.2. Non-Coding RNAs and RNA Editing
4.3.3. Cis-Elements (AU Rich Elements and CA Repeat Elements)
4.4. Cytoplasmic Granules
5. Interplay between the Different Stages of mRNA Life-Cycle and Coupling to Other Vital Cellular Processes
6. Therapeutic Approaches Targeting RBPs and Related mRNA
6.1. Which Targets?
6.2. Which Tools?
7. Conclusions and Take-Home Messages
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| aa | Amino acids |
| APA | Alternative polyadenylation |
| ARE | AU rich element |
| AREBP | ARE-binding protein |
| ASOs | Anti-sense oligonucleotides |
| CARE | CA repeat element |
| CBC | Cap-binding complex |
| CRD | Carbohydrate recognition domain |
| hnRNPs | Heterogeneous nuclear ribonucleoproteins |
| HuR | Human antigen R |
| IDR | Intrinsically Disordered Region |
| LC | Low complexity disordered region |
| miR | MicroRNA |
| mRBP | mRNA-binding protein |
| NMD | Nonsense mediated decay |
| NPC | Nuclear pore complex |
| Nt | Nucleotides |
| PABP | poly(A) binding protein |
| PBs | Processing-Bodies |
| PT | post-transcriptional |
| RBD | RNA-binding domain |
| RBP | RNA-binding protein |
| RNP | Ribonucleprotein |
| SGs | Stress granules |
| SR proteins | Serine arginine rich proteins |
| TTP | Tristetrapolin |
| UTR | Untranslated Transcribed Region |
References
- Singh, G.; Pratt, G.; Yeo, G.W.; Moore, M.J. The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu. Rev. Biochem. 2015, 84, 325–354. [Google Scholar] [CrossRef] [PubMed]
- Calabretta, S.; Richard, S. Emerging Roles of Disordered Sequences in RNA-Binding Proteins. Trends Biochem. Sci. 2015, 40, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef] [PubMed]
- Gerstberger, S.; Hafner, M.; Ascano, M.; Tuschl, T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv. Exp. Med. Biol. 2014, 825, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Hong, S. RNA Binding Protein as an Emerging Therapeutic Target for Cancer Prevention and Treatment. J. Cancer Prev. 2017, 22, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, R.; Babu, A.; Amreddy, N.; Srivastava, A.; Chen, A.; Zhao, Y.D.; Kompella, U.B.; Munshi, A.; Ramesh, R. Tumor-targeted Nanoparticle Delivery of HuR siRNA Inhibits Lung Tumor Growth In Vitro and In Vivo By Disrupting the Oncogenic Activity of the RNA-binding Protein HuR. Mol. Cancer Ther. 2017, 16, 1470–1486. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Peng, W.; Furuuchi, N.; Gerhart, J.; Rhodes, K.; Mukherjee, N.; Jimbo, M.; Gonye, G.E.; Brody, J.R.; Getts, R.C.; et al. Delivery of Therapeutics Targeting the mRNA-Binding Protein HuR Using 3DNA Nanocarriers Suppresses Ovarian Tumor Growth. Cancer Res. 2016, 76, 1549–1559. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lan, L.; Wilson, D.M.; Marquez, R.T.; Tsao, W.-C.; Gao, P.; Roy, A.; Turner, B.A.; McDonald, P.; Tunge, J.A.; et al. Identification and validation of novel small molecule disruptors of HuR-mRNA interaction. ACS Chem. Biol. 2015, 10, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- David, C.J.; Chen, M.; Assanah, M.; Canoll, P.; Manley, J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010, 463, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Huang, Y.; Seckl, M.J.; Pardo, O.E. Emerging roles of hnRNPA1 in modulating malignant transformation. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.L.; Wächter, K.; Mühleck, B.; Pazaitis, N.; Köhn, M.; Lederer, M.; Hüttelmaier, S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell. Mol. Life Sci. CMLS 2013, 70, 2657–2675. [Google Scholar] [CrossRef] [PubMed]
- Palanichamy, J.K.; Tran, T.M.; Howard, J.M.; Contreras, J.R.; Fernando, T.R.; Sterne-Weiler, T.; Katzman, S.; Toloue, M.; Yan, W.; Basso, G.; et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J. Clin. Investig. 2016, 126, 1495–1511. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Lu, D.; Liu, L.; Cai, J.; Zhou, Y.; Yang, Y.; Zhang, Y.; Zhang, J. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) promotes lung tumorigenesis via attenuating p53 stability. Oncotarget 2017, 8, 93672–93687. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ji, Q.; Jiao, C.; Ren, L.; Zhao, Y.; Chen, Y.; Shi, R.; Feng, Y. IGF2BP3 as a potential tissue marker for the diagnosis of esophageal high-grade intraepithelial neoplasia. Oncotargets Ther. 2017, 10, 3861–3866. [Google Scholar] [CrossRef] [PubMed]
- Frisone, P.; Pradella, D.; Di Matteo, A.; Belloni, E.; Ghigna, C.; Paronetto, M.P. SAM68: Signal Transduction and RNA Metabolism in Human Cancer. BioMed Res. Int. 2015, 2015, 528954. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J.; Livermore, K.; Rajan, P.; Elliott, D.J. RNA splicing and splicing regulator changes in prostate cancer pathology. Hum. Genet. 2017, 136, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Locatelli, A.; Lofgren, K.A.; Daniel, A.R.; Castro, N.E.; Lange, C.A. Mechanisms of HGF/Met signaling to Brk and Sam68 in breast cancer progression. Horm. Cancer 2012, 3, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Linder, P. Dead-box proteins: A family affair—Active and passive players in RNP-remodeling. Nucleic Acids Res. 2006, 34, 4168–4180. [Google Scholar] [CrossRef] [PubMed]
- Russell, R. Unwinding the mechanisms of a DEAD-box RNA helicase in cancer. J. Mol. Biol. 2015, 427, 1797–1800. [Google Scholar] [CrossRef] [PubMed]
- Andreou, A.Z.; Klostermeier, D. The DEAD-box helicase eIF4A: Paradigm or the odd one out? RNA Biol. 2013, 10, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Mao, Y.; Zhou, J.; Zhao, Y.; Cao, Y.; Chen, X. Multifunctional DDX3: Dual roles in various cancer development and its related signaling pathways. Am. J. Cancer Res. 2016, 6, 387–402. [Google Scholar] [PubMed]
- Lee, T.; Pelletier, J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget 2016, 7, 42716–42739. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, P.; Stepniak-Konieczna, E.; Sobczak, K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014, 42, 10873–10887. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Jhuang, Y.-L.; Chen, Y.-L.; Jeng, Y.-M.; Yuan, R.-H. Paradoxical overexpression of MBNL2 in hepatocellular carcinoma inhibits tumor growth and invasion. Oncotarget 2016, 7, 65589–65601. [Google Scholar] [CrossRef] [PubMed]
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell 2013, 50, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Järvelin, A.I.; Noerenberg, M.; Davis, I.; Castello, A. The new (dis)order in RNA regulation. Cell Commun. Signal. CCS 2016, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Bahadur, R.P. A structural perspective of RNA recognition by intrinsically disordered proteins. Cell. Mol. Life Sci. CMLS 2016, 73, 4075–4084. [Google Scholar] [CrossRef] [PubMed]
- Hudson, W.H.; Ortlund, E.A. The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 2014, 15, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Srikantan, S.; Gorospe, M. UneCLIPsing HuR nuclear function. Mol. Cell 2011, 43, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Hocine, S.; Singer, R.H.; Grünwald, D. RNA processing and export. Cold Spring Harb. Perspect. Biol. 2010, 2, a000752. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.F.; Parker, R. Principles and properties of eukaryotic mRNPs. Mol. Cell 2014, 54, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Kedersha, N.; Ivanov, P. Stress granules, P-bodies and cancer. Biochim. Biophys. Acta 2015, 1849, 861–870. [Google Scholar] [CrossRef] [PubMed]
- García-Mauriño, S.M.; Rivero-Rodríguez, F.; Velázquez-Cruz, A.; Hernández-Vellisca, M.; Díaz-Quintana, A.; de la Rosa, M.A.; Díaz-Moreno, I. RNA Binding Protein Regulation and Cross-Talk in the Control of AU-rich mRNA Fate. Front. Mol. Biosci. 2017, 4, 71. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K. Post-translational modifications of RNA-binding proteins and their roles in RNA granules. Curr. Protein Pept. Sci. 2012, 13, 331–336. [Google Scholar] [PubMed]
- Anderson, P.; Kedersha, N. RNA granules. J. Cell Biol. 2006, 172, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Müller-McNicoll, M.; Neugebauer, K.M. How cells get the message: Dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013, 14, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Blech-Hermoni, Y.; Ladd, A.N. RNA binding proteins in the regulation of heart development. Int. J. Biochem. Cell Biol. 2013, 45, 2467–2478. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.H.; Jansen, R.-P. A jack of all trades: The RNA-binding protein vigilin. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Conrad, T.; Albrecht, A.-S.; de Melo Costa, V.R.; Sauer, S.; Meierhofer, D.; Ørom, U.A. Serial interactome capture of the human cell nucleus. Nat. Commun. 2016, 7, 11212. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, E.C.; van Nostrand, E.L.; Yeo, G.W. Advances and challenges in the detection of transcriptome-wide protein-RNA interactions. Wiley Interdiscip. Rev. RNA 2017. [Google Scholar] [CrossRef] [PubMed]
- Adeli, K. Translational control mechanisms in metabolic regulation: Critical role of RNA binding proteins, microRNAs, and cytoplasmic RNA granules. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E1051–E1064. [Google Scholar] [CrossRef] [PubMed]
- Dahan, N.; Choder, M. The eukaryotic transcriptional machinery regulates mRNA translation and decay in the cytoplasm. Biochim. Biophys. Acta 2013, 1829, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Haimovich, G.; Choder, M.; Singer, R.H.; Trcek, T. The fate of the messenger is pre-determined: A new model for regulation of gene expression. Biochim. Biophys. Acta 2013, 1829, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ortín, J.E.; Alepuz, P.; Chávez, S.; Choder, M. Eukaryotic mRNA decay: Methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 2013, 425, 3750–3775. [Google Scholar] [CrossRef] [PubMed]
- Parent, A.; Bisaillon, M. Synergy between transcription and mRNA processing events. Med. Sci. MS 2006, 22, 626–632. [Google Scholar] [CrossRef]
- Bentley, D.L. The union of transcription and mRNA processing: 20 years of coupling. RNA 2015, 21, 569–570. [Google Scholar] [CrossRef] [PubMed]
- Lenzken, S.C.; Loffreda, A.; Barabino, S.M.L. RNA splicing: A new player in the DNA damage response. Int. J. Cell Biol. 2013, 2013, 153634. [Google Scholar] [CrossRef] [PubMed]
- Montecucco, A.; Biamonti, G. Pre-mRNA processing factors meet the DNA damage response. Front. Genet. 2013, 4, 102. [Google Scholar] [CrossRef] [PubMed]
- Naro, C.; Bielli, P.; Pagliarini, V.; Sette, C. The interplay between DNA damage response and RNA processing: The unexpected role of splicing factors as gatekeepers of genome stability. Front. Genet. 2015, 6, 142. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Xiong, J.; Wang, D.; Fu, X.-D. Pre-mRNA splicing: Where and when in the nucleus. Trends Cell Biol. 2011, 21, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, H.; Saulière, J.; Wang, Z. The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 2016, 17, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Sibley, C.R. Regulation of gene expression through production of unstable mRNA isoforms. Biochem. Soc. Trans. 2014, 42, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kornblihtt, A.R.; Schor, I.E.; Alló, M.; Dujardin, G.; Petrillo, E.; Muñoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.-D.; Ares, M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Moehle, E.A.; Braberg, H.; Krogan, N.J.; Guthrie, C. Adventures in time and space: Splicing efficiency and RNA polymerase II elongation rate. RNA Biol. 2014, 11, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, V.; Pereira, J.F.S.; Jordan, P. Signaling Pathways Driving Aberrant Splicing in Cancer Cells. Genes 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Ohno, K.; Takeda, J.-I.; Masuda, A. Rules and tools to predict the splicing effects of exonic and intronic mutations. Wiley Interdiscip. Rev. RNA 2017. [Google Scholar] [CrossRef] [PubMed]
- Scotti, M.M.; Swanson, M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016, 17, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Pandit, S.; Zhou, Y.; Shiue, L.; Coutinho-Mansfield, G.; Li, H.; Qiu, J.; Huang, J.; Yeo, G.W.; Ares, M.; Fu, X.-D. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell 2013, 50, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Kędzierska, H.; Piekiełko-Witkowska, A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 2017, 396, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Robb, G.B.; Chan, S.-H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef] [PubMed]
- Eckmann, C.R.; Rammelt, C.; Wahle, E. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA 2011, 2, 348–361. [Google Scholar] [CrossRef] [PubMed]
- Jalkanen, A.L.; Coleman, S.J.; Wilusz, J. Determinants and implications of mRNA poly(A) tail size—Does this protein make my tail look big? Semin. Cell Dev. Biol. 2014, 34, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Weill, L.; Belloc, E.; Bava, F.-A.; Méndez, R. Translational control by changes in poly(A) tail length: Recycling mRNAs. Nat. Struct. Mol. Biol. 2012, 19, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Curinha, A.; Oliveira Braz, S.; Pereira-Castro, I.; Cruz, A.; Moreira, A. Implications of polyadenylation in health and disease. Nucl. Austin Tex 2014, 5, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Carmody, S.R.; Wente, S.R. mRNA nuclear export at a glance. J. Cell Sci. 2009, 122, 1933–1937. [Google Scholar] [CrossRef] [PubMed]
- Björk, P.; Wieslander, L. Integration of mRNP formation and export. Cell. Mol. Life Sci. CMLS 2017, 74, 2875–2897. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, V.O.; Laskey, R.A. Control of mammalian gene expression by selective mRNA export. Nat. Rev. Mol. Cell Biol. 2015, 16, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Katahira, J. mRNA export and the TREX complex. Biochim. Biophys. Acta 2012, 1819, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Vera, M.; Pani, B.; Griffiths, L.A.; Muchardt, C.; Abbott, C.M.; Singer, R.H.; Nudler, E. The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. eLife 2014, 3, e03164. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, N.; Borden, K.L.B. mRNA export and cancer. Wiley Interdiscip. Rev. RNA 2012, 3, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Hurt, J.A.; Silver, P.A. mRNA nuclear export and human disease. Dis. Model. Mech. 2008, 1, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Mor, A.; White, M.A.; Fontoura, B.M.A. Nuclear trafficking in health and disease. Curr. Opin. Cell Biol. 2014, 28, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Hautbergue, G.M. RNA Nuclear Export: From Neurological Disorders to Cancer. Adv. Exp. Med. Biol. 2017, 1007, 89–109. [Google Scholar] [CrossRef] [PubMed]
- Eliscovich, C.; Singer, R.H. RNP transport in cell biology: The long and winding road. Curr. Opin. Cell Biol. 2017, 45, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Brook, M.; Gray, N.K. The role of mammalian poly(A)-binding proteins in co-ordinating mRNA turnover. Biochem. Soc. Trans. 2012, 40, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Decker, C.J.; Parker, R. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Karam, R.; Zhou, Y.; Su, F.; Ji, Y.; Li, G.; Xu, G.; Lu, L.; Wang, C.; Song, M.; et al. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat. Med. 2014, 20, 596–598. [Google Scholar] [CrossRef] [PubMed]
- Nasif, S.; Contu, L.; Mühlemann, O. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol. 2017, 75, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Mangus, D.A.; Evans, M.C.; Jacobson, A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003, 4, 223. [Google Scholar] [CrossRef] [PubMed]
- Gorgoni, B.; Gray, N.K. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: A developmental perspective. Brief. Funct. Genom. Proteom. 2004, 3, 125–141. [Google Scholar] [CrossRef] [PubMed]
- Jerbi, S.; Jolles, B.; Bouceba, T.; Jean-Jean, O. Studies on human eRF3-PABP interaction reveal the influence of eRF3a N-terminal glycin repeat on eRF3-PABP binding affinity and the lower affinity of eRF3a 12-GGC allele involved in cancer susceptibility. RNA Biol. 2016, 13, 306–315. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Detassis, S.; Grasso, M.; del Vescovo, V.; Denti, M.A. microRNAs Make the Call in Cancer Personalized Medicine. Front. Cell Dev. Biol. 2017, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Shan, H.; Su, Y.; Xia, K.; Zou, R.; Shao, Q. Let-7a inhibits migration, invasion and tumor growth by targeting AKT2 in papillary thyroid carcinoma. Oncotarget 2017, 8, 69746–69755. [Google Scholar] [CrossRef] [PubMed]
- Ratnadiwakara, M.; Mohenska, M.; Änkö, M.-L. Splicing factors as regulators of miRNA biogenesis—Links to human disease. Semin. Cell Dev. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.; Harries, L.W. Circular RNAs (circRNAs) in Health and Disease. Genes 2017, 8, 353. [Google Scholar] [CrossRef]
- Han, Y.-N.; Xia, S.-Q.; Zhang, Y.-Y.; Zheng, J.-H.; Li, W. Circular RNAs: A novel type of biomarker and genetic tools in cancer. Oncotarget 2017, 8, 64551–64563. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.-P.; He, Y.-J.; Hou, J.-C.; Chen, X.; Zhou, S.-Y.; Yang, S.-J.; Li, J.; Zhang, H.-D.; Hu, J.-H.; Zhong, S.-L.; et al. The role of circRNAs in cancers. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yang, L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gatsiou, A.; Vlachogiannis, N.; Lunella, F.F.; Sachse, M.; Stellos, K. Adenosine-to-Inosine RNA Editing in Health and Disease. Antioxid. Redox Signal. 2017. [Google Scholar] [CrossRef] [PubMed]
- Barreau, C.; Paillard, L.; Osborne, H.B. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 2005, 33, 7138–7150. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, C.J.; Wilusz, J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. TIG 2004, 20, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Srikantan, S.; Tominaga, K.; Gorospe, M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 2012, 13, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Brennan, S.E.; Kuwano, Y.; Alkharouf, N.; Blackshear, P.J.; Gorospe, M.; Wilson, G.M. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009, 69, 5168–5176. [Google Scholar] [CrossRef] [PubMed]
- Jafarifar, F.; Yao, P.; Eswarappa, S.M.; Fox, P.L. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011, 30, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Anderson, P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007, 431, 61–81. [Google Scholar] [CrossRef] [PubMed]
- Rajgor, D.; Shanahan, C.M. RNA granules and cytoskeletal links. Biochem. Soc. Trans. 2014, 42, 1206–1210. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Fuxreiter, M. The Structure and Dynamics of Higher-Order Assemblies: Amyloids, Signalosomes, and Granules. Cell 2016, 165, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Adjibade, P.; Mazroui, R. Control of mRNA turnover: Implication of cytoplasmic RNA granules. Semin. Cell Dev. Biol. 2014, 34, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Shigunov, P.; Dallagiovanna, B. Stem Cell Ribonomics: RNA-Binding Proteins and Gene Networks in Stem Cell Differentiation. Front. Mol. Biosci. 2015, 2, 74. [Google Scholar] [CrossRef] [PubMed]
- DeGracia, D.J.; Jamison, J.T.; Szymanski, J.J.; Lewis, M.K. Translation arrest and ribonomics in post-ischemic brain: Layers and layers of players. J. Neurochem. 2008, 106, 2288–2301. [Google Scholar] [CrossRef] [PubMed]
- Jønson, L.; Vikesaa, J.; Krogh, A.; Nielsen, L.K.; Hansen, T.; Borup, R.; Johnsen, A.H.; Christiansen, J.; Nielsen, F.C. Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell. Proteom. MCP 2007, 6, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.C.; Ephrussi, A. mRNA localization: Gene expression in the spatial dimension. Cell 2009, 136, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Elvira, G.; Wasiak, S.; Blandford, V.; Tong, X.-K.; Serrano, A.; Fan, X.; Sánchez-Carbente, M.; del, R.; Servant, F.; Bell, A.W.; Boismenu, D.; et al. Characterization of an RNA Granule from Developing Brain. Mol. Cell. Proteom. 2006, 5, 635–651. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Parker, R. Hypo- and Hyper-Assembly Diseases of RNA-Protein Complexes. Trends Mol. Med. 2016, 22, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Dahan, O.; Gingold, H.; Pilpel, Y. Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet. TIG 2011, 27, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Navarro, S.; Hurt, E. Linking gene regulation to mRNA production and export. Curr. Opin. Cell Biol. 2011, 23, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Haudek, K.C.; Patterson, R.J.; Wang, J.L. SR proteins and galectins: What’s in a name? Glycobiology 2010, 20, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H.; Qiu, D.-C.; Chang, W.-H.; Yeh, Y.-Q.; Jeng, U.-S.; Liu, F.-T.; Huang, J.-R. The intrinsically disordered N-terminal domain of galectin-3 dynamically mediates multisite self-association of the protein through fuzzy interactions. J. Biol. Chem. 2017, 292, 17845–17856. [Google Scholar] [CrossRef] [PubMed]
- Coppin, L.; Vincent, A.; Frénois, F.; Duchêne, B.; Lahdaoui, F.; Stechly, L.; Renaud, F.; Villenet, C.; Van Seuningen, I.; Leteurtre, E.; et al. Galectin-3 is a non-classic RNA binding protein that stabilizes the mucin MUC4 mRNA in the cytoplasm of cancer cells. Sci. Rep. 2017, 7, 43927. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, K.; Mernberger, M.; Nist, A.; Stiewe, T.; Brehm, A.; Jacob, R. Galectin-3 interacts with components of the nuclear ribonucleoprotein complex. BMC Cancer 2016, 16, 502. [Google Scholar] [CrossRef] [PubMed]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.-Y.; Wang, P.; Han, J.; Rosenfeld, M.G.; Fu, X.-D. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell 2009, 35, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Haudek, K.C.; Voss, P.G.; Wang, J.L.; Patterson, R.J. A 10S galectin-3-U1 snRNP complex assembles into active spliceosomes. Nucleic Acids Res. 2016, 44, 6391–6397. [Google Scholar] [CrossRef] [PubMed]
- Magescas, J.; Sengmanivong, L.; Viau, A.; Mayeux, A.; Dang, T.; Burtin, M.; Nilsson, U.J.; Leffler, H.; Poirier, F.; Terzi, F.; et al. Spindle pole cohesion requires glycosylation-mediated localization of NuMA. Sci. Rep. 2017, 7, 1474. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.S.; Fernandes, V.C.; Nepomuceno, T.C.; Rodrigues, D.C.; Woods, N.T.; Suarez-Kurtz, G.; Chammas, R.; Monteiro, A.N.; Carvalho, M.A. Characterization of LGALS3 (galectin-3) as a player in DNA damage response. Cancer Biol. Ther. 2014, 15, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Dumic, J.; Dabelic, S.; Flögel, M. Galectin-3: An open-ended story. Biochim. Biophys. Acta 2006, 1760, 616–635. [Google Scholar] [CrossRef] [PubMed]
- Newlaczyl, A.U.; Yu, L.-G. Galectin-3—A jack-of-all-trades in cancer. Cancer Lett. 2011, 313, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Cornejo-García, J.A.; Romano, A.; Guéant-Rodríguez, R.M.; Oussalah, A.; Blanca-López, N.; Gaeta, F.; Tramoy, D.; Josse, T.; Doña, I.; Torres, M.J.; et al. A non-synonymous polymorphism in galectin-3 lectin domain is associated with allergic reactions to beta-lactam antibiotics. Pharmacogenom. J. 2016, 16, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.C.; Zheng, Y.; Yan, J.; Zhou, Y.; Tai, G.; Mayo, K.H. Novel polysaccharide binding to the N-terminal tail of galectin-3 is likely modulated by proline isomerization. Glycobiology 2017, 27, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Dagher, S.F.; Wang, J.L.; Patterson, R.J. Identification of galectin-3 as a factor in pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 1995, 92, 1213–1217. [Google Scholar] [CrossRef] [PubMed]
- Haudek, K.C.; Voss, P.G.; Locascio, L.E.; Wang, J.L.; Patterson, R.J. A mechanism for incorporation of galectin-3 into the spliceosome through its association with U1 snRNP. Biochemistry 2009, 48, 7705–7712. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zeng, Z.; Wei, H.; Wang, Z. Alternative splicing in cancers: From aberrant regulation to new therapeutics. Semin. Cell Dev. Biol. 2017, 75, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Sabò, A.; Guccione, E. Targeting MYC in cancer therapy: RNA processing offers new opportunities. BioEssays News Rev. Mol. Cell. Dev. Biol. 2016, 38, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Rana, S.; Espinosa-Diez, C.; Anand, S. MicroRNAs in Cancer: Challenges and opportunities in early detection, disease monitoring, and therapeutic agents. Curr. Pathobiol. Rep. 2017, 5, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Curtin, C.M.; Castaño, I.M.; O’Brien, F.J. Scaffold-Based microRNA Therapies in Regenerative Medicine and Cancer. Adv. Healthc. Mater. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Goblirsch, M.; Richtig, G.; Slaby, O.; Berindan-Neagoe, I.; Gerger, A.; Pichler, M. MicroRNAs as a tool to aid stratification of colorectal cancer patients and to guide therapy. Pharmacogenomics 2017, 18, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Shirjang, S.; Baradaran, B. MicroRNAs in the Diagnosis and Treatment of Cancer. Immunol. Investig. 2017, 46, 880–897. [Google Scholar] [CrossRef] [PubMed]
- Moles, R. MicroRNAs-based Therapy: A Novel and Promising Strategy for Cancer Treatment. MicroRNA Shariqah UAE 2017, 6, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Mukohyama, J.; Shimono, Y.; Minami, H.; Kakeji, Y.; Suzuki, A. Roles of microRNAs and RNA-Binding Proteins in the Regulation of Colorectal Cancer Stem Cells. Cancers 2017, 9, 143. [Google Scholar] [CrossRef] [PubMed]
- Gomez, I.G.; Nakagawa, N.; Duffield, J.S. MicroRNAs as novel therapeutic targets to treat kidney injury and fibrosis. Am. J. Physiol. Renal Physiol. 2016, 310, F931–F944. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Sood, A.K.; Dang, C.V.; Zhang, L. The role of long noncoding RNAs in cancer: The dark matter matters. Curr. Opin. Genet. Dev. 2017, 48, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Jandura, A.; Krause, H.M. The New RNA World: Growing Evidence for Long Noncoding RNA Functionality. Trends Genet. TIG 2017, 33, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Huang, M.; Guo, W.W.; Huang, Q.; Zhang, L.Z.; Jiang, G. Nano-based delivery of RNAi in cancer therapy. Mol. Cancer 2017, 16, 134. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, R.; Van Roosbroeck, K.; Calin, G.A. Cell-to-cell communication: MicroRNAs as hormones. Mol. Oncol. 2017, 11, 1673–1686. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Keto, J.; Fabbri, M. Exosomal MicroRNAs in Breast Cancer towards Diagnostic and Therapeutic Applications. Cancers 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheong, C.-G.; Hall, T.M.T.; Wang, Z. Engineering splicing factors with designed specificities. Nat. Methods 2009, 6, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Montiel, N.; Anaya-Ruiz, M.; Pérez-Santos, M.; Martínez-Contreras, R.D. Alternative Splicing in Breast Cancer and the Potential Development of Therapeutic Tools. Genes 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Marcelino Meliso, F.; Hubert, C.G.; Favoretto Galante, P.A.; Penalva, L.O. RNA processing as an alternative route to attack glioblastoma. Hum. Genet. 2017, 136, 1129–1141. [Google Scholar] [CrossRef] [PubMed]
- Kranz, L.M.; Diken, M.; Haas, H.; Kreiter, S.; Loquai, C.; Reuter, K.C.; Meng, M.; Fritz, D.; Vascotto, F.; Hefesha, H.; et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Berg, T. Small-molecule modulators of c-Myc/Max and Max/Max interactions. Curr. Top. Microbiol. Immunol. 2011, 348, 139–149. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coppin, L.; Leclerc, J.; Vincent, A.; Porchet, N.; Pigny, P. Messenger RNA Life-Cycle in Cancer Cells: Emerging Role of Conventional and Non-Conventional RNA-Binding Proteins? Int. J. Mol. Sci. 2018, 19, 650. https://doi.org/10.3390/ijms19030650
Coppin L, Leclerc J, Vincent A, Porchet N, Pigny P. Messenger RNA Life-Cycle in Cancer Cells: Emerging Role of Conventional and Non-Conventional RNA-Binding Proteins? International Journal of Molecular Sciences. 2018; 19(3):650. https://doi.org/10.3390/ijms19030650
Chicago/Turabian StyleCoppin, Lucie, Julie Leclerc, Audrey Vincent, Nicole Porchet, and Pascal Pigny. 2018. "Messenger RNA Life-Cycle in Cancer Cells: Emerging Role of Conventional and Non-Conventional RNA-Binding Proteins?" International Journal of Molecular Sciences 19, no. 3: 650. https://doi.org/10.3390/ijms19030650
APA StyleCoppin, L., Leclerc, J., Vincent, A., Porchet, N., & Pigny, P. (2018). Messenger RNA Life-Cycle in Cancer Cells: Emerging Role of Conventional and Non-Conventional RNA-Binding Proteins? International Journal of Molecular Sciences, 19(3), 650. https://doi.org/10.3390/ijms19030650




