Translocator Protein (TSPO) as a Potential Biomarker in Human Cancers
Abstract
1. Introduction
2. Results
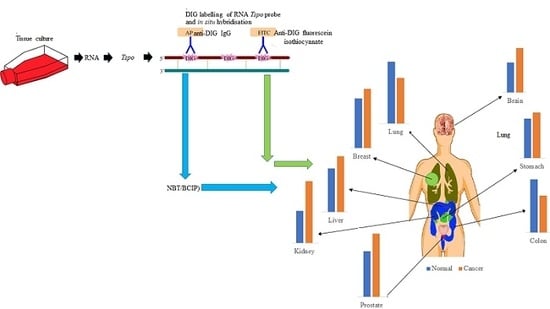
2.1. Tspo mRNA Transcription Is Observed within Specific Cell Types in Normal and Cancer Tissues of Different Organs
2.1.1. Prostate
2.1.2. Stomach
2.1.3. Colon
2.1.4. Liver
2.1.5. Lung
2.1.6. Kidney
2.1.7. Breast
2.1.8. Brain
2.2. The Level of Tspo mRNA Transcription Is Different in Healthy and Cancerous Tissue
3. Discussion
4. Materials and Methods
4.1. Human Tissue Arrays
4.2. Cell Culture
4.3. RNA Probe Synthesis
4.4. In Situ Hybridization
4.5. Image and Statistical Analysis
4.6. Real-Time Polymerase Chain Reaction
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| TSPO | translocator protein |
| CNS | central nervous system |
| VDAC | voltage-dependent anion channel |
| ANC | adenine nucleotide carrier |
References
- Braestrup, C.; Squires, R.F. Specific benzodiazepine receptors in rat brain characterized by high-affinity (3H)diazepam binding. Proc. Natl. Acad. Sci. USA 1977, 74, 3805–3809. [Google Scholar] [CrossRef] [PubMed]
- Benavides, J.; Dubois, A.; Dennis, T.; Hamel, E.; Scatton, B. Omega 3 (peripheral type benzodiazepine binding) site distribution in the rat immune system: An autoradiographic study with the photoaffinity ligand [3H]PK 14105. J. Pharmacol. Exp. Ther. 1989, 249, 333–339. [Google Scholar] [PubMed]
- Anholt, R.R.; Pedersen, P.L.; De Souza, E.B.; Snyder, S.H. The peripheral-type benzodiazepine receptor. Localization to the mitochondrial outer membrane. J. Boil. Chem. 1986, 261, 576–583. [Google Scholar]
- Antkiewicz-Michaluk, L.; Guidotti, A.; Krueger, K.E. Molecular characterization and mitochondrial density of a recognition site for peripheral-type benzodiazepine ligands. Mol. Pharmacol. 1988, 34, 272–278. [Google Scholar] [PubMed]
- Anholt, R.R.; De Souza, E.B.; Oster-Granite, M.L.; Snyder, S.H. Peripheral-type benzodiazepine receptors: Autoradiographic localization in whole-body sections of neonatal rats. J. Pharmacol. Exp. Ther. 1985, 233, 517–526. [Google Scholar] [PubMed]
- Papadopoulos, V.; Berkovich, A.; Krueger, K.E.; Costa, E.; Guidotti, A. Diazepam binding inhibitor and its processing products stimulate mitochondrial steroid biosynthesis via an interaction with mitochondrial benzodiazepine receptors. Endocrinology 1991, 129, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, V.; Guarneri, P.; Kreuger, K.E.; Guidotti, A.; Costa, E. Pregnenolone biosynthesis in C6-2B glioma cell mitochondria: Regulation by a mitochondrial diazepam binding inhibitor receptor. Proc. Natl. Acad. Sci. USA 1992, 89, 5113–5117. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, V.; Amri, H.; Boujrad, N.; Cascio, C.; Culty, M.; Garnier, M.; Hardwick, M.; Li, H.; Vidic, B.; Brown, A.S.; et al. Peripheral benzodiazepine receptor in cholesterol transport and steroidogenesis. Steroids 1997, 62, 21–28. [Google Scholar] [CrossRef]
- Li, H.; Papadopoulos, V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology 1998, 139, 4991–4997. [Google Scholar] [CrossRef] [PubMed]
- Gavish, M.; Bachman, I.; Shoukrun, R.; Katz, Y.; Veenman, L.; Weisinger, G.; Weizman, A. Enigma of the Peripheral Benzodiazepine Receptor. Pharmacol. Rev. 1999, 51, 629–650. [Google Scholar] [PubMed]
- Papadopoulos, V.; Baraldi, M.; Guilarte, T.R.; Knudsen, T.B.; Lacapere, J.J.; Lindemann, P.; Norenberg, M.D.; Nutt, D.; Weizman, A.; Zhang, M.R.; et al. Translocator protein (18kDa): New nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci. 2006, 27, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.J.; Middleton, R.J.; Hatty, C.R.; Kam, W.W.; Chan, R.; Pham, T.; Harrison-Brown, M.; Dodson, E.; Veale, K.; Banati, R.B. The 18 kDa translocator protein, microglia and neuroinflammation. Brain Pathol. 2014, 24, 631–653. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Liu, J.; Liu, N.; Kuhn, L.A.; Garavito, R.M.; Ferguson-Miller, S. Translocator Protein 18 kDa (TSPO): An Old Protein with New Functions? Biochemistry 2016, 55, 2821–2831. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, V.; Stocco, D.M. Letter to the Editor: Dubious conclusions on TSPO function. Endocrinology 2018. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Zirkin, B.; Papadopoulos, V. Response to Letter to the Editor: “Dubious conclusions on TSPO function”. Endocrinology 2018. [Google Scholar] [CrossRef] [PubMed]
- McEnery, M.W.; Snowman, A.M.; Trifiletti, R.R.; Snyder, S.H. Isolation of the mitochondrial benzodiazepine receptor: Association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc. Natl. Acad. Sci. USA 1992, 89, 3170–3174. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.M.; Ciliax, B.J.; Mancini, W.R.; Young, A.B. Presence of peripheral-type benzodiazepine binding sites on human erythrocyte membranes. Eur. J. Pharmacol. 1988, 152, 47–53. [Google Scholar] [CrossRef]
- O’Beirne, G.B.; Woods, M.J.; Williams, D.C. Two subcellular locations for peripheral-type benzodiazepine acceptors in rat liver. Eur. J. Biochem. 1990, 188, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, M.; Fertikh, D.; Culty, M.; Li, H.; Vidic, B.; Papadopoulos, V. Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: Correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res. 1999, 59, 831–842. [Google Scholar] [PubMed]
- De Souza, E.B.; Anholt, R.R.; Murphy, K.M.; Snyder, S.H.; Kuhar, M.J. Peripheral-type benzodiazepine receptors in endocrine organs: Autoradiographic localization in rat pituitary, adrenal, and testis. Endocrinology 1985, 116, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Gehlert, D.R.; Yamamura, H.I.; Wamsley, J.K. Autoradiographic localization of “peripheral-type” benzodiazepine binding sites in the rat brain, heart and kidney. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1985, 328, 454–460. [Google Scholar] [CrossRef]
- Awad, M.; Gavish, M. Peripheral-type benzodiazepine receptors in human cerebral cortex, kidney, and colon. Life Sci. 1991, 49, 1155–1161. [Google Scholar] [CrossRef]
- Bribes, E.; Carrière, D.; Goubet, C.; Galiègue, S.; Casellas, P.; Joêlle, S.-L. Immunohistochemical Assessment of the Peripheral Benzodiazepine Receptor in Human Tissues. J. Histochem. Cytochem. 2004, 52, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, H.; Kononen, J.; Haapasalo, H.; Helén, P.; Sallinen, P.; Harjuntausta, T.; Helin, H.; Alho, H. Expression of Peripheral-Type Benzodiazepine Receptor and Diazepam Binding Inhibitor in Human Astrocytomas: Relationship to Cell Proliferation. Cancer Res. 1995, 55, 2691–2695. [Google Scholar] [PubMed]
- Miyazawa, N.; Hamel, E.; Diksic, M. Assessment of the peripheral benzodiazepine receptors in human gliomas by two methods. J. Neuro-Oncol. 1998, 38, 19–26. [Google Scholar] [CrossRef]
- Fafalios, A.; Akhavan, A.; Parwani, A.V.; Bies, R.R.; McHugh, K.J.; Pflug, B.R. Translocator protein blockade reduces prostate tumor growth. Clin. Cancer Res. 2009, 15, 6177–6184. [Google Scholar] [CrossRef] [PubMed]
- Katz, Y.; Eitan, A.; Gavish, M. Increase in peripheral benzodiazepine binding sites in colonic adenocarcinoma. Oncology 1990, 47, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Maaser, K.; Grabowski, P.; Sutter, A.P.; Hopfner, M.; Foss, H.-D.; Stein, H.; Berger, G.; Gavish, M.; Zeitz, M.; Scherubl, H. Overexpression of the Peripheral Benzodiazepine Receptor Is a Relevant Prognostic Factor in Stage III Colorectal Cancer. Clin. Cancer Res. 2002, 8, 3205–3209. [Google Scholar] [PubMed]
- Konigsrainer, I.; Vogel, U.F.; Beckert, S.; Sotlar, K.; Coerper, S.; Braun, A.; Lembert, N.; Steurer, W.; Konigsrainer, A.; Kupka, S. Increased translocator protein (TSPO) mRNA levels in colon but not in rectum carcinoma. Eur. Surg. Res. 2007, 39, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Beinlich, A.; Strohmeier, R.; Kaufmann, M.; Kuhl, H. Relation of cell proliferation to expression of peripheral benzodiazepine receptors in human breast cancer cell lines. Biochem. Pharmacol. 2000, 60, 397–402. [Google Scholar] [CrossRef]
- Galiegue, S.; Casellas, P.; Kramar, A.; Tinel, N.; Simony-Lafontaine, J. Immunohistochemical Assessment of the Peripheral Benzodiazepine Receptor in Breast Cancer and Its Relationship with Survival. Clin. Cancer Res. 2004, 10, 2058–2064. [Google Scholar] [CrossRef] [PubMed]
- Sutter, A.P.; Maaser, K.; Hopfner, M.; Barthel, B.; Grabowski, P.; Faiss, S.; Carayon, P.; Zeitz, M.; Scherubl, H. Specific ligands of the peripheral benzodiazepine receptor induce apoptosis and cell cycle arrest in human esophageal cancer cells. Int. J. Cancer 2002, 102, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Batra, S.; Iosif, C.S. Peripheral benzodiazepine receptor in human endometrium and endometrial carcinoma. Anticancer Res. 2000, 20, 463–466. [Google Scholar] [PubMed]
- Venturini, I.; Alho, H.; Podkletnova, I.; Corsi, L.; Rybnikova, E.; Pellicci, R.; Baraldi, M.; Pelto-Huikko, M.; Helen, P.; Zeneroli, M.L. Increased expression of peripheral benzodiazepine receptors and diazepam binding inhibitor in human tumors sited in the liver. Life Sci. 1999, 65, 2223–2231. [Google Scholar] [CrossRef]
- Nagler, R.; Ben-Izhak, O.; Savulescu, D.; Krayzler, E.; Akrish, S.; Leschiner, S.; Otradnov, I.; Zeno, S.; Veenman, L.; Gavish, M. Oral cancer, cigarette smoke and mitochondrial 18kDa translocator protein (TSPO)—In vitro, in vivo, salivary analysis. Biochim. Biophys. Acta 2010, 1802, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Janczar, K.; Su, Z.; Raccagni, I.; Anfosso, A.; Kelly, C.; Durrenberger, P.F.; Gerhard, A.; Roncaroli, F. The 18-kDa mitochondrial translocator protein in gliomas: From the bench to bedside. Biochem. Soc. Trans. 2015, 43, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Ruksha, T.; Aksenenko, M.; Papadopoulos, V. Role of translocator protein in melanoma growth and progression. Arch. Dermatol. Res. 2012, 304, 839–845. [Google Scholar]
- Chen, M.K.; Guilarte, T.R. Translocator protein 18 kDa (TSPO): Molecular sensor of brain injury and repair. Pharmacol. Ther. 2008, 118, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Giatzakis, C.; Papadopoulos, V. Differential utilization of the promoter of peripheral-type benzodiazepine receptor by steroidogenic versus nonsteroidogenic cell lines and the role of Sp1 and Sp3 in the regulation of basal activity. Endocrinology 2004, 145, 1113–1123. [Google Scholar] [CrossRef] [PubMed]
- Kruczek, C.; Gorg, B.; Keitel, V.; Pirev, E.; Kroncke, K.D.; Schliess, F.; Haussinger, D. Hypoosmotic swelling affects zinc homeostasis in cultured rat astrocytes. Glia 2009, 57, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Slack, R.S.; Li, W.; Papadopoulos, V. Expression of peripheral benzodiazepine receptor (PBR) in human tumors: Relationship to breast, colorectal, and prostate tumor progression. J. Recept. Signal Transduct. Res. 2003, 23, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Tantawy, M.N.; Charles Manning, H.; Peterson, T.E.; Colvin, D.C.; Gore, J.C.; Lu, W.; Chen, Z.; Chad Quarles, C. Translocator Protein PET Imaging in a Preclinical Prostate Cancer Model. Mol. Imaging Boil. Mib. 2018, 20, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Carmel, I.; Fares, F.A.; Leschiner, S.; Scherübl, H.; Weisinger, G.; Gavish, M. Peripheral-type benzodiazepine receptors in the regulation of proliferation of MCF-7 human breast carcinoma cell line. Biochem. Pharmacol. 1999, 58, 273–278. [Google Scholar] [CrossRef]
- Wang, H.J.; Fan, J.; Papadopoulos, V. Translocator protein (Tspo) gene promoter-driven green fluorescent protein synthesis in transgenic mice: An in vivo model to study Tspo transcription. Cell Tissue Res. 2012, 350, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.L.; Unterrainer, M.; Fleischmann, D.F.; Lindner, S.; Vettermann, F.; Brunegraf, A.; Vomacka, L.; Brendel, M.; Wenter, V.; Wetzel, C.; et al. TSPO PET for glioma imaging using the novel ligand (18)F-GE-180: First results in patients with glioblastoma. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 2230–2238. [Google Scholar] [CrossRef] [PubMed]
- Veenman, L.; Gavish, M. The peripheral-type benzodiazepine receptor and the cardiovascular system. Implications for drug development. Pharmacol. Ther. 2006, 110, 503–524. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, V.; Lecanu, L. Translocator protein (18 kDa) TSPO: An emerging therapeutic target in neurotrauma. Exp. Neurol. 2009, 219, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Rupprecht, R.; Papadopoulos, V.; Rammes, G.; Baghai, T.C.; Fan, J.; Akula, N.; Groyer, G.; Adams, D.; Schumacher, M. Translocator protein (18 kDa) (TSPO) as a therapeutic target for neurological and psychiatric disorders. Nat. Rev. Drug Discov. 2010, 9, 971–988. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Cordero, R.; Gupta, A.; Jayakumar, A.R.; Ciancio, G.; Nielsen, G.P.; Jorda, M. Adrenal Oncocytic Neoplasm with Paradoxical Loss of Important Mitochondrial Steroidogenic Protein: The 18 kDA Translocator Protein. Case Rep. Endocrinol. 2017, 2017, 6734695. [Google Scholar] [CrossRef] [PubMed]
- Megger, D.A.; Rosowski, K.; Ahrens, M.; Bracht, T.; Eisenacher, M.; Schlaak, J.F.; Weber, F.; Hoffmann, A.C.; Meyer, H.E.; Baba, H.A.; et al. Tissue-based quantitative proteome analysis of human hepatocellular carcinoma using tandem mass tags. Biomarkers 2017, 22, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Woods, M.J.; Williams, D.C. Multiple forms and locations for the peripheral-type benzodiazepine receptor. Biochem. Pharmacol. 1996, 52, 1805–1814. [Google Scholar] [CrossRef]
- Gazouli, M.; Yao, Z.X.; Boujrad, N.; Corton, J.C.; Culty, M.; Papadopoulos, V. Effect of peroxisome proliferators on Leydig cell peripheral-type benzodiazepine receptor gene expression, hormone-stimulated cholesterol transport, and steroidogenesis: Role of the peroxisome proliferator-activator receptor alpha. Endocrinology 2002, 143, 2571–2583. [Google Scholar] [CrossRef] [PubMed]
- Mak, J.C.; Barnes, P.J. Peripheral type benzodiazepine receptors in human and guinea pig lung: Characterization and autoradiographic mapping. J. Pharmacol. Exp. Ther. 1990, 252, 880–885. [Google Scholar] [PubMed]
- Wu, X.; Gallo, K.A. The 18-kDa translocator protein (TSPO) disrupts mammary epithelial morphogenesis and promotes breast cancer cell migration. PLoS ONE 2013, 8, e71258. [Google Scholar] [CrossRef] [PubMed]
- Butlen, D. Benzodiazepine receptors along the nephron: [3H]PK 11195 binding in rat tubules. FEBS Lett. 1984, 169, 138–142. [Google Scholar] [CrossRef]
- Bribes, E.; Casellas, P.; Vidal, H.; Dussossoy, D.; Casellas, D. Peripheral benzodiazepine receptor mapping in rat kidney. Effects of angiotensin II-induced hypertension. J. Am. Soc. Nephrol. 2002, 13, 1–9. [Google Scholar] [PubMed]
- Ishiguro, K.; Taft, W.C.; DeLorenzo, R.J.; Sartorelli, A.C. The role of benzodiazepine receptors in the induction of differentiation of HL-60 leukemia cells by benzodiazepines and purines. J. Cell. Physiol. 1987, 131, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Canat, X.; Guillaumont, A.; Bouaboula, M.; Poinot-Chazel, C.; Derocq, J.M.; Carayon, P.; LeFur, G.; Casellas, P. Peripheral benzodiazepine receptor modulation with phagocyte differentiation. Biochem. Pharmacol. 1993, 46, 551–554. [Google Scholar] [CrossRef]
- Taketani, S.; Kohno, H.; Okuda, M.; Furukawa, T.; Tokunaga, R. Induction of peripheral-type benzodiazepine receptors during differentiation of mouse erythroleukemia cells. A possible involvement of these receptors in heme biosynthesis. J. Boil. Chem. 1994, 269, 7527–7531. [Google Scholar]
- Landau, M.; Weizman, A.; Zoref-Shani, E.; Beery, E.; Wasseman, L.; Landau, O.; Gavish, M.; Brenner, S.; Nordenberg, J. Antiproliferative and differentiating effects of benzodiazepine receptor ligands on B16 melanoma cells. Biochem. Pharmacol. 1998, 56, 1029–1034. [Google Scholar] [CrossRef]
- Stoebner, P.E.; Carayon, P.; Penarier, G.; Frechin, N.; Barneon, G.; Casellas, P.; Cano, J.P.; Meynadier, J.; Meunier, L. The expression of peripheral benzodiazepine receptors in human skin: The relationship with epidermal cell differentiation. Br. J. Dermatol. 1999, 140, 1010–1016. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Mukherjee, S. Role of peripheral benzodiazepine receptors on secretion of surfactant in guinea pig alveolar type II cells. Biosci. Rep. 1999, 19, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Wade, F.M.; Wakade, C.; Mahesh, V.B.; Brann, D.W. Differential expression of the peripheral benzodiazepine receptor and gremlin during adipogenesis. Obes. Res. 2005, 13, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Campioli, E.; Carnevale, G.; Avallone, R.; Guerra, D.; Baraldi, M. Morphological and receptorial changes in the epididymal adipose tissue of rats subjected to a stressful stimulus. Obesity 2011, 19, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Alenfall, J.; Batra, S. Modulation of peripheral benzodiazepine receptor density by testosterone in Dunning G prostatic adenocarcinoma. Life Sci. 1995, 56, 1897–1902. [Google Scholar] [CrossRef]
- Hardwick, M.; Cavalli, L.R.; Barlow, K.D.; Haddad, B.R.; Papadopoulos, V. Peripheral-type benzodiazepine receptor (PBR) gene amplification in MDA-MB-231 aggressive breast cancer cells. Cancer Genet. Cytogenet. 2002, 139, 48–51. [Google Scholar] [CrossRef]
- Austin, C.J.; Kahlert, J.; Kassiou, M.; Rendina, L.M. The translocator protein (TSPO): A novel target for cancer chemotherapy. Int. J. Biochem. Cell Boil. 2013, 45, 1212–1216. [Google Scholar] [CrossRef] [PubMed]
- Mammen, J.M.; Matthews, J.B. Mucosal repair in the gastrointestinal tract. Crit. Care Med. 2003, 31, S532–S537. [Google Scholar] [CrossRef] [PubMed]
- Casellas, P.; Galiegue, S.; Basile, A.S. Peripheral benzodiazepine receptors and mitochondrial function. Neurochem. Int. 2002, 40, 475–486. [Google Scholar] [CrossRef]
- Batarseh, A.; Papadopoulos, V. Regulation of translocator protein 18 kDa (TSPO) expression in health and disease states. Mol. Cell. Endocrinol. 2010, 327, 1–12. [Google Scholar] [CrossRef] [PubMed]









| Prostate | ||
|---|---|---|
| Tissue Type | Increase Decrease | Localisation |
| Normal |
| |
| Grade II adenocarcinoma | –Insignificant increase |
|
| Grade III adenocarcinoma |
| |
| Stomach | ||
| Epiploon |
| |
| Stomach |
| |
| Grade III stomach squamous cell carcinoma | –Insignificant increase |
|
| Colon | ||
| Normal |
| |
| Grade I adenocarcinoma | –Significant decrease |
|
| Grade II adenocarcinoma |
| |
| Grade III adenocarcinoma |
| |
| Liver | ||
| Normal |
| |
| Grade II Hepato-cellular carcinoma | –Significant increase |
|
| Grade III Hepato-cellular carcinoma |
| |
| Lung | ||
| Normal |
| |
| Grade III lung adenocarcinoma | –Significant decrease |
|
| Grade III lung squamous cell carcinoma |
| |
| small cell carcinoma |
| |
| Kidney | ||
| Normal |
| |
| Grade III chromophobe renal cell carcinoma | –Significant increase |
|
| Clear cell renal carcinoma |
| |
| Breast | ||
| Normal |
| |
| Grade III invasive carcinoma (NST) |
| |
| Brain | ||
| White matter |
| |
| Grey matter |
| |
| Grade II astrocytoma | –Significant increase |
|
| CNS lymphoma |
| |
| Ependymoma |
| |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhoola, N.H.; Mbita, Z.; Hull, R.; Dlamini, Z. Translocator Protein (TSPO) as a Potential Biomarker in Human Cancers. Int. J. Mol. Sci. 2018, 19, 2176. https://doi.org/10.3390/ijms19082176
Bhoola NH, Mbita Z, Hull R, Dlamini Z. Translocator Protein (TSPO) as a Potential Biomarker in Human Cancers. International Journal of Molecular Sciences. 2018; 19(8):2176. https://doi.org/10.3390/ijms19082176
Chicago/Turabian StyleBhoola, Nimisha H., Zukile Mbita, Rodney Hull, and Zodwa Dlamini. 2018. "Translocator Protein (TSPO) as a Potential Biomarker in Human Cancers" International Journal of Molecular Sciences 19, no. 8: 2176. https://doi.org/10.3390/ijms19082176
APA StyleBhoola, N. H., Mbita, Z., Hull, R., & Dlamini, Z. (2018). Translocator Protein (TSPO) as a Potential Biomarker in Human Cancers. International Journal of Molecular Sciences, 19(8), 2176. https://doi.org/10.3390/ijms19082176