Abstract
Periodontal ligament mesenchymal stem cells (hPDLSCs), as well as all mesenchymal stem cells, show self-renewal, clonogenicity, and multi-tissue differentiation proprieties and can represent a valid support for regenerative medicine. We treated hPDLSCs with a combination of Moringin (MOR) and Cannabidiol (CBD), in order to understand if treatment could improve their survival and their in vitro differentiation capacity. Stem cells survival is fundamental to achieve a successful therapy outcome in the re-implanted tissue of patients. Through NGS transcriptome analysis, we found that combined treatment increased hPDLSCs survival, by inhibition of apoptosis as demonstrated by enhanced expression of anti-apoptotic genes and reduction of pro-apoptotic ones. Moreover, we investigated the possible involvement of PI3K/Akt/mTOR pathway, emphasizing a differential gene expression between treated and untreated cells. Furthermore, hPDLSCs were cultured for 48 h in the presence or absence of CBD and MOR and, after confirming the cellular viability through MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazoliumbromide) assay, we examined the presence of neuronal markers, through immunofluorescence analysis. We found an increased expression of Nestin and GAP43 (growth associated protein 43) in treated cells. In conclusion, hPDLSCs treated with Moringin and Cannabidiol showed an improved survival capacity and neuronal differentiation potential.
1. Introduction
Human adult mesenchymal stem cells (MSCs) retain the characteristic of all stem cells to generate different mature cell lines and self-renewal propriety. These cells can be obtained from different tissue sources such as bone marrow, adipose tissue, and peripheral blood [1]. Another source of adult stem cells, which involve less invasive extraction techniques, is represented by oral and dental tissues, such as apical papilla [2], gingiva, dental pulp [3], exfoliated deciduous teeth [4], dental follicle, and periodontal ligament (hPDLSCs) [5,6,7,8]. Mesenchymal stem cells derived from the oral cavity are able to differentiate in vitro, also at long-term passage, resulting easy to manipulate and to use in laboratory practice [9]. Self-renewal capacity, differentiation, and immunomodulatory proprieties of human MSCs provide new avenues for cell-based therapy in regenerative medicine [10,11,12,13].
A pretreatment of stem cells with substances that show antioxidant, anti-inflammatory and anticancer proprieties can improve their survival in the re-implanted tissue of patients and can influence a preferential differentiation into a specific cell lineage. Our research group has extensively investigated the effects of two phytocompounds, Moringin (4-(α-l-rhamnosyloxy)-benzyl isothiocyanate-MOR) and Cannabidiol (CBD), on stem cell culture. MOR is the principal active constituent obtained from the extract of Moringa oleifera (fam. Moringaceae) seeds. We have conducted numerous studies which demonstrated anti-inflammatory proprieties of this compound both in vivo [14,15] and in vitro experiments [16], and recently, we have demonstrated its capacity to improve differentiation of periodontal ligament stem cells to osteocytes, chondrocytes, adipocytes, and neuronal cells [17]. Also CBD, the active phytocannabinoid extract from Cannabis sativa, was largely studied for its anti-inflammatory and antioxidant proprieties. Our research in anti-inflammatory activity of CBD was conducted both in macrophage culture [18] and in human gingiva-derived mesenchymal stem cells (hGMSCs) [19]. The effects of these two compounds have always been investigated when they are used alone and not in combination. Our only research in which the effects of combined treatment had been investigated, had been performed on macrophages rather than on stem cells. In this previous research, we had seen the anti-apoptotic effect of combined use of MOR and CBD on LPS-stimulated macrophages, by immunohistochemical analysis for detection of Bax and Bcl-2 expression, noting a decreased level of Bax, Bcl-2, and caspase-3 [18].
In the present study, we have investigated more accurately the anti-apoptotic or proliferative effect of combined treatment with MOR and CBD on hPDLSCs, analyzing their transcriptomic profile by Next Generation Sequencing analysis (NGS). In particular, we investigated the possible involvement of PI3K/Akt/mTOR pathway in stem cell survival, by enhanced expression of anti-apoptotic genes and contemporary inhibition of pro-apoptotic genes. Moreover, a longer survival time in the transplanted tissue becomes important both for the success of the cell-therapy and for the increased possibility to differentiate into a specific tissue lineage. Furthermore, we investigated if the treatment of hPDLSCs with CBD and MOR could influence differentiation to neuronal lineage, providing a valid tool for stem-based therapy in the neurodegenerative diseases treatment.
2. Results
2.1. Cytofluorimetric and Morphological Analysis of hPDLSCs
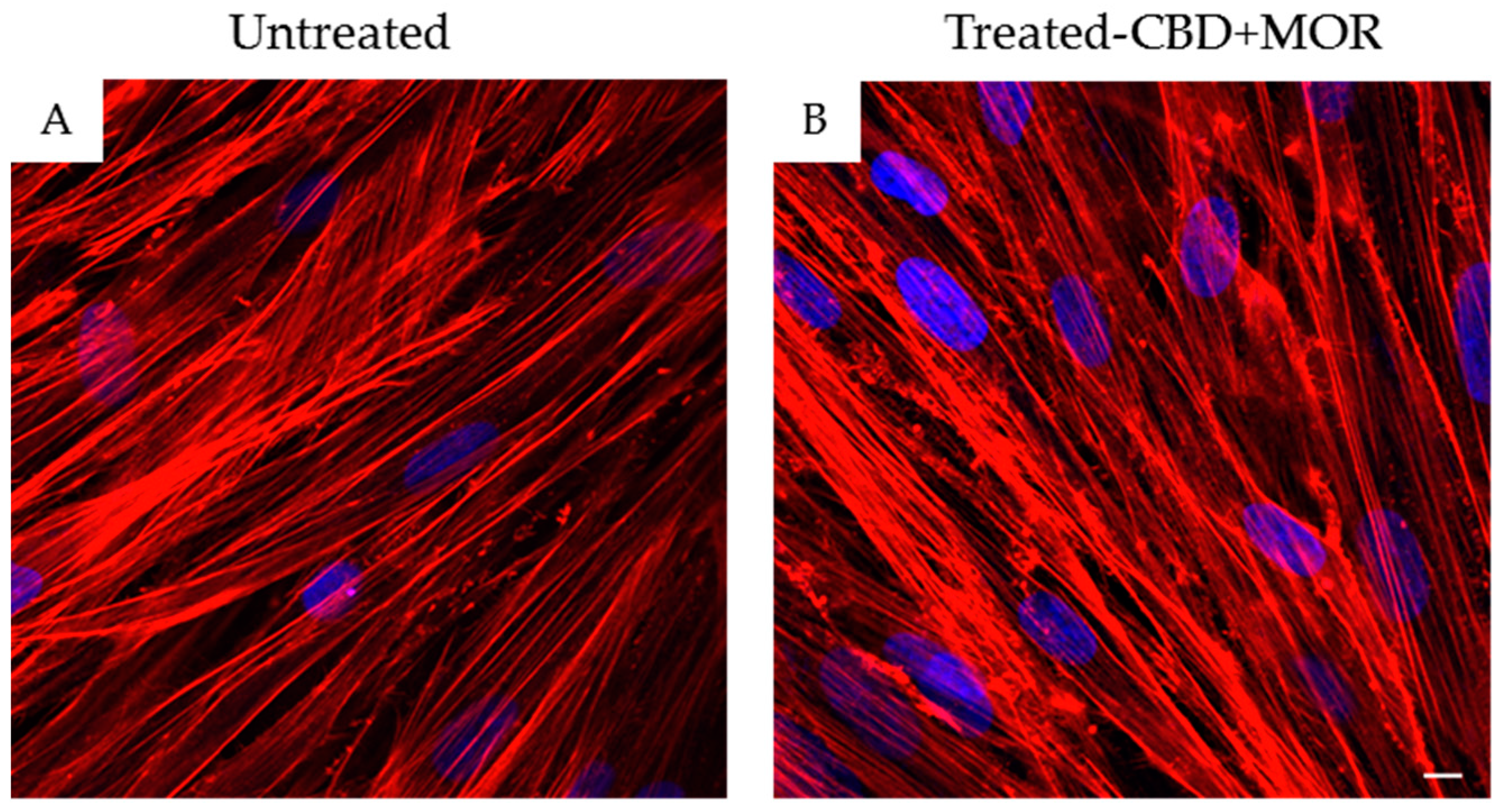
In order to confirm hPDLSCs phenotype, flow cytometry analysis was conducted on stem cells at the second passage. Ex vivo expanded hPDLSCs showed positivity for Oct3/4, Sox-2, SSEA-4, CD29, CD44, CD73, CD90, and CD105. On the contrary, the hematopoietic stem cell markers as CD14, CD34, and CD45 were negative (Table 1). Confocal laser morphological observation of untreated hPDLSCs, maintained in standard culture conditions, displayed an elongated spindle-shaped structure (Figure 1A), while hPDLSCs, treated with a mixture of CBD and MOR, showed a cytoskeleton rearrangement, in fact many cells showed a roundish structure (Figure 1B).

Table 1.
Cytofluorimetric analysis of hPDLSCs. In the table, were reported: cell typology, specific markers, and percentage of expression of each marker. All markers of stem cells and mesenchymal stem cells were expressed on the surface, while hematopoietic cell markers were not expressed.
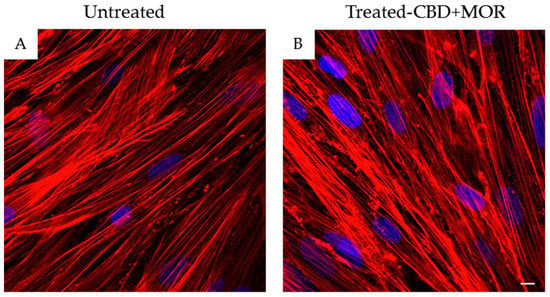
Figure 1.
Morphology of hPDLSCs. (A) Confocal laser scanning observations of untreated hPDLSCs. (B) Confocal laser scanning observations of hPDLSCs treated with the mixture CBD+MOR. Red fluorescence: F-actin; blue fluorescence: nuclei. Scale bar: 5 µm.
2.2. Cell Viability Assays
To demonstrate that treatment with CBD and MOR was not toxic for cells and to evaluate cell viability, MTT assay and trypan blue exclusion test were performed.
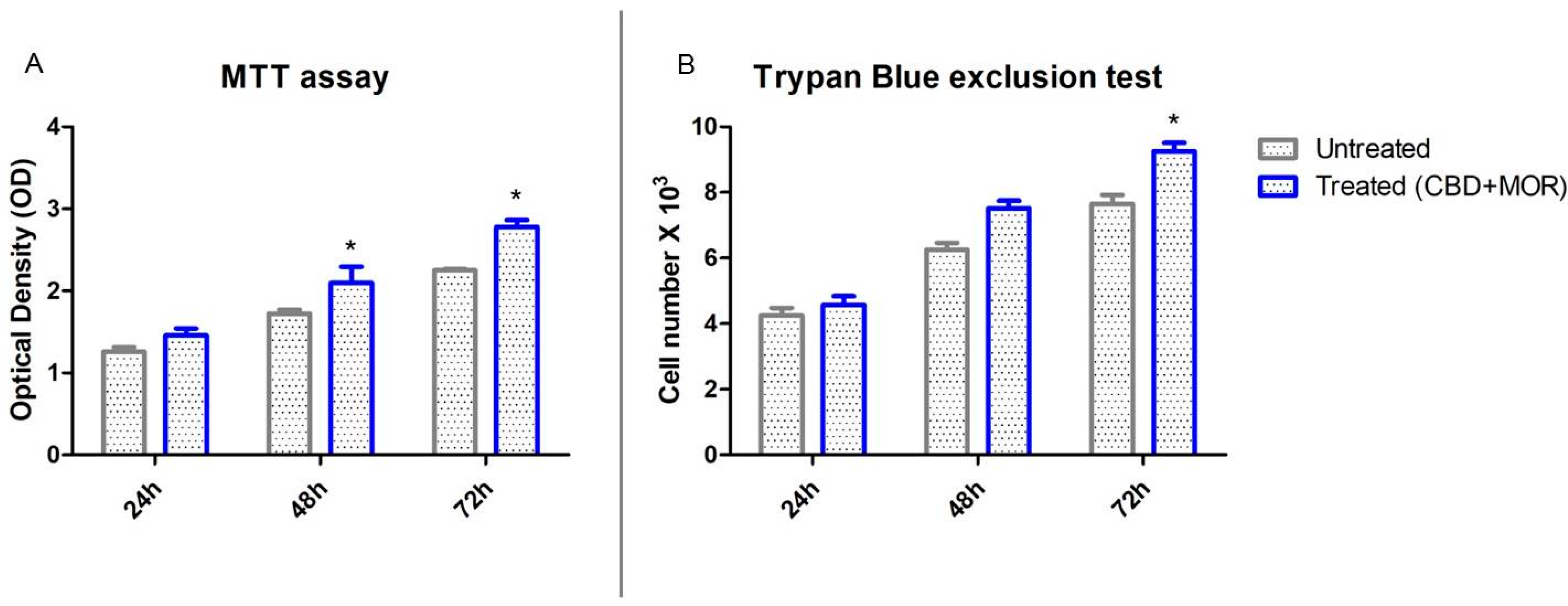
The mixture composed by CBD and MOR showed no toxicity. Figure 2 showed the influence of CBD and MOR on hPDLSCs viability. The obtained results indicated that treated hPDLSCs maintained a viability similar to untreated cells and also showed a slow increase in the growth capacity (Figure 2A). Moreover, trypan blue exclusion test showed the number of the viable treated cells was slowly increased compared with the untreated (Figure 2B).
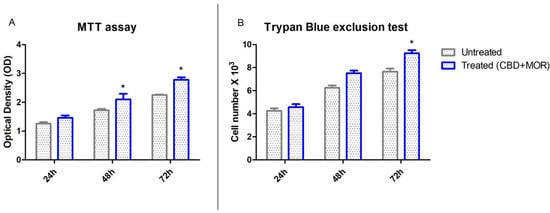
Figure 2.
Cell viability assays. (A) Viability rate of untreated and treated (CBD+MOR) hPDLSCs at different endpoint (24, 48, and 72 h) evaluated using MTT assay. Data are expressed as the mean of optical density. (B) Viability, as determined by Trypan Blue exclusion, of treated and untreated hPDLSCs. Data are expressed as the mean (n = 3) number of cells with 95% confidence limits. * p < 0.05.
2.3. Immunofluorescence Analysis
In order to evaluate neuronal differentiation of hPDLSCs after combined treatment, we performed immunofluorescence analysis.
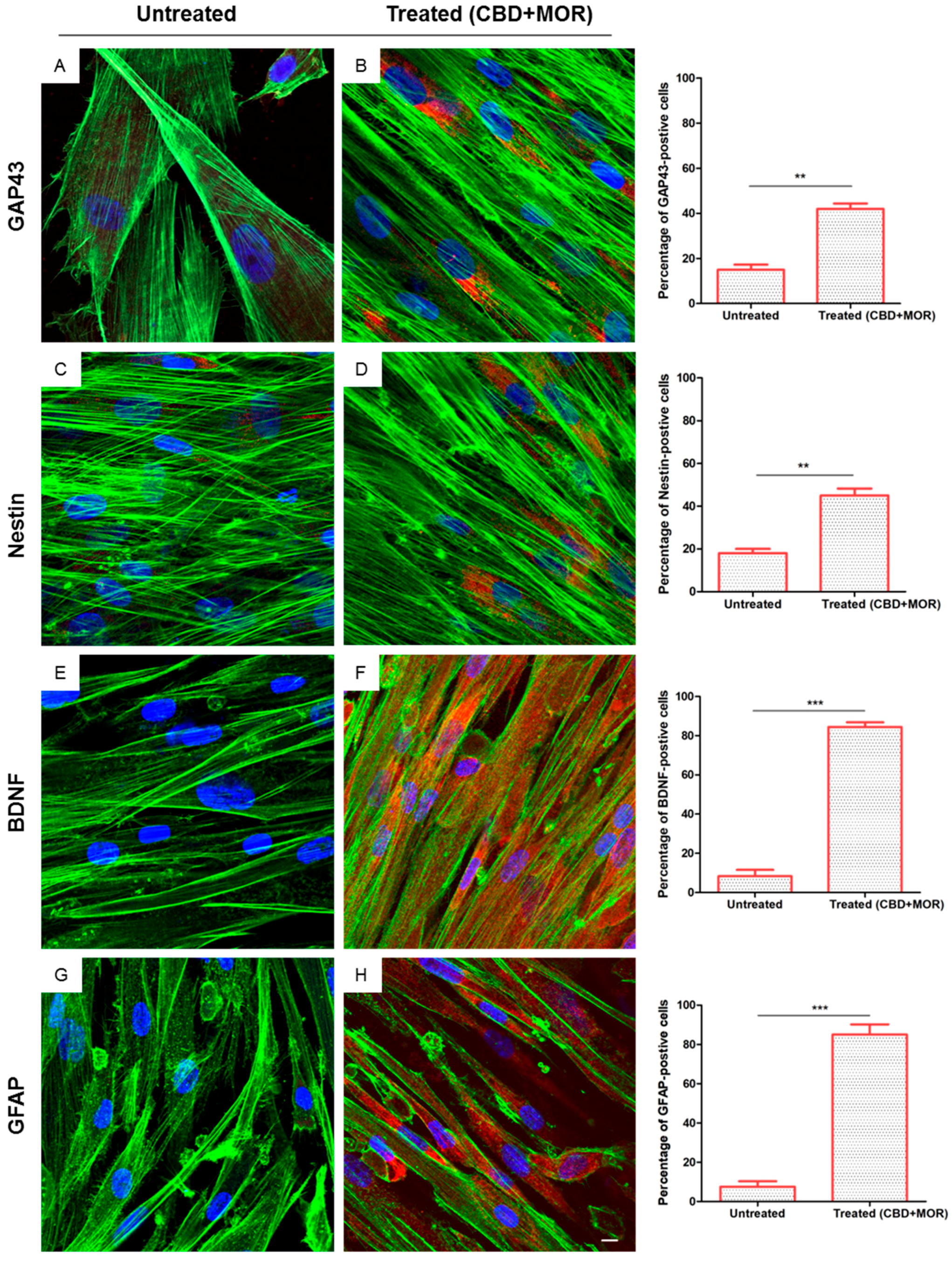
After 48 h of incubation with CBD+MOR, treated hPDLSCs showed a cytoskeletal remodeling, analyzed through F-actin assembly expression. A qualitative analysis of fluorescent photomicrographs, showed a slow cytoplasmatic expression of GAP43 (growth associated protein 43) and NES (Nestin) in treated hPDLSCs when compared with untreated hPDLSCs, maintained in the same culture conditions (>40%, Figure 3B,D). On the other hand treated hPDLSCs showed a high positivity for BDNF (brain derived neurotrophic factor) and GFAP (glial fibrillary acidic protein), which are well-recognized markers of neuronal and glial cells. As showed in Figure 3F,H, more than 80% of cells were positive for BDNF and GFAP markers.
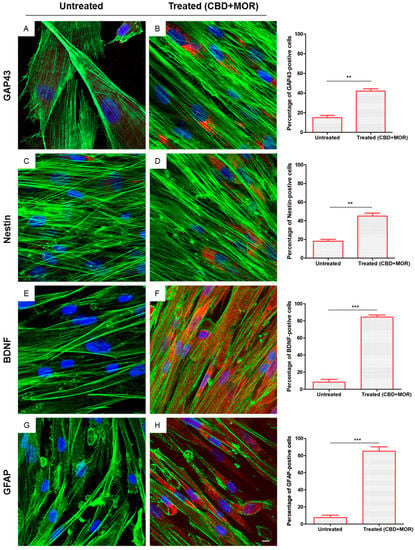
Figure 3.
Immunofluorescence analysis. Immunolabeling with GAP43 in (A) untreated hPDLSCs and (B) treated (CBD+MOR) hPDLSCs. Immunolabeling with neuron-specific NES in (C) untreated hPDLSCs and (D) treated (CBD+MOR) hPDLSCs. Immunolabeling with neuron-specific BDNF in (E) untreated hPDLSCs and (F) treated (CBD+MOR) hPDLSCs. Immunolabeling with neuron-specific GFAP in (G) untreated hPDLSCs and (H) treated (CBD+MOR) hPDLSCs. Histograms represent the percentage of positive cells for the specific markers. ** p < 0.01, *** p < 0.001 significant difference of hPDLSCs treated with CBD and MOR compared to untreated cells. Green fluorescence: F-actin; red fluorescence: specific markers; blue fluorescence: nuclei. Scale bar: 5 µm.
2.4. NGS Analysis
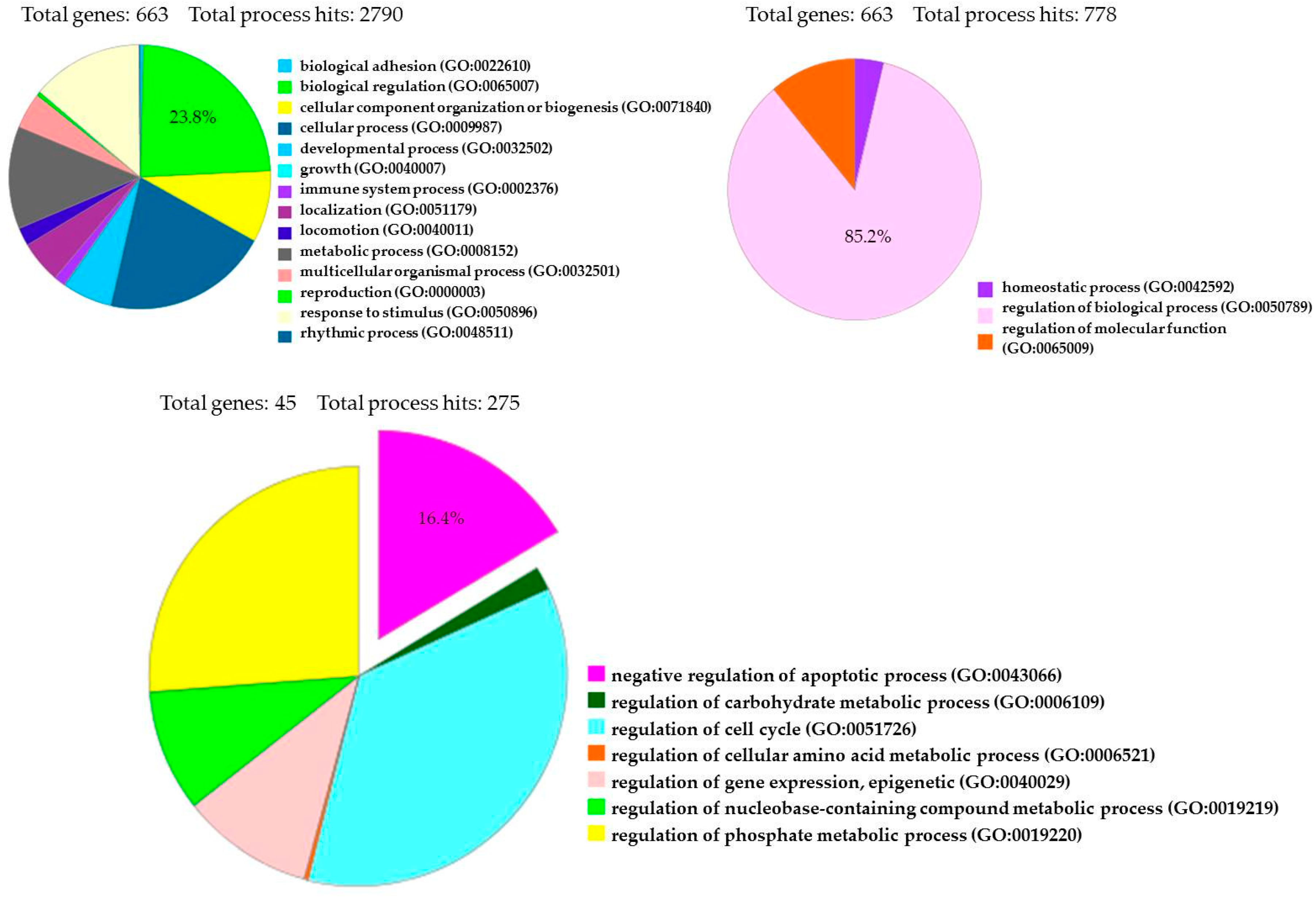
The transcriptome of treated hPDLSCs (MOR+CBD) and untreated cells (CTR) was carried out using NGS Technology (Illumina, San Diego, CA, USA) and was conducted in triplicate. We identified a total of 6843 genes statistically significant (q value < 0.05) and differentially expressed in two experimental groups. More precisely, 3439 genes were upregulated (Log2-fold change between 0.045 and 19.37), while 3404 genes were downregulated (Log2-fold change between −0.055 and −29.32). The fold change indicates the differential gene expression between CTR (untreated-hPDLSCs) and sample (hPDLSCs treated with a combination of MOR and CBD). We investigated the anti-apoptotic effect of treatment with combination of MOR and CBD, by PI3K/Akt/mTOR pathway involvement, through the use of database, such as Gene Ontology and KEGG. Among 6843 genes differentially expressed in our analysis, Gene Ontology identified 663 genes (23.8%) involved in GO: “biological regulation”, among 2790 genes implicated in the regulation of different biological processes, and 663 genes (85.2%) among 778 genes involved in GO: “regulation of biological process”. Moreover, Gene Ontology identified 45 genes (16.4%) involved in GO: “negative regulation of apoptotic process” (Figure 4). Furthermore, Gene Ontology identified 670 genes implicated in GO: “signal transduction” and among them identified 21 genes (3.1%) involved in GO: “PI3 kinase pathway (P00048)”.
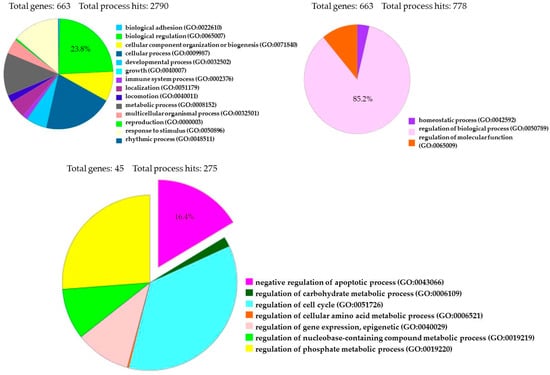
Figure 4.
Gene Ontology Analysis of 6843 genes differentially expressed between treated hPDLSCs (MOR+CBD) and untreated cells (CTR).
The simultaneous consultation of websites as NCBI and KEGG and literature, led us to find a larger number of genes involved in the inhibition of apoptosis (63 genes, Table 2 and Table 3), in death signaling (31 genes, Table 4), in mTOR pathway (63 genes, Table 5), and finally 38 genes belonging to PI3K/Akt/mTOR pathway (Table 6).

Table 2.
Upregulated genes involved in “Inhibition of Apoptosis”.

Table 3.
Downregulated genes involved in “Inhibition of Apoptosis”.

Table 4.
Up- and downregulated genes, involved in the “Cell death receptor signaling” pathway.

Table 5.
Up- and downregulated genes, involved in “mTOR pathway”.

Table 6.
Up- and downregulated genes, involved in “PI3K/Akt/mTOR pathway”.
In each table, we have reported: gene nomenclature, the whole name of the genes, their expression value (calculated as FPKM, Fragments Per Kilobase of exon model per Million mapped fragments) both in control cells (CTR) and treated cells (MOR+CBD), the corresponding values of Log2-fold change and the statistical significance, indicated by q value (FDR, False Discovery Rate) < 0.05. All genes are placed on tables, following descending order of Log2-fold change. Fold change represents the ratio between the value of gene expression in the treated sample compared to the untreated one, calculated as Log2. A positive Log2-fold change value indicates that gene is more expressed in treated sample than in control. A negative value is correlated to a major gene expression in CTR. We used a Log-2 fold change value of −3.5 as cutoff and showed in tables all genes with higher values. In Table 2, we reported all genes involved in “apoptosis inhibition” with a positive value of fold change, while in Table 3 those with negative fold change value. In the table footer of Table 2, main genes showing anti-apoptotic or pro-apoptotic proprieties have been listed.
3. Discussion
Stem cell-based therapies for the repair and regeneration of various tissues and organs could provide alternative therapeutic solutions for a number of diseases and neurodegenerative pathologies [20]. hPDLSCs retain all the features of MSCs, such as self-renewal, immunomodulatory, clonogenicity, and multi-tissue differentiation potential [21,22]. Improve and extend survival time of stem cells in transplantation, is one of the main objectives of research in regenerative medicine. A pretreatment with antioxidant, anti-inflammatory, or anticancer compounds can improve their survival and their differentiation capacity. Phytocompounds, such as Cannabidiol and Moringin, were widely studied for their antioxidant [18], anti-inflammatory [16,19], and anticancer [23] proprieties and so a treatment with MOR and CBD of mesenchymal cells can improve their survival. Mesenchymal stem cells can spontaneously differentiate to different cell lineage, showing osteogenic, adipogenic, chondrogenic, and neurogenic differentiation capabilities. We previously demonstrated that a treatment with MOR of hPDLSCs improve their transformation into nervous cell lineage [17]. In this research, we first investigated the effects of combined treatment of CBD and MOR on survival of hPDLSCs and then on neuronal differentiation. Through NGS analysis, we could evaluate the expression of thousands of genes implicated in apoptosis and cell proliferation pathways.
3.1. Inhibition of Apoptosis
De novo gene expression and gene rearrangement are necessary to the apoptotic machinery. Although the complete subset of genes that is induced by survival factor is not yet known, several inducible genes, such as BAX (BCL2 associated X, apoptosis regulator) and BAK1 (BCL2 antagonist/killer 1), which are members of pro-apoptotic Bcl-2 family, have been identified as critical components of the apoptotic machinery. Apoptotic process can occur by following extrinsic and intrinsic pathways. The extrinsic pathway is mediated by death receptors such as TNFR (Tumor Necrosis Factor receptors), Fas (Fas cell surface death receptor), and TRAIL (TNFSF10, TNF superfamily member 10). Activation of these receptors leads to the recruitment and activation of initiator of caspases cascade (caspases 8 and 10) and finally to activation of an effector caspase, typically caspase 3, that is responsible for the cleavage of a number of death substrates causing membrane blebbing (LMNA, lamin A/C and ROCK1, Rho associated coiled-coil containing protein kinase 1), and DNA fragmentation (ACIN1, apoptotic chromatin condensation inducer 1 and PARP1, poly(ADP-ribose) polymerase 1). The intrinsic pathway is largely centered on mitochondria that release cytochrome c, in response to pro-apoptotic members of the Bcl-2 protein family, promoting the formation of the apoptosome, composed of dATP, APAF1 (apoptosis protease activation factor-1), and activated caspase 9, leading to activation of caspase 3. After cytochrome c release, apoptosis can be still inhibited by IAP (Inhibitor Apoptosis Proteins) family, which binds and inhibits caspases [24]. On the other hand, IAP proteins are inhibited by DIABLO (diablo IAP-binding mitochondrial protein), promoting cell death [25]. Our analysis showed a reduced expression of gene coding for receptor TNFR-1, (TNFRSF1A, TNF receptor superfamily member 1A, fold change = −1.32, q value = 0.0001) and also transducer of signaling such as TRADD (TRADD, TNFRSF1A associated via death domain, fold change = −0.72, q value = 0.0318) are considerably reduced in treated sample. Gene coding for RIP (RIPK1, receptor interacting serine/threonine kinase 1, fold change = −0.71, q value = 0.0001), a pro-apoptotic protein that enhanced the association between TNFR-1 and TRADD, also had a reduced value of expression, thanks to the high expression value of the TNFAIP3 gene (TNF alpha induced protein 3, fold change = +2.06, q value = 0.0001), coding for A20 protein that exerts an inhibitory activity on RIP. According to Kim et al. (2000), pro-apoptotic C-terminal fragment of RIP inhibited NF-κB (nuclear factor kappa light chain enhancer of activated B cells) activation by suppressing the activity of IKKb (IKBKB, I-kB kinase b), which phosphorylates I-Kb (NFKBIB, NFKB inhibitor β), an inhibitor of NF-κB [26]. We found that IKBKB had a decreasing value of expression (fold change = −0.57, q value = 0.0001), while NFKBIA (NFKB inhibitor α, fold change = +0.36, q value = 0.0025) was increased. Inhibition of NF-κB was confirmed by the reduced value of its expression (NFKB1, nuclear factor kappa B subunit 1, fold change = −0.65, q value = 0.0001). NF-κB, after translocation into the nucleus, regulates the expression of FLIP protein (CFLAR, CASP8, and FADD like apoptosis regulator, fold change = −1.59, q value = 0.0001), which is structurally similar to caspase 8 and so is a competitor for FADD (Fas associated via death domain) linkage, inhibiting caspase 8 and apoptosis. Moreover, NF-κB regulates upexpression of TNFAIP3, BCL2A1 (BCL2 related protein A1, fold change = +14.74, q value = 0.0001), BCLXL (BCL2L1, BCL2-like 1, fold change = +1.94, q value = 0.0001), and XIAP (X-linked inhibitor of apoptosis, fold change: +0.71, q value = 0.0001), belonging to IAP family. XIAP is regulated by DIABLO, which appears downregulated (fold change= −0.37, q value = 0.0018). Caspase 8 (CASP8, fold change = −0.903, q value = 0.0001), caspase 9 (CASP9, fold change = −1.59, q value = 0.0002), and caspase 10 (CASP10, fold change = −1.55, q value = 0.0024) are initiators of caspases cascade, while caspase 6 (CASP6, fold change = −1.36, q value = 0.0032), caspase 7 (CASP7, fold change = −1.98, q value = 0.0001), and caspase 3 are effector caspases. Note that all caspases, except caspase 3, that which not appear in our analysis, had negative values of fold change. Following increased expression of pro-apoptotic factors, caspase 9 is activated by apoptosome formation and cytochrome c is released from mitochondrion. BCL2A1 and BCL2L1 are anti-apoptotic genes that regulate cytochrome c release from the outer mitochondrial membrane. When BCL2L1 is over expressed, pores are nonpermeable to pro-apoptotic molecules and the cell survives, while if it is sequestered away by Bim or Bad, proteins such as Bax and Bak are activated and cytochrome c is released, leading to initiation of caspase cascade and apoptotic events. BAX gene has a reduced expression in treated cells if compared to untreated cells (fold change = −1.18). Despite this, BCL2L1 increased and BAX and BAD (BCL2 associated agonist of cell death, fold change = −0.778, q value = 0.0139) decreased in our analysis, the gene coding for cytochrome c (CYC1, fold change = +0.66, q value = 0.0001) had an increased value of the expression. As mentioned before, also after release of cytochrome c, apoptosis can be still inhibited by over expression of genes coding for inhibitors, such as XIAP, HSPB1 (heat shock protein family B-small-member 1), HSP90AA1 (heat shock protein 90 alpha family class A member 1), and HSPA4 (heat shock protein family A (Hsp70) member 4), which showed increased values of fold change. In the mitochondrion, p53 induces Bax and Bak oligomerization and physically interacts with Bcl-xL and Bcl-2, antagonizing their anti-apoptotic effects [27]. In our analysis, we found a decreased value of expression of TP53 (tumor protein p53, fold change = −1.301, q value = 0.0001), coding for p53 protein, confirming the anti-apoptotic effect on hPDLSCs cells of combined treatment. The pro-apoptotic BAD gene is also downregulated by Akt (serine/threonine kinase 1). GSK3 gene can carry out both pro-apoptotic and anti-apoptotic role GSK3B (glycogen synthase kinase 3 beta, fold change = +0.310, q value = 0.0035) promotes cell death by the mitochondrial intrinsic apoptotic pathway and inhibits the death receptor-mediated extrinsic apoptotic signaling pathway [28] and GSK3A (glycogen synthase kinase 3 alpha, fold change = +0.253, q value = 0.0231) also plays an important role in the WNT and PI3K signaling pathways.
3.2. PI3K/Akt/mTOR Pathway
Akt is also a negative regulator of mTORC1 (mechanistic target of rapamycin complex 1). The mTORC1 complex is composed by the phosphoinositide 3 kinase-related protein kinase (PIKK) mTOR, and the subunits RAPTOR and mLST8 are implicated in cell growth processes such as protein synthesis, lipogenesis, glucose metabolism, proliferation, and survival. Akt inhibits TSC2 (TSC complex subunit 2), a protein associated with hamartin (TSC1) forming a cytosolic complex. This complex negatively regulates mTORC1 signaling, through inhibition of GTPase RHEB, an activator of mTORC1. In our analysis TSC2 gene (fold change = −0.53, q value = 0.0001) is downregulated and RHEB (Ras homolog, mTORC1 binding) is upregulated (fold change = +1.62, q value = 0.0001). RHEB regulates the expression of MTOR gene that increases in treated cells (fold change = +0.18, q value = 0.0271). The best known substrates of mTORC1 are the eIF4E-binding protein 1 (EIF4EBP1, fold change = +0.70, q value = 0.0032) and the ribosomal S6 kinase 1 (S6K1) which regulates RPS6 gene (ribosomal protein S6), coding for ribosomal protein S6. Phosphorylation of EIF4EBP1 destabilizes the 4EBP1–eIF4E complex and activates cap-dependent translation. Also, RPS6 is upregulated (fold change = +0.076, q value = 0.0354) and, together with eIF4E, promotes multiple aspects of protein synthesis. PI3K/Akt and mTOR pathways are related to apoptosis and cell survival and are also largely studied for their activation after cannabinoids administration, through linkage to the CB1 receptor [29,30]. Akt1 upstream regulates cell survival, influencing different biological processes such as apoptosis, through modulation of BCL2A1, BCL2L1, BAD, and BAX gene expression; protein synthesis, through activation of genes EIF4E and RPS6; growth and proliferation, through SGK1 (serum/glucocorticoid regulated kinase 1, fold change = +4.507, q value = 0.0001) activation; cellular glucose metabolism by enhanced expression of GYS1 (glycogen synthase 1, fold change = +1.022, q value = 0.0001); cell cycle progression through upexpression of MYC (MYC proto-oncogene, bHLH transcription factor, fold change = +1.89, q value = 0.0001), CCND1 (cyclin D1, fold change = +2.33, q value = 0.0001), and CDK1 (cyclin dependent kinase 1, fold change = +2.64, q value = 0.0001). Although genes coding for PI3K (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha/beta, PIK3CA and PIK3CB) are downregulated (fold change = −0.514, q value = 0.0001; and −0.534, q value = 0.0060, respectively) and AKT gene (AKT serine/threonine kinase 1) is not present in our analysis, all genes downstream regulated and implicated in cell survival are substantially overexpressed, confirming the activation of PI3K/Akt pathway. Moreover, proteins that negatively regulate AKT signaling pathway, such as PTEN in our analysis was down regulated (PTEN, phosphatase, and tensin homolog, fold change = −1.711, q value = 0.0001). The PI3K/Akt signaling pathway plays a crucial role also in the survival process of different neuronal cell types as reported by numerous studies [31,32], and its activation is necessary for survival of sympathetic neurons supported by NGF (nerve growth factor), both in the cell body and in the distal axons [33]. Also mesenchymal stem cells, stimulated with BDNF to promote neuronal differentiation, showed an improved survival by apoptosis inhibition, through WNT and PI3K/Akt-dependent signaling pathways [34]. In line with the activation of this pathway, the MTT and Trypan blue assay also showed increased cell viability after the treatment with CBD and MOR. Periodontal ligament stem cells (hPDLSCs), treated with a MOR and CBD combination, showed PI3K/Akt signaling pathway activated and, as previously described, treatment with only MOR stimulated hPDLSCs differentiation to neuronal lineage [17].
3.3. Neuronal Differentiation
In immunofluorescent analysis, we observed that treated hPDLSCs had an increased expression of Nestin and GAP43. Nestin is considered a marker of neuronal progenitor cells [35], while the positivity for GAP43 is associated with electrical excitability of stem cells [36]. These results lead us to conclude that the treatment of stem cells with the combination of MOR and CBD activates in vitro neuronal differentiation of totipotent cells.
We have also shown that combined treatment with MOR and CBD leads to an increase in BDNF and the appearance of the neuronal marker GFAP [37].
Also, the cytoskeletal structure of treated cells (CBD+MOR) showed a similar-neuron organization, observed by F-actin labeling (Figure 3).
4. Materials and Methods
4.1. Ethic Statement
The protocol for isolation and culture of stem cells from human periodontal ligament was approved by the Ethical Committee at the Medical School, “G. d’Annunzio” University, Chieti, Italy (266-17/04/14) and informed consent was given by the patients.
4.2. CBD and Moringin Extraction
Pure CBD (>99%) was isolated from Carmagnola, an Italian variety of industrial hemp, provided by greenhouse cultivation (CREA-CIN; Rovigo, Italy), in accordance with their legal status (Authorization SP/106 23/05/2013 of the Ministry of Health, Rome, Italy) following the standardized protocol of the cannabinoid purification to avoid any trace of THC [38,39]. MOR was isolated from powdered seeds of M. oleifera seeds (provided by Indena India Pvt. Ltd., Bangalore, India) at the Bologna laboratory (CREA-AA; previously CIN) following methods previously described [40], and the structure was confirmed by NMR (nuclear magnetic resonance) spectroscopic analyses.
4.3. hPDLSCs Culture
Periodontal ligament tissue was collected from three healthy patients enrolled in the study, after signing the informed consent, as previously described [41]. hPDLSCs were cultured in MSCGM-CD medium (Lonza, Basel, Switzerland) at 37 °C in a 5% of CO2 atmosphere. Cells were treated with the mixture of CBD and MOR at the first passages and when 80% of the confluence was reached. After 48 h of treatment, cells were collected for RNA extraction. Also untreated cells, that were cultured as a control (CTR), were collected at second passage. The experiment was conducted in triplicate. CBD and MOR have been used as a mixture 1:1 (0.5 μM concentration) for a time of incubation of 48 h.
4.4. Cytofluorimetric Analysis of hPDLSCs
Evaluation of hPDLSCs phenotype was conducted by flow cytometry, as previously described [17,19]. Briefly, 2.5 × 105 cells were incubated for 30 min with following antibodies: anti-CD44-FITC, anti-CD105-FITC, anti-CD29-PE, and anti-CD45-FITC (Ancell Corporation, Bayport, MN, USA); anti-CD14-FITC (Miltenyi Biotec, Bergisch Gladbach, Germany); OCT3/4-PE, CD73-PE, SOX2-Alexa488, SSEA4-FITC, CD90-FITC (Becton Dickinson, San Jose, CA, USA), and CD34-PE (Beckman Coulter, Fullerton, CA, USA). After incubation with appropriate secondary antibodies, fixation in 1 mL PBS 0.5% paraformaldehyde and washing, cells were analyzed using a FACStar-plus flow cytometry system and the FlowJo™ software v10.0.7 (TreeStar, Ashland, OR, USA).
4.5. Cell Viability Assays
In order to evaluate the cellular proliferation and viability, MTT test and Trypan Blue exclusion assay were performed. MTT method is a colorimetric assay based on the use of 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazoliumbromide solution (Promega, Milan, Italy). MTT assay was performed to assess the effects of the treatment of the mixture (CBD+MOR) on cell proliferation. Treated and untreated 2nd passage hPDLSCs were plated into 96-well plates at a density of 5 × 104 cells per well in 200 μL culture medium. At the designated end points (24, 48, and 72 h) 20 μL PBS MTT solution was added to each well and the plates were incubated at 37 °C for an additional 3 h. The plates were read at 650 nm using a microplate reader (Synergy HT, BioTek Instruments, Winooski, VT, USA) [42]. Moreover, to test the viability of treated and untreated hPDLSCs they were washed with culture medium, incubated for 3 min with trypsin-EDTA to obtain a single cell suspension, and washed with culture medium, and the number of living cells was determined by using Trypan Blue exclusion assay. Experiments have been carried out in triplicates on cells derived from three different donors.
4.6. Immunofluorescence Detection
hPDLSCs, at the second passage, treated and untreated with the mixture (CBD and MOR) were fixed and processed for immunofluorescence staining as reported by Diomede et al. [43]. Cells were incubated with mouse primary monoclonal antibody anti-GAP43 1:200 (Sigma Aldrich, Milan, Italy), rabbit anti-Nestin 1:200 (Santa Cruz Biotechnology, Inc., Dallas, TX, USA), rabbit anti-BDNF 1:100 (Santa Cruz Biotechnology), and mouse anti-GFAP 1:100 (Santa Cruz Biotechnology) and mouse anti-GFAP 1:100 (Santa Cruz Biotechnology), followed by anti-mouse Alexa Fluor 488 (Molecular Probes, Life Technologies, Monza, Italy) and anti-rabbit Alexa Fluor 568 (Molecular Probes), respectively.
All samples were incubated with Alexa Fluor 568 phalloidin red fluorescence conjugate (1:400), as a marker of the cytoskeleton actin and with TOPRO to highlight the nuclei [44]. Samples were observed using a CLSM (Zeiss LSM800 META, Zeiss, Jena, Germany). After treatment the percentages of GAP43/Nestin/BDNF/GFAP-positive cells were quantified based on the 15 images collected randomly. Experiments have been carried out in triplicates on cells derived from three different donors.
4.7. RNA Libraries Preparation for NGS Analysis
Experiments have been carried out in triplicates on cells derived from three different donors. Total RNA extraction was performed using Reliaprep RNA Cell Miniprep System (Promega Corporation, Madison, WI, USA) and quantified by QuantiFluor® RNA System kit and Quantus Fluorometer (Promega). Sequencing was done in triplicate. As previously described [17], and in accordance with TruSeq RNA Access library kit protocol (Illumina, San Diego, CA, USA), about 50 ng of total RNA has been fragmented at 94 °C for 8 min, obtaining fragments >200 nt. Using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) a first strand of cDNA has been synthesized. The complementary strand of cDNA has been synthesized thanks to Second Strand Marking Master Mix. Two successive steps of adapter hybridization/PCR amplification are necessary for the identification of each sample and ligation to the flow cell. The first reaction of hybridization uses exome capture probes in order to select and to enrich specific cDNA library regions. A quantity of 200 ng of resulting indexed library was used as input of a second hybridization/PCR amplification. Finally, the library has been qualitatively validated using a Bioanalyzer instrument (Agilent High Sensitivity DNA Kit, Richardson, TX, USA) and quantified using QuantiFluor® ONE dsDNA System kit and Quantus Fluorometer (Promega).
The obtained library has been denatured by adding 2 M NaOH and diluted until final concentration of 14 pM. MiSeq Reagent Kit v3 (150 cycles) has been used for sequencing on the MiSeq Instrument (Illumina, San Diego, CA, USA).
4.8. Data Analysis
The CASAVA software version 1.8 (Illumina, San Diego, CA, USA) was used to generate the “Fastq.File”. The fastq.files have been aligned to the “homo sapiens UCSC hg19” reference sequence, using the STAR, Open Source software (software distributed under GPLv3 license and can be downloaded from http://code.google.com/p/rna-star/). For statistical analysis, we used Cufflinks Assembly & DE package version 2.0.0. (software of Illumina propriety; cole-trapnell-lab.github.io/cufflinks/), in order to evaluate the statistically relevant data. Gene expression values have been calculated as FPKM (fragments per kilobase of exon per million fragments mapped) by using the Cufflinks algorithm, while differential gene expression values between the two different experimental groups (treated and untreated hPDLSCs) have been calculated using Cuffdiff algorithm. To normalize and to compare all samples, the FPKM value was calculated by applying the mathematical formula: (1000× read count)/(number of gene covered bases × number of mapped fragments in million). Only statistically significant data with q value (FDR, False Discovery Rate) <0.05 have been considered for the analysis. A scatter plot of the Log2 of the FPKM was used in order to compare the two experimental groups. The NCBI website (http://www.ncbi.nlm.nih.gov/gene), Gene Ontology (http://www.geneontology.org/), and KEGG database (http://www.genome.jp/kegg/) have been used to investigate and classify the differentially expressed genes between the two experimental groups, related to cellular processes such as: cell division, proliferation, and regulation of apoptosis. In particular, we focused on involvement of PI3K/Akt/mTOR pathway in apoptosis inhibition, by upregulation of anti-apoptotic genes and contemporary downregulation of pro-apoptotic ones.
5. Conclusions
Stem cells represent an encouraging perspective for regenerative medicine and periodontal ligament MSCs retain self-renewal, clonogenicity, and multi-tissue differentiation potential. The possibility to elongate their survival is essential to the success of stem cell-based therapy. A pretreatment of stem cells with phytocompounds could improve their survival in the re-implanted tissue and their differentiation capacity. Through NGS transcriptome analysis, we demonstrated that hPDLSCs treated with a combination of MOR and CBD showed improved survival characteristics compared to untreated cells. Prolonged survival is accompanied by inhibition of apoptosis, through activation of the PI3K/Akt/mTOR pathway, as demonstrated by differential gene expression between treated and untreated cells. Moreover, we demonstrated that hPDLSCs treated with MOR and CBD for 48 h showed an increased expression of the neuronal progenitor cell marker Nestin, and of the neuronal markers GAP43, GFAP, and BDNF evaluated by immunofluorescence analysis, confirming an improved neurogenic differentiation. However, we only performed in vitro studies, for this reason in vivo models are needed to evaluate the effects of CBD and MOR on MSCs after transplantation in animals.
Author Contributions
V.L.C. performed cDNA library preparation, analysis of data, and wrote the manuscript; D.S. performed sequencing and data analysis; A.R. has made the tables; R.I. provided MOR; F.P. provided CBG; F.D. performed in vitro experiments on stem cells; P.B. revised the manuscript and acquired funding; O.T. conceived the experiments about stem cells; E.M. conceived the experiments about sequencing and critically revised the manuscript.
Funding
This work was supported by current research funds 2018 of IRCCS “Centro Neurolesi Bonino Pulejo”.
Acknowledgments
This study was supported by current research funds 2018 of IRCCS “Centro Neurolesi Bonino-Pulejo”, Messina, Italy. The authors would like to thank Appendino Giovanni (University of Eastern Piedmont “Amedeo Avogadro”, Novara, Italy), for his technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| hPDLSCs | Human Periodontal Ligament Stem Cells |
| MOR | Moringin |
| CBD | Cannabidiol |
| NGS | Next Generation Sequencing |
| MSCs | Mesenchymal Stem Cells |
| CTR | Control |
| MTT | 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide |
References
- Hass, R.; Kasper, C.; Bohm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal. 2011, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Sonoyama, W.; Liu, Y.; Yamaza, T.; Tuan, R.S.; Wang, S.; Shi, S.; Huang, G.T. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: A pilot study. J. Endod. 2008, 34, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Gronthos, S.; Brahim, J.; Li, W.; Fisher, L.W.; Cherman, N.; Boyde, A.; DenBesten, P.; Robey, P.G.; Shi, S. Stem cell properties of human dental pulp stem cells. J. Dent. Res. 2002, 81, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Miura, M.; Gronthos, S.; Zhao, M.; Lu, B.; Fisher, L.W.; Robey, P.G.; Shi, S. Shed: Stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. USA 2003, 100, 5807–5812. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.T.; Gronthos, S.; Shi, S. Mesenchymal stem cells derived from dental tissues vs. Those from other sources: Their biology and role in regenerative medicine. J. Dent. Res. 2009, 88, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Trubiani, O.; Giacoppo, S.; Ballerini, P.; Diomede, F.; Piattelli, A.; Bramanti, P.; Mazzon, E. Alternative source of stem cells derived from human periodontal ligament: A new treatment for experimental autoimmune encephalomyelitis. Stem Cell Res. Ther. 2016, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Gugliandolo, A.; Diomede, F.; Cardelli, P.; Bramanti, A.; Scionti, D.; Bramanti, P.; Trubiani, O.; Mazzon, E. Transcriptomic analysis of gingival mesenchymal stem cells cultured on 3D bioprinted scaffold: A promising strategy for neuroregeneration. J. Biomed. Mater. Res. Part A 2018, 106, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Diomede, F.; Caputi, S.; Merciaro, I.; Frisone, S.; D’Arcangelo, C.; Piattelli, A.; Trubiani, O. Pro-inflammatory cytokine release and cell growth inhibition in primary human oral cells after exposure to endodontic sealer. Int. Endod. J. 2014, 47, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Diomede, F.; Rajan, T.S.; Gatta, V.; D’Aurora, M.; Merciaro, I.; Marchisio, M.; Muttini, A.; Caputi, S.; Bramanti, P.; Mazzon, E.; et al. Stemness maintenance properties in human oral stem cells after long-term passage. Stem Cells Int. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Granero-Molto, F.; Weis, J.A.; Longobardi, L.; Spagnoli, A. Role of mesenchymal stem cells in regenerative medicine: Application to bone and cartilage repair. Expert Opin. Biol. Ther. 2008, 8, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.M.; Kalamegam, G.; Pushparaj, P.N.; Matta, C.; Memic, A.; Khademhosseini, A.; Mobasheri, R.; Poletti, F.L.; Hoyland, J.A.; Mobasheri, A. Mesenchymal stem cells in regenerative medicine: Focus on articular cartilage and intervertebral disc regeneration. Methods 2016, 99, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Ballerini, P.; Diomede, F.; Petragnani, N.; Cicchitti, S.; Merciaro, I.; Cavalcanti, M.; Trubiani, O. Conditioned medium from relapsing-remitting multiple sclerosis patients reduces the expression and release of inflammatory cytokines induced by lps-gingivalis in THP-1 and MO3.13 cell lines. Cytokine 2017, 96, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Giacoppo, S.; Thangavelu, S.R.; Diomede, F.; Bramanti, P.; Conti, P.; Trubiani, O.; Mazzon, E. Anti-inflammatory effects of hypoxia-preconditioned human periodontal ligament cell secretome in an experimental model of multiple sclerosis: A key role of IL-37. FASEB J. 2017, 31, 5592–5608. [Google Scholar] [CrossRef] [PubMed]
- Galuppo, M.; Giacoppo, S.; De Nicola, G.R.; Iori, R.; Navarra, M.; Lombardo, G.E.; Bramanti, P.; Mazzon, E. Antiinflammatory activity of glucomoringin isothiocyanate in a mouse model of experimental autoimmune encephalomyelitis. Fitoterapia 2014, 95, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Giacoppo, S.; Iori, R.; Bramanti, P.; Mazzon, E. Topical moringin-cream relieves neuropathic pain by suppression of inflammatory pathway and voltage-gated ion channels in murine model of multiple sclerosis. Mol. Pain 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Giacoppo, S.; Rajan, T.S.; Iori, R.; Rollin, P.; Bramanti, P.; Mazzon, E. The alpha-cyclodextrin complex of the moringa isothiocyanate suppresses lipopolysaccharide-induced inflammation in raw 264.7 macrophage cells through AKT and p38 inhibition. Inflamm. Res. 2017, 66, 487–503. [Google Scholar] [CrossRef] [PubMed]
- Romeo, L.; Diomede, F.; Gugliandolo, A.; Scionti, D.; Lo Giudice, F.; Lanza Cariccio, V.; Iori, R.; Bramanti, P.; Trubiani, O.; Mazzon, E. Moringin induces neural differentiation in the stem cell of the human periodontal ligament. Sci. Rep. 2018, 8, 9153. [Google Scholar] [CrossRef] [PubMed]
- Rajan, T.S.; Giacoppo, S.; Iori, R.; De Nicola, G.R.; Grassi, G.; Pollastro, F.; Bramanti, P.; Mazzon, E. Anti-inflammatory and antioxidant effects of a combination of cannabidiol and moringin in lps-stimulated macrophages. Fitoterapia 2016, 112, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Libro, R.; Scionti, D.; Diomede, F.; Marchisio, M.; Grassi, G.; Pollastro, F.; Piattelli, A.; Bramanti, P.; Mazzon, E.; Trubiani, O. Cannabidiol modulates the immunophenotype and inhibits the activation of the inflammasome in human gingival mesenchymal stem cells. Front. Physiol. 2016, 7, 559. [Google Scholar] [CrossRef] [PubMed]
- Rajan, T.S.; Giacoppo, S.; Diomede, F.; Ballerini, P.; Paolantonio, M.; Marchisio, M.; Piattelli, A.; Bramanti, P.; Mazzon, E.; Trubiani, O. The secretome of periodontal ligament stem cells from MS patients protects against EAE. Sci. Rep. 2016, 6, 38743. [Google Scholar] [CrossRef] [PubMed]
- Cianci, E.; Recchiuti, A.; Trubiani, O.; Diomede, F.; Marchisio, M.; Miscia, S.; Colas, R.A.; Dalli, J.; Serhan, C.N.; Romano, M. Human periodontal stem cells release specialized proresolving mediators and carry immunomodulatory and prohealing properties regulated by lipoxins. Stem Cells Transl. Med. 2016, 5, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Diomede, F.; Zini, N.; Gatta, V.; Fulle, S.; Merciaro, I.; D’Aurora, M.; La Rovere, R.M.L.; Traini, T.; Pizzicannella, J.; Ballerini, P.; et al. Human periodontal ligament stem cells cultured onto cortico-cancellous scaffold drive bone regenerative process. Eur. Cells Mater. 2016, 32, 181–201. [Google Scholar] [CrossRef]
- Rajan, T.S.; De Nicola, G.R.; Iori, R.; Rollin, P.; Bramanti, P.; Mazzon, E. Anticancer activity of glucomoringin isothiocyanate in human malignant astrocytoma cells. Fitoterapia 2016, 110, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Deveraux, Q.L.; Reed, J.C. Iap family proteins—Suppressors of apoptosis. Genes Dev. 1999, 13, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Verhagen, A.M.; Ekert, P.G.; Pakusch, M.; Silke, J.; Connolly, L.M.; Reid, G.E.; Moritz, R.L.; Simpson, R.J.; Vaux, D.L. Identification of diablo, a mammalian protein that promotes apoptosis by binding to and antagonizing iap proteins. Cell 2000, 102, 43–53. [Google Scholar] [CrossRef]
- Kim, J.W.; Choi, E.J.; Joe, C.O. Activation of death-inducing signaling complex (disc) by pro-apoptotic c-terminal fragment of rip. Oncogene 2000, 19, 4491–4499. [Google Scholar] [CrossRef] [PubMed]
- Wolff, S.; Erster, S.; Palacios, G.; Moll, U.M. P53’s mitochondrial translocation and momp action is independent of puma and bax and severely disrupts mitochondrial membrane integrity. Cell Res. 2008, 18, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Beurel, E.; Jope, R.S. The paradoxical pro- and anti-apoptotic actions of gsk3 in the intrinsic and extrinsic apoptosis signaling pathways. Prog. Neurobiol. 2006, 79, 173–189. [Google Scholar] [CrossRef] [PubMed]
- Giacoppo, S.; Pollastro, F.; Grassi, G.; Bramanti, P.; Mazzon, E. Target regulation of PI3K/AKT/mTOR pathway by cannabidiol in treatment of experimental multiple sclerosis. Fitoterapia 2017, 116, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Ozaita, A.; Puighermanal, E.; Maldonado, R. Regulation of PI3K/AKT/GSK-3 pathway by cannabinoids in the brain. J. Neurochem. 2007, 102, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Brunet, A.; Datta, S.R.; Greenberg, M.E. Transcription-dependent and -independent control of neuronal survival by the pi3k-akt signaling pathway. Cur. Opin. Neurobiol. 2001, 11, 297–305. [Google Scholar] [CrossRef]
- Dudek, H.; Datta, S.R.; Franke, T.F.; Birnbaum, M.J.; Yao, R.; Cooper, G.M.; Segal, R.A.; Kaplan, D.R.; Greenberg, M.E. Regulation of neuronal survival by the serine-threonine protein kinase AKT. Science 1997, 275, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Kuruvilla, R.; Ye, H.; Ginty, D.D. Spatially and functionally distinct roles of the pi3-k effector pathway during ngf signaling in sympathetic neurons. Neuron 2000, 27, 499–512. [Google Scholar] [CrossRef]
- Lim, J.Y.; Park, S.I.; Oh, J.H.; Kim, S.M.; Jeong, C.H.; Jun, J.A.; Lee, K.S.; Oh, W.; Lee, J.K.; Jeun, S.S. Brain-derived neurotrophic factor stimulates the neural differentiation of human umbilical cord blood-derived mesenchymal stem cells and survival of differentiated cells through MAPK/ERK and PI3K/AKT-dependent signaling pathways. J. Neurosci. Res. 2008, 86, 2168–2178. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, T.; Ozawa, A.; Katsuura, M.; Nomura, S.; Satoh, Y. Effects of muscarinic acetylcholine receptor stimulation on the differentiation of mouse induced pluripotent stem cells into neural progenitor cells. Clin. Exp. Pharmacol. Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Van Inzen, W.G.; Peppelenbosch, M.P.; Van Den Brand, M.W.; Tertoolen, L.G.; De Laat, S.W. Neuronal differentiation of embryonic stem cells. Biochim. Biophys. Acta 1996, 1312, 21–26. [Google Scholar] [CrossRef]
- Zemelko, V.I.; Kozhucharova, I.V.; Kovaleva, Z.V.; Domnina, A.P.; Pugovkina, N.A.; Fridlyanskaya, I.I.; Puzanov, M.V.; Anisimov, S.V.; Grinchuk, T.M.; Nikolsky, N.N. Brain-derived neurotrofic factor (BDNF) secretion of human mesenchymal stem cells isolated from bone marrow, endometrium and adipose tissue. Cell Tissue Biol. 2014, 8, 283–291. [Google Scholar] [CrossRef]
- Rajan, T.S.; Scionti, D.; Diomede, F.; Grassi, G.; Pollastro, F.; Piattelli, A.; Cocco, L.; Bramanti, P.; Mazzon, E.; Trubiani, O. Gingival stromal cells as an in vitro model: Cannabidiol modulates genes linked with amyotrophic lateral sclerosis. J. Cell Biochem. 2017, 118, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Taglialatela-Scafati, O.; Pagani, A.; Scala, F.; De Petrocellis, L.; Di Marzo, V.; Grassi, G.; Appendino, G. Cannabimovone, a cannabinoid with a rearranged terpenoid skeleton from hemp. Eur. J. Org. Chem. 2010, 2010, 2067–2072. [Google Scholar] [CrossRef]
- Muller, C.; van Loon, J.; Ruschioni, S.; De Nicola, G.R.; Olsen, C.E.; Iori, R.; Agerbirk, N. Taste detection of the non-volatile isothiocyanate moringin results in deterrence to glucosinolate-adapted insect larvae. Phytochemistry 2015, 118, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Diomede, F.; Merciaro, I.; Martinotti, S.; Cavalcanti, M.F.; Caputi, S.; Mazzon, E.; Trubiani, O. Mir-2861 is involved in osteogenic commitment of human periodontal ligament stem cells grown onto 3D scaffold. J. Biol. Regul. Homeost. Agents 2016, 30, 1009–1018. [Google Scholar] [PubMed]
- Cavalcanti, M.F.; Maria, D.A.; de Isla, N.; Leal-Junior, E.C.; Joensen, J.; Bjordal, J.M.; Lopes-Martins, R.A.; Diomede, F.; Trubiani, O.; Frigo, L. Evaluation of the proliferative effects induced by low-level laser therapy in bone marrow stem cell culture. Photomed. Laser Surg. 2015, 33, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Diomede, F.; Zingariello, M.; Cavalcanti, M.; Merciaro, I.; Pizzicannella, J.; De Isla, N.; Caputi, S.; Ballerini, P.; Trubiani, O. MyD88/ERK/NFkB pathways and pro-inflammatory cytokines release in periodontal ligament stem cells stimulated by porphyromonas gingivalis. Eur. J. Histochem. 2017, 61, 2791. [Google Scholar] [CrossRef] [PubMed]
- Orciani, M.; Trubiani, O.; Guarnieri, S.; Ferrero, E.; Di Primio, R. Cd38 is constitutively expressed in the nucleus of human hematopoietic cells. J. Cell. Biochem. 2008, 105, 905–912. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).