Genome-Wide Analysis of Watermelon HSP20s and Their Expression Profiles and Subcellular Locations under Stresses
Abstract
:1. Introduction
2. Results
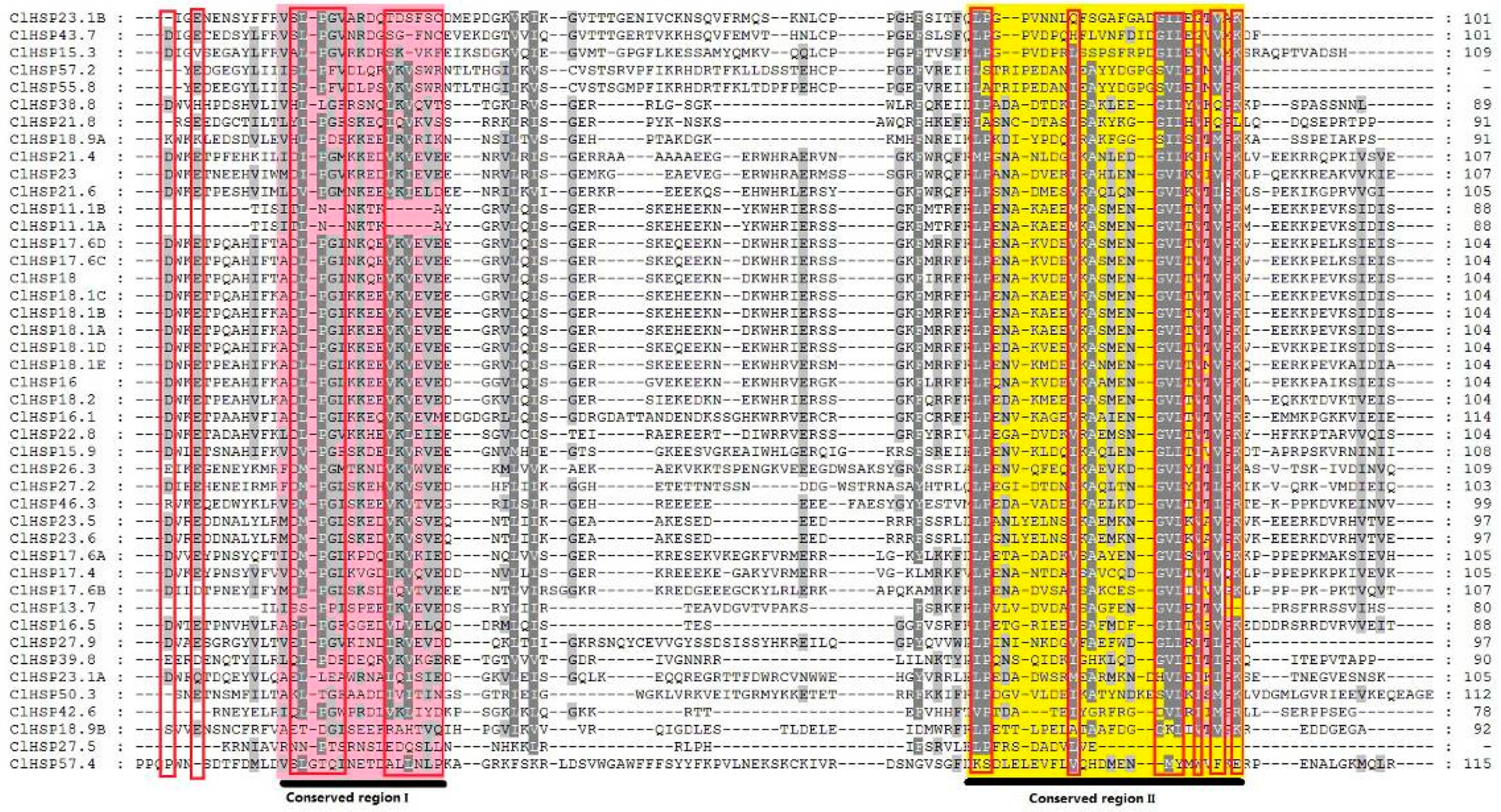
2.1. Identification of HSP20 Genes in Watermelon and Cucumber
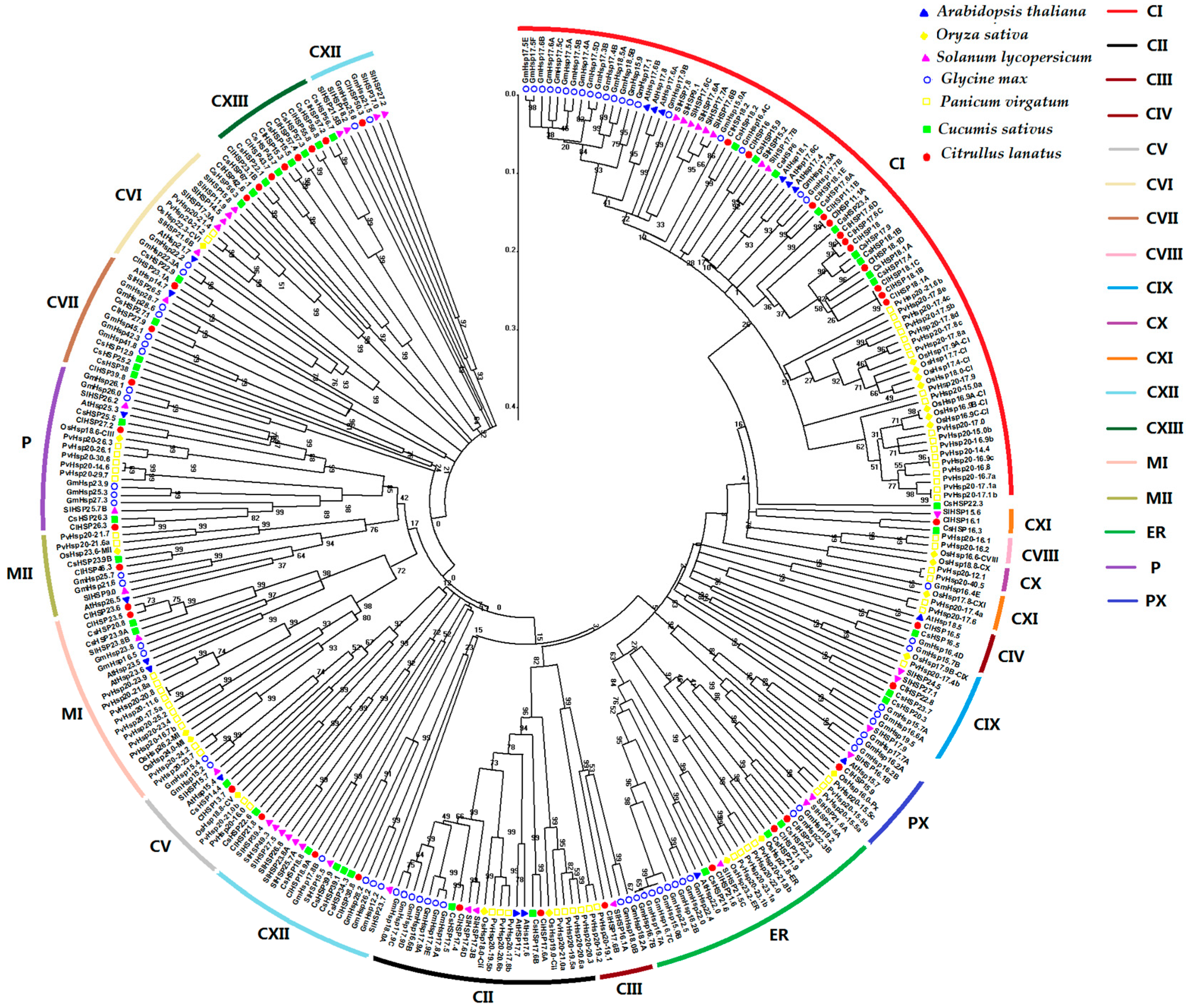
2.2. Phylogenetic Relationship of Plant HSP20 Members
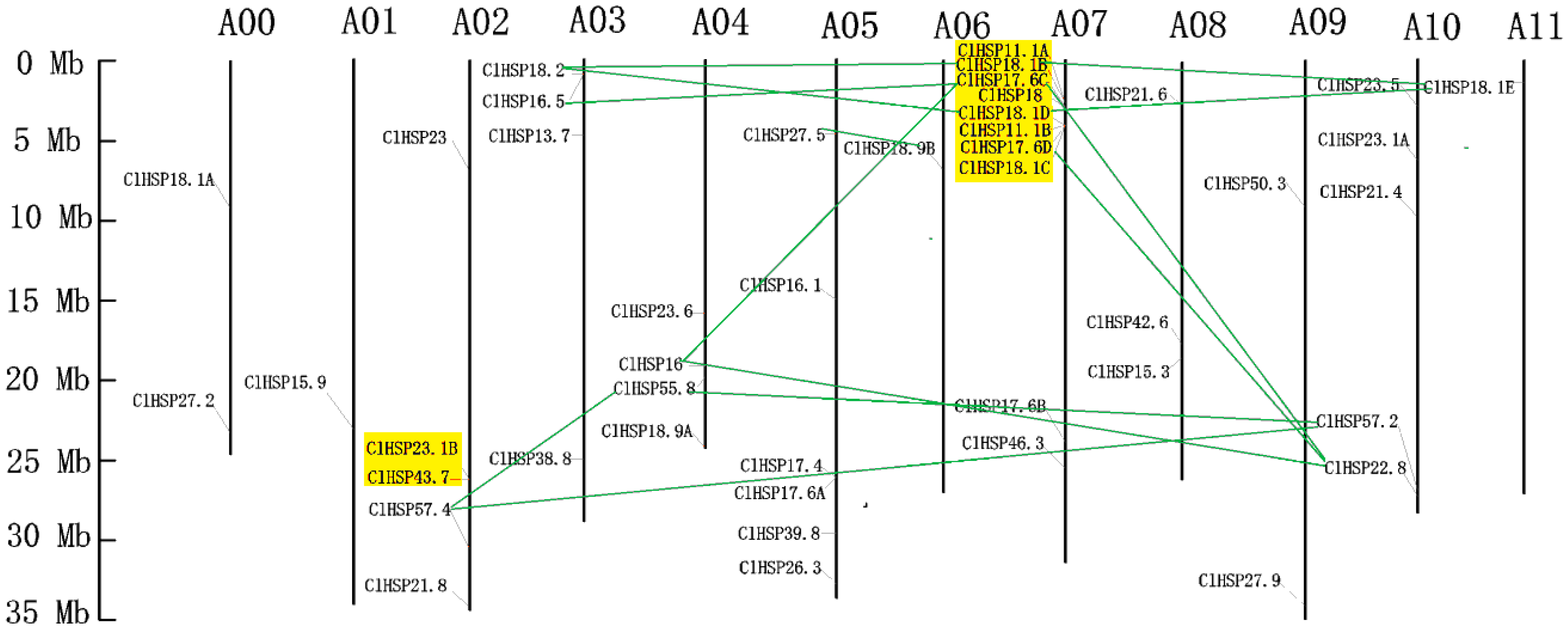
2.3. Genomic Distribution and Evolutionary Analysis of Watermelon HSP20s
2.4. Analysis of Putative cis-Acting Elements in ClHSP20 Promoters
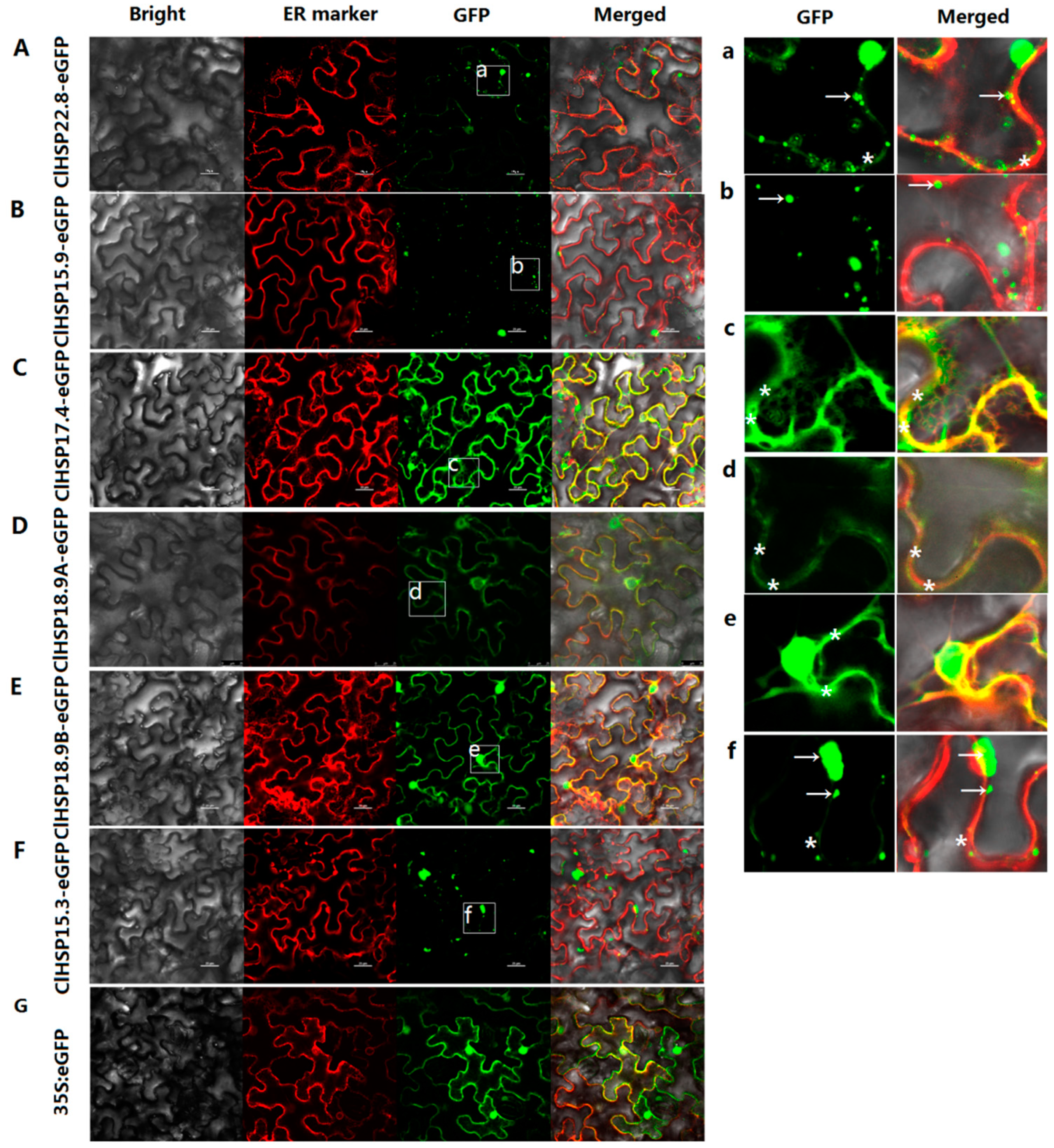
2.5. Subcellular Localizations of HSP20 Proteins in Watermelon
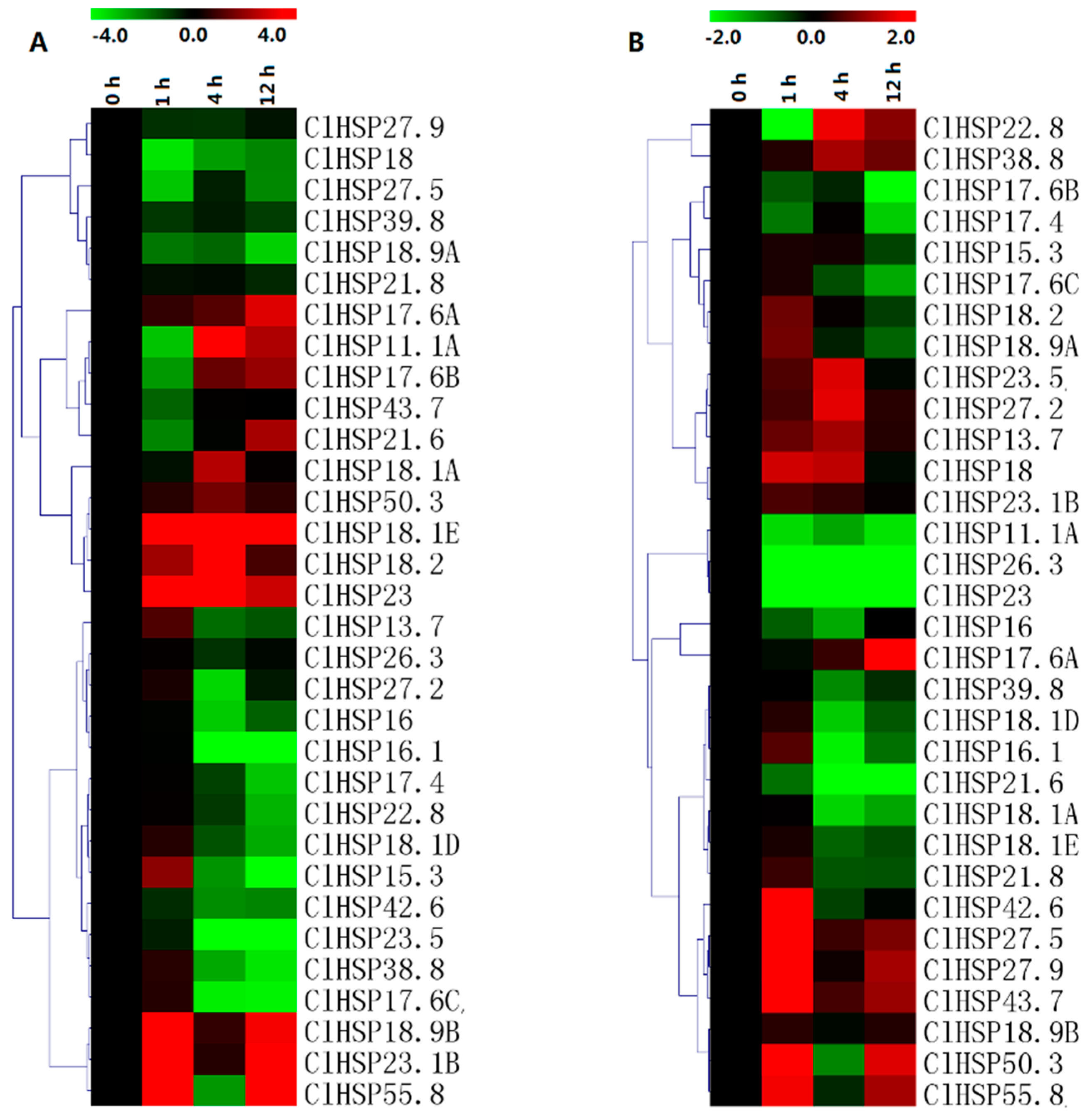
2.6. Expression Analyses of ClHSP20 Genes in Response to Abscisic Acid and Melatonin
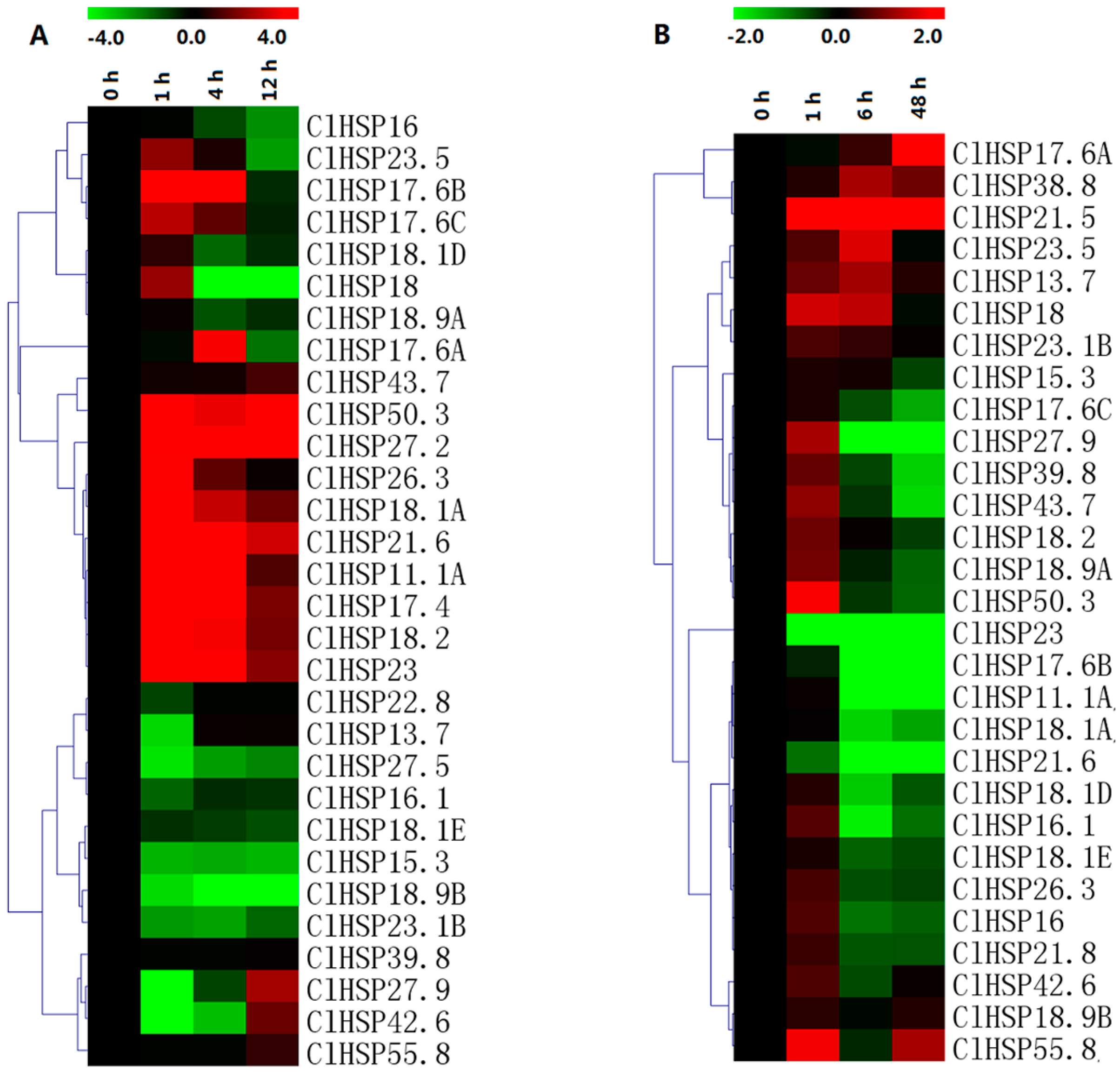
2.7. Expression Analyses of ClHSP20 Genes in Response to Heat and CGMMV
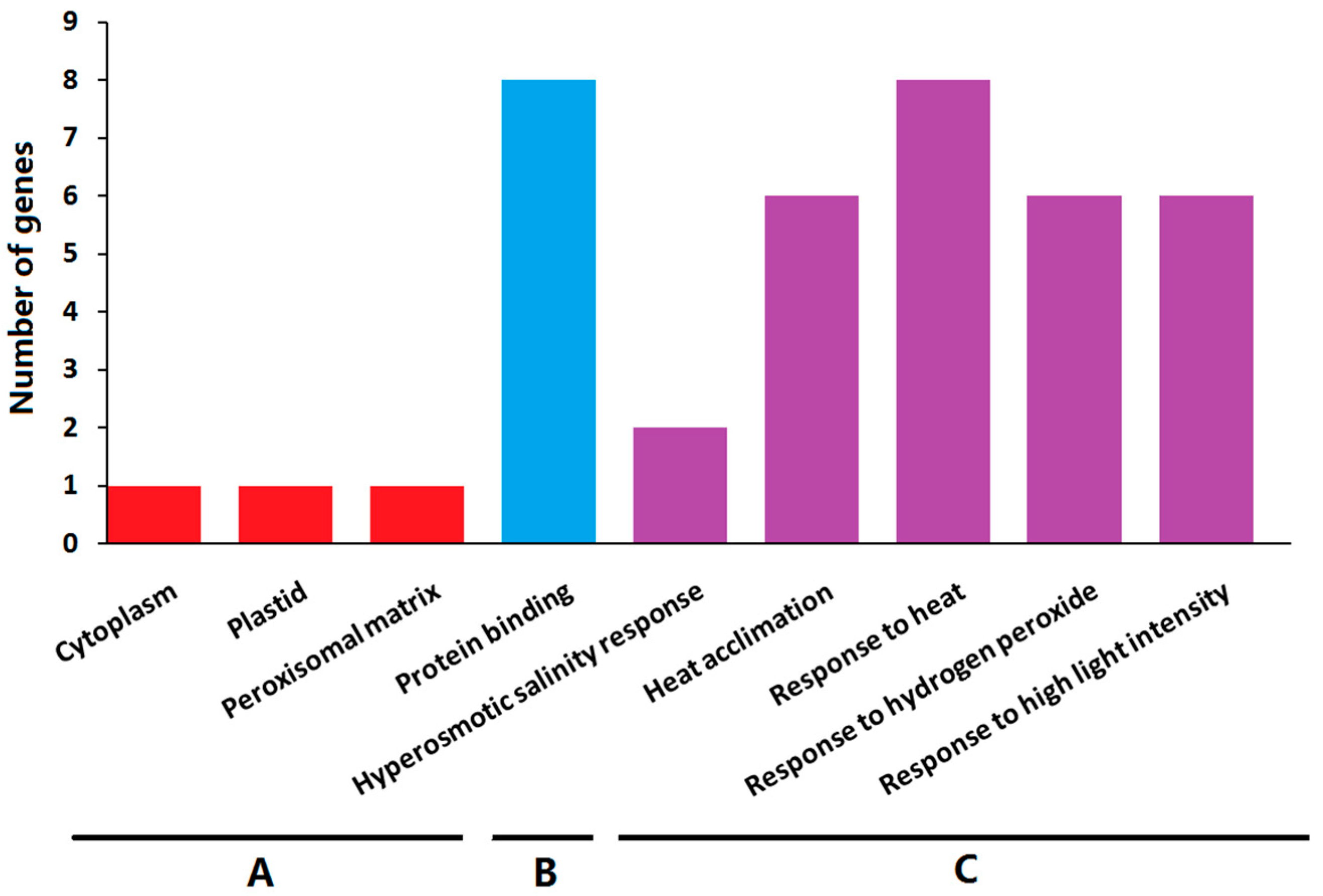
2.8. Gene Ontology Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Identification of HSP20s in Watermelon and Cucumber
4.2. Phylogenetic Analysis, Gene Structure Construction, and Motif Analysis
4.3. Chromosomal Localization, Gene Duplication, and Evolutionary Analysis
4.4. Analysis of cis-Acting Elements in HSP20 Putative Promoter Regions in Watermelon
4.5. Watermelon Plant Growth and Treatments
4.6. RNA Isolation and qRT–PCR
4.7. Subcellular Localization Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Asea, A.A.A.; Kaur, P.; Calderwood, S.K. Heat Shock Proteins and Plants; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Sarkar, N.K.; Kim, Y.K.; Grover, A. Rice sHSP genes: Genomic organization and expression profiling under stress and development. BMC Genom. 2009, 10, 393. [Google Scholar] [CrossRef] [PubMed]
- Charng, Y.Y.; Liu, H.C.; Liu, N.Y.; Hsu, F.C.; Ko, S.S. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 2006, 140, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Matsuba, S.; Funatsuki, H.; Kawaguchi, K.; Saruyama, H.; Tanida, M.; Sato, Y. Overexpression of a small heat shock protein, sHSP17.7, confers both heat tolerance and UV-B resistance to rice plants. Mol. Breed. 2004, 13, 165–175. [Google Scholar] [CrossRef]
- Sun, L.P.; Liu, Y.; Kong, X.P.; Zhang, D.; Pan, J.W.; Zhou, Y.; Wang, L.; Li, D.Q.; Yang, X.H. ZmHSP16.9, a cytosolic class I small heat shock protein in maize (Zea mays), confers heat tolerance in transgenic tobacco. Plant Cell Rep. 2012, 31, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Khurana, N.; Chauhan, H.; Khurana, P. Wheat chloroplast targeted sHSP26 promoter confers heat and abiotic stress inducible expression in transgenic Arabidopsis plants. PLoS ONE 2013, 8, e54418. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, S.N.; Heckathorn, S.; Luthe, D.S. Analysis of gene sequences indicates that quantity not quality of chloroplast small HSPs improves thermotolerance in C4 and CAM plants. Plant Cell Rep. 2012, 31, 1943–1957. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Liu, C.F.; Liu, A.L.; Zou, D.; Chen, X.B. Overexpression of OsHsp17.0 and OsHsp23.7 enhances drought and salt tolerance in rice. J. Plant Physiol. 2012, 169, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Petla, B.P.; Kamble, N.U.; Singh, A.; Rao, V.; Salvi, P.; Ghosh, S.; Majee, M. Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress. Front. Plant Sci. 2015, 6, 713. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Khurana, N.; Nijhavan, A.; Khurana, J.P.; Khurana, P. The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 2012, 35, 1912–1931. [Google Scholar] [CrossRef] [PubMed]
- Mu, C.J.; Zhang, S.J.; Yu, G.Z.; Chen, N.; Li, X.F.; Liu, H. Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS ONE 2013, 8, e82264. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ji, L.; Jia, Z.; Yang, X.; Meng, Q.; Guo, S. Constitutive expression of CaHsp22.5 enhances chilling tolerance in transgenic tobacco by promoting the activity of antioxidative enzymes. Funct. Plant Biol. 2018, 45, 575–585. [Google Scholar] [CrossRef]
- Sun, X.; Sun, C.; Li, Z.; Hu, Q.; Han, L.; Luo, H. AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant Cell Environ. 2016, 39, 1320–1337. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Seo, Y.S. Heat shock proteins: A review of the molecular chaperones for plant immunity. Plant Pathol. J. 2015, 31, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Mandadi, K.K.; Scholthof, K.B.G. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef] [PubMed]
- Maimbo, M.; Ohnishi, K.; Hikichi, Y.; Yoshioka, H.; Kiba, A. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol. 2007, 145, 1588–1599. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, G.; Lukasik, E.; Van Den Burg, H.A.; Vossen, J.H.; Cornelissen, B.J.C.; Takken, F.L.W. The small heat shock protein 20 RSI2 interacts with and is required for stability and function of tomato resistance protein I-2. Plant J. 2010, 63, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.Q.; Zhu, B.Z.; Luo, Y.B.; Fu, D.Q. Unraveling the protein network of tomato fruit in response to necrotrophic phytopathogenic Rhizopus nigricans. PLoS ONE 2013, 8, e73034. [Google Scholar] [CrossRef]
- Yogendra, K.N.; Kumar, A.; Sarkar, K.; Li, Y.L.; Pushpa, D.; Mosa, K.A.; Duggavathi, R.; Kushalappa, A.C. Transcription factor StWRKY1 regulates phenylpropanoid metabolites conferring late blight resistance in potato. J. Exp. Bot. 2015, 66, 7377–7389. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Pedersen, C.; Schultz-Larsen, T.; Kawaaitaal, M.; Jørgensen, H.J.L.; Thrdal-Christensen, H. The barley powdery mildew candidate secreted effector protein csep0105 inhibits the chaperone activity of a small heat shock protein. Plant Physiol. 2015, 168, 321–333. [Google Scholar] [CrossRef]
- Lopes-Caitar, V.S.; de Carvalho, M.C.; Darben, L.M.; Kuwahara, M.K.; Nepomuceno, A.L.; Dias, W.P.; Abdelnoor, R.V.; Marcelino-Guimarães, F.C. Genome-wide analysis of the Hsp20 gene family in soybean: Comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genom. 2013, 14, 577. [Google Scholar] [CrossRef]
- Bricchi, I.; Bertea, C.M.; Occhipinti, A.; Paponov, I.A.; Maffei, M.E. Dynamics of membrane potential variation and gene expression induced by Spodoptera littoralis, Myzus persicae, and Pseudomonas syringae in Arabidopsis. PLoS ONE 2012, 7, e46673. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xiang, C.Y.; Yang, J.; Chen, J.P.; Zhang, H.M. Interaction of HSP20 with a viral RdRp changes its sub-cellular localization and distribution pattern in plants. Sci. Rep. UK 2015, 5, 14016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, L.; Du, Z.; Qin, M.; Wang, P.; Lan, H.; Niu, X.; Jia, D.; Xie, L.; Lin, Q.; Xie, L.; et al. Pc4, a putative movement protein of Rice stripe virus, interacts with a type I DnaJ protein and a small Hsp of rice. Virus Genes 2009, 38, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Scharf, K.D.; Siddique, M.; Vierling, E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (ACD proteins). Cell Stress Chaperones 2001, 6, 225–237. [Google Scholar] [CrossRef]
- Yu, J.H.; Cheng, Y.; Feng, K.; Ruan, M.Y.; Ye, Q.J.; Wang, R.Q.; Li, Z.M.; Zhou, G.Z.; Yao, Z.P.; Yang, Y.J.; et al. Genome-wide identification and expression profiling of tomato HSP20 gene family in response to biotic and abiotic stresses. Front Plant Sci. 2016, 7, 1215. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.; Kaur, A.; Gupta, O.P.; Sharma, I.; Sharma, P. Identification of HSP20 gene family in wheat and barley and their differential expression profiling under heat stress. Appl. Biochem. Biotechnol. 2015, 175, 2427–2446. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.H.; Lu, J.P.; Zhai, Y.F.; Wang, H.; Gong, Z.H.; Wang, S.B.; Lu, M.H. Genome-wide analysis of the CaHsp20 gene family in pepper: Comprehensive sequence and expression profile analysis under heat stress. Front Plant Sci. 2015, 6, 806. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhang, A.; Chen, J.; He, X.; Xu, B.; Xie, G.; Miao, Z.; Zhang, X.; Huang, L. Genome-Wide Analysis of the PvHsp20 Family in Switchgrass: Motif, Genomic Organization, and Identification of Stress or Developmental-Related Hsp20s. Front. Plant Sci. 2017, 8, 1024. [Google Scholar] [CrossRef]
- De Jong, W.W.; Caspers, G.J.; Leunissen, J.A. Genealogy of the acrystallin–small heat-shock protein superfamily. Int. J. Biol. Macromol. 1998, 22, 151–162. [Google Scholar] [CrossRef]
- Bondino, H.G.; Valle, E.M.; Ten, H.A. Evolution and functional diversification of the small heat shock protein/α-crystallin family in higher plants. Planta 2012, 235, 1299–1313. [Google Scholar] [CrossRef]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, P.; Li, Y.; Liu, Y.; Yang, N.; Yu, J.; Ma, X.; Sun, S.; Xia, R.; Liu, X.; et al. An overlooked paleo-tetraploidization in Cucurbitaceae. Mol. Biol. Evol. 2018, 35, 16. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.R.; Aevermann, B.D.; Sanders-Reed, Z. Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperones 2008, 13, 127–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, Y.; Chen, J.; Xie, W.; Wang, L.; Zhang, Q. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol. Biol. 2009, 70, 341–357. [Google Scholar] [CrossRef]
- Yu, Y.J.; Liang, Y.; Lv, M.L.; Wu, J.; Lu, G.; Cao, J.S. Genome-wide identification and characterization of polygalacturonase genes in Cucumis sativus and Citrullus lanatus. Plant Physiol. Biochem. 2014, 74, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Huang, X.; Bao, Y.; Wang, B.; Zeng, H.; Cheng, W.; Tang, M.; Li, Y.; Ren, J.; Sun, Y. Genome-wide identification of SAUR genes in watermelon (Citrullus lanatus). Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2017, 23, 619. [Google Scholar] [CrossRef]
- He, Y.; Liu, X.; Zou, T.; Pan, C.; Qin, L.; Chen, L.; Lu, G. Genome-Wide Identification of Two-Component System Genes in Cucurbitaceae Crops and Expression Profiling Analyses in Cucumber. Front. Plant Sci. 2016, 7, 899. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef]
- Sun, W.; Van Montagu, M.; Verbruggen, N. Small heat shock proteins and stress tolerance in plants. Biochim. Biophys. Acta 2002, 1577, 1–9. [Google Scholar] [CrossRef]
- Dombrovsky, A.; Tran-Nguyen, L.T.T.; Jones, R.A.C. Cucumber green mottle mosaic virus: Rapidly increasing global distribution, etiology, epidemiology, and management. Annu. Rev. Phytopathol. 2017, 55, 231–256. [Google Scholar] [CrossRef] [PubMed]
- Whitham, S.A.; Quan, S.; Chang, H.S.; Cooper, B.; Zhu, T.; Wang, X.; Hou, Y. Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 2003, 33, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; An, M.; Xia, Z.; Bai, X.; Wu, Y. Transcriptome analysis of watermelon (Citrullus lanatus) fruits in response to Cucumber green mottle mosaic virus (CGMMV) infection. Sci. Rep. UK 2017, 7, 16747. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernández-Ruiz, J. Functions of melatonin in plants: A review. J. Pineal Res. 2015, 59, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.Y.; Zhang, Y.; Xu, Y.P.; Qi, Z.Y.; Li, M.Q.; Ahammed, G.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Reiter, R.J.; Yu, J.Q.; Zhou, J. HsfA1a upregulates melatonin biosynthesis to confer cadmium tolerance in tomato plants. J. Pineal Res. 2017, 62, e12387. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Cai, S.; Zhang, Y.; Wang, Y.; Ahammed, G.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Reiter, R.J.; Zhou, J. Melatonin enhances thermotolerance by promoting cellular protein protection in tomato plants. J. Pineal Res. 2016, 61, 457–469. [Google Scholar] [CrossRef]
- Basha, E.; O Neill, H.; Vierling, E. Small heat shock proteins and a-crystallins: Dynamic proteins with flexible functions. Trends Biochem. Sci. 2012, 37, 106–117. [Google Scholar] [CrossRef]
- Piotrowska, J.; Hansen, S.J.; Park, N.; Jamka, K.; Sarnow, P.; Gustin, K.E. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 2010, 84, 3654–3665. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Hu, L.F.; Liu, S.Q. Genome-wide analysis of the MADS-box gene family in cucumber. Genet. Mol. Res. 2013, 12, 4317–4331. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Feng, L.; Zhu, Y.; Li, Y.; Yan, H.; Xiang, Y. Comparative genomic analysis of the WRKY III gene family in populus, grape, Arabidopsis, and rice. Biol. Direct 2015, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chang, J.; Zheng, J.; Dong, Y.; Liu, Q.; Yang, X.; Wei, C.; Zhang, Y.; Ma, J.; Zhang, X. Local melatonin application induces cold tolerance in distant organs of Citrullus lanatus L. via long distance transport. Sci. Rep. UK 2017, 7, 40858. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O.; Rivas, S.; Mestre, P.; Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003, 33, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Yuan, J.; Gao, L.Y.; Zhao, S.; Jiang, W.; Huang, Y.; Bie, Z. Identification of suitable reference genes for gene expression normalization in qRT-PCR analysis in watermelon. PLoS ONE 2014, 9, e90612. [Google Scholar] [CrossRef] [PubMed]



| Species | The Number of HSP20s | Reference |
|---|---|---|
| Arabidopsis thaliana | 19 | [25] |
| Oryzasativa | 23 | [2] |
| Solanum lycopersicum | 42 | [26] |
| Triticum aestivum | 27 | [27] |
| Hordeum vulgare | 13 | [27] |
| Capsicum annuum | 35 | [28] |
| Glycine max | 51 | [21] |
| Panicum virgatum | 63 | [29] |
| Cucumis sativus | 45 | This work |
| Citrullus lanatus | 44 | This work |
| Duplicated Gene Pairs | Ks | Ka | Ka/Ks | Duplicated Type | Purify Selection | Time * (MYA) |
|---|---|---|---|---|---|---|
| ClHSP22.8/ClHSP17.6D | 19.7650 | 0.5612 | 0.0284 | Segmental | Yes | 1520.38 |
| ClHSP22.8/ClHSP17.6C | 19.7777 | 0.5611 | 0.0284 | Segmental | Yes | 1521.36 |
| ClHSP22.8/ClHSP16 | 12.8876 | 0.4231 | 0.0328 | Segmental | Yes | 991.35 |
| ClHSP18.2/ClHSP18.1B | 2.4385 | 0.2044 | 0.0838 | Segmental | Yes | 187.58 |
| ClHSP18.2/ClHSP18.1D | 1.6708 | 0.1864 | 0.1115 | Segmental | Yes | 128.52 |
| ClHSP18.1E/ClHSP18.1D | 2.4264 | 0.1057 | 0.0435 | Segmental | Yes | 186.65 |
| ClHSP18.1E/ClHSP18.1B | 2.5427 | 0.1239 | 0.0487 | Segmental | Yes | 195.59 |
| ClHSP18.9B/ClHSP27.5 | 3.4679 | 0.2528 | 0.0729 | Segmental | Yes | 266.76 |
| ClHSP55.8/ClHSP57.2 | 1.5394 | 0.1181 | 0.0767 | Segmental | Yes | 118.42 |
| ClHSP55.8/ClHSP57.4 | 3.2918 | 0.1509 | 0.0458 | Segmental | Yes | 253.22 |
| ClHSP57.2/ClHSP57.4 | 3.0423 | 0.1941 | 0.0638 | Segmental | Yes | 234.02 |
| ClHSP16/ClHSP17.6C | 9.3352 | 0.1704 | 0.0183 | Segmental | Yes | 718.09 |
| ClHSP16.5/ClHSP17.6C | 2.7813 | 0.5545 | 0.1994 | Segmental | Yes | 213.95 |
| ClHSP11.1A/ClHSP18.1B | 0.3202 | 0.2266 | 0.7076 | Tandem | Yes | 24.63 |
| ClHSP11.1A/ClHSP17.6C | 1.1953 | 0.2548 | 0.2131 | Tandem | Yes | 91.95 |
| ClHSP11.1A/ClHSP18 | 1.2543 | 0.2635 | 0.2101 | Tandem | Yes | 96.48 |
| ClHSP18.1B/ClHSP17.6C | 0.8027 | 0.0525 | 0.0653 | Tandem | Yes | 61.75 |
| ClHSP18.1B/ClHSP18 | 0.8097 | 0.0517 | 0.0639 | Tandem | Yes | 62.28 |
| ClHSP17.6C/ClHSP18 | 0.0865 | 0.0152 | 0.1756 | Tandem | Yes | 6.65 |
| ClHSP18.1D/ClHSP11.1B | 0.7433 | 0.2644 | 0.3557 | Tandem | Yes | 57.18 |
| ClHSP18.1D/ClHSP18.1C | 0.4589 | 0.0427 | 0.0931 | Tandem | Yes | 35.30 |
| ClHSP18.1D/ClHSP17.6D | 0.6050 | 0.0565 | 0.0934 | Tandem | Yes | 46.54 |
| ClHSP11.1B/ClHSP18.1C | 0.3202 | 0.2266 | 0.7076 | Tandem | Yes | 24.63 |
| ClHSP11.1B/ClHSP17.6D | 1.1953 | 0.2548 | 0.2131 | Tandem | Yes | 91.95 |
| ClHSP18.1C/ClHSP17.6D | 0.8027 | 0.0525 | 0.0653 | Tandem | Yes | 61.75 |
| ClHSP43.7/ClHSP23.1B | 1.7552 | 0.4791 | 0.2730 | Tandem | Yes | 135.02 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Fan, M.; Sun, Y.; Li, L. Genome-Wide Analysis of Watermelon HSP20s and Their Expression Profiles and Subcellular Locations under Stresses. Int. J. Mol. Sci. 2019, 20, 12. https://doi.org/10.3390/ijms20010012
He Y, Fan M, Sun Y, Li L. Genome-Wide Analysis of Watermelon HSP20s and Their Expression Profiles and Subcellular Locations under Stresses. International Journal of Molecular Sciences. 2019; 20(1):12. https://doi.org/10.3390/ijms20010012
Chicago/Turabian StyleHe, Yanjun, Min Fan, Yuyan Sun, and Lili Li. 2019. "Genome-Wide Analysis of Watermelon HSP20s and Their Expression Profiles and Subcellular Locations under Stresses" International Journal of Molecular Sciences 20, no. 1: 12. https://doi.org/10.3390/ijms20010012





