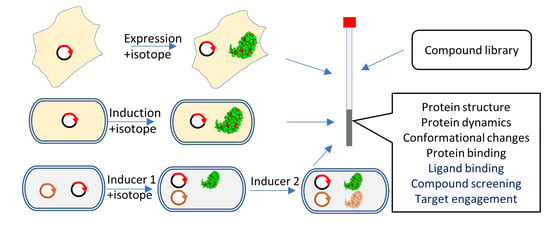
Applications of In-Cell NMR in Structural Biology and Drug Discovery
Abstract
:1. Introduction
2. In-Cell NMR
2.1. Cells Used in In-Cell NMR
2.2. Isotopic Incorporation
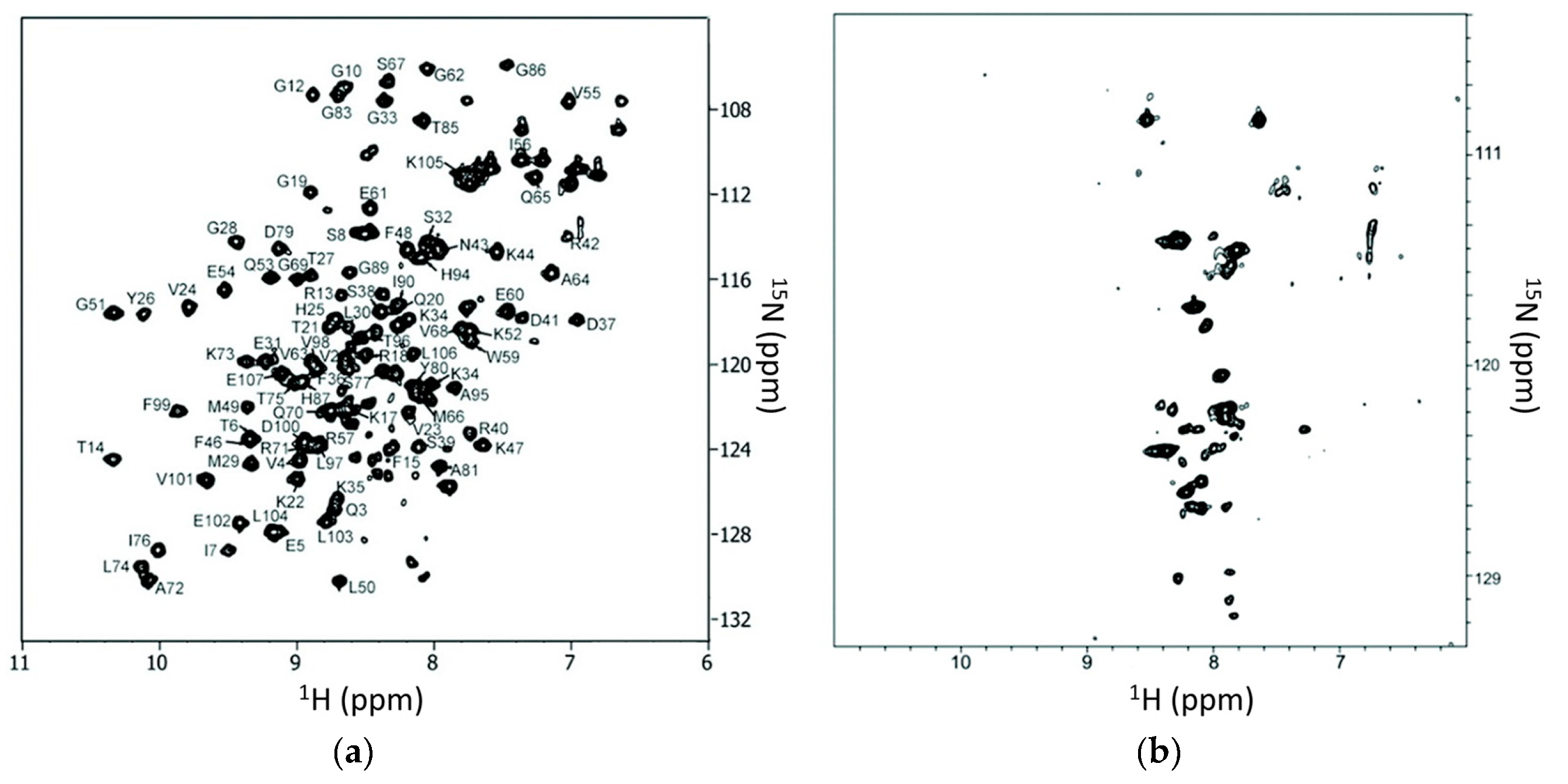
2.3. NMR Experiments for In-Cell NMR Studies
2.4. Challenges in In-Cell NMR
3. In-Cell NMR in Different Cells
3.1. In-Cell NMR in Bacterial Cells
3.2. In-Cell NMR in Yeast
3.3. In-Cell NMR in Oocytes of Xenopus laevis
3.4. In-Cell NMR in Insect Cells
3.5. In-Cell NMR in Human Cells
4. In-Cell NMR in Probing Protein–Protein Interactions
5. In-Cell NMR in Drug Discovery
5.1. Application of In-Cell NMR in Ligand Screening
5.2. Application of In-Cell NMR in Target Engagement
6. Perspective
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NMR | Nuclear magnetic resonance |
| Cryo-EM | cryogenic electron microscopy |
| STD | Saturation-transfer difference |
| SOFAST-HMBC | Band-Selective Optimized Flip Angle Short Transient-heteronuclear multiple quantum coherence) |
| Mia40 | Mitochondrial intermembrane space import and assembly protein 40 |
| NOESY | nuclear Overhauser effect spectroscopy) |
| XT-GB1 | SV40 regulatory domain-GB1 |
| Cot17 | cytochrome c oxidase copper chaperone |
| Tβ4 | thymosin β4 |
| Bcl-2 | B-cell lymphoma 2 |
| GB1 | the B domain of G protein |
| HSQC | heteronuclear single quantum coherence |
| UIM | ubiquitin interacting motif |
| hSOD1 | human copper, zinc superoxide dismutase 1 |
| SVD | Single Value Decomposition |
| ADK | adenylate kinase |
| FBDD | Fragment-based drug discovery |
| CPP | cell-penetrating peptide |
| PCS | pseudo-contact shifts |
| RDC | Residue dipolar coupling |
| PFN1 | human protein profilin 1 |
| FKBP12 | FK506 binding protein 12 |
| FRB | the 100-residue FKBP-rapamycin binding domain from the mammalian target of rapamycin |
| Pup | prokaryotic ubiquitin like protein |
| Mpa | mycobacterial protease ATPase |
| HDH | Histidinol Dehydrogensase |
| GFP | Green Fluorescence Protein |
References
- Billeter, M.; Wagner, G.; Wuthrich, K. Solution NMR structure determination of proteins revisited. J. Biomol. NMR 2008, 42, 155–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanske, J.; Sadian, Y.; Muller, C.W. The cryo-EM resolution revolution and transcription complexes. Curr. Opin. Struct. Biol. 2018, 52, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Campbell, I.D. The march of structural biology. Nat. Rev. Mol. Cell Biol. 2002, 3, 377. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. A Glimpse of Structural Biology through X-Ray Crystallography. Cell 2014, 159, 995–1014. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Rubinstein, J.L. Cryo-EM of ATP synthases. Curr. Opin. Struct. Biol. 2018, 52, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; MacKinnon, R. Cryo-EM Structure of the Open Human Ether-a-go-go-Related K(+) Channel hERG. Cell 2017, 169, 422–430.e10. [Google Scholar] [CrossRef] [PubMed]
- Madl, T.; Gabel, F.; Sattler, M. NMR and small-angle scattering-based structural analysis of protein complexes in solution. J. Struct. Biol. 2011, 173, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yong, H.Y.; Panutdaporn, N.; Liu, C.; Tang, K.; Luo, D. High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5. Nucleic Acids Res. 2015, 43, 1216–1230. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, C.; Gotze, M.; Ihling, C.H.; Piotrowski, C.; Arlt, C.; Schafer, M.; Hage, C.; Schmidt, R.; Sinz, A. A cross-linking/mass spectrometry workflow based on MS-cleavable cross-linkers and the MeroX software for studying protein structures and protein-protein interactions. Nat. Protoc. 2018, 13, 2864. [Google Scholar] [CrossRef] [PubMed]
- Pielak, G.J.; Li, C.; Miklos, A.C.; Schlesinger, A.P.; Slade, K.M.; Wang, G.F.; Zigoneanu, I.G. Protein nuclear magnetic resonance under physiological conditions. Biochemistry 2009, 48, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.J.; Marshall, C.B.; Theillet, F.X.; Binolfi, A.; Selenko, P.; Ikura, M. Real-time NMR monitoring of biological activities in complex physiological environments. Curr. Opin. Struct. Biol. 2015, 32, 39–47. [Google Scholar] [CrossRef]
- Riek, R.; Pervushin, K.; Wuthrich, K. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem. Sci. 2000, 25, 462–468. [Google Scholar] [CrossRef]
- Hyberts, S.G.; Arthanari, H.; Robson, S.A.; Wagner, G. Perspectives in magnetic resonance: NMR in the post-FFT era. J. Magn. Reson. 2014, 241, 60–73. [Google Scholar] [CrossRef] [Green Version]
- Tugarinov, V.; Choy, W.Y.; Orekhov, V.Y.; Kay, L.E. Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proc. Natl. Acad. Sci. USA 2005, 102, 622–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugarinov, V.; Kay, L.E. An isotope labeling strategy for methyl TROSY spectroscopy. J. Biomol. NMR 2004, 28, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Tugarinov, V.; Kay, L.E. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J. Am. Chem. Soc. 2003, 125, 13868–13878. [Google Scholar] [CrossRef] [PubMed]
- Barrett, P.J.; Chen, J.; Cho, M.K.; Kim, J.H.; Lu, Z.; Mathew, S.; Peng, D.; Song, Y.; Van Horn, W.D.; Zhuang, T.; et al. The quiet renaissance of protein nuclear magnetic resonance. Biochemistry 2013, 52, 1303–1320. [Google Scholar] [CrossRef]
- Kay, L.E. New Views of Functionally Dynamic Proteins by Solution NMR Spectroscopy. J. Mol. Biol. 2016, 428 Pt A, 323–331. [Google Scholar] [CrossRef]
- Gayen, S.; Li, Q.; Kang, C. Solution NMR study of the transmembrane domain of single-span membrane proteins: Opportunities and strategies. Curr. Protein Pept. Sci. 2012, 13, 585–600. [Google Scholar] [CrossRef]
- Kang, C.; Li, Q. Solution NMR study of integral membrane proteins. Curr. Opin. Chem. Biol. 2011, 15, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Sanders, C.R.; Sonnichsen, F. Solution NMR of membrane proteins: Practice and challenges. Magn. Reson. Chem. 2006, 44, S24–S40. [Google Scholar] [CrossRef] [PubMed]
- Radoicic, J.; Lu, G.J.; Opella, S.J. NMR structures of membrane proteins in phospholipid bilayers. Q. Rev. Biophys. 2014, 47, 249–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, B.; Tamm, L.K. NMR as a tool to investigate the structure, dynamics and function of membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 468–474. [Google Scholar] [CrossRef] [Green Version]
- Clark, L.; Dikiy, I.; Rosenbaum, D.M.; Gardner, K.H. On the use of Pichia pastoris for isotopic labeling of human GPCRs for NMR studies. J. Biomol. NMR 2018, 71, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Opella, S.J.; Marassi, F.M. Applications of NMR to membrane proteins. Arch. Biochem. Biophys. 2017, 628, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Oxenoid, K.; Chou, J.J. The present and future of solution NMR in investigating the structure and dynamics of channels and transporters. Curr. Opin. Struct. Biol. 2013, 23, 547–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, I.; Ueda, T.; Kofuku, Y.; Eddy, M.T.; Wuthrich, K. GPCR drug discovery: Integrating solution NMR data with crystal and cryo-EM structures. Nat. Rev. Drug Discov. 2018. [Google Scholar] [CrossRef]
- Hennig, J.; Sattler, M. The dynamic duo: Combining NMR and small angle scattering in structural biology. Protein Sci. 2014, 23, 669–682. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, Z.; Phoo, W.W.; Loh, Y.R.; Li, R.; Yang, H.Y.; Jansson, A.E.; Hill, J.; Keller, T.H.; Nacro, K.; et al. Structural Insights into the Inhibition of Zika Virus NS2B-NS3 Protease by a Small-Molecule Inhibitor. Structure 2018, 26, 555–564. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Y.; Loh, Y.R.; Phoo, W.W.; Hung, A.W.; Kang, C.; Luo, D. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 2016, 354, 1597–1600. [Google Scholar] [CrossRef]
- Phoo, W.W.; Li, Y.; Zhang, Z.; Lee, M.Y.; Loh, Y.R.; Tan, Y.B.; Ng, E.Y.; Lescar, J.; Kang, C.; Luo, D. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat. Commun. 2016, 7, 13410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantsyzov, A.B.; Shen, Y.; Lee, J.H.; Hummer, G.; Bax, A. MERA: A webserver for evaluating backbone torsion angle distributions in dynamic and disordered proteins from NMR data. J. Biomol. NMR 2015, 63, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Bax, A. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol. Biol. 2015, 1260, 17–32. [Google Scholar]
- Shen, Y.; Delaglio, F.; Cornilescu, G.; Bax, A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Bax, A. Homology modeling of larger proteins guided by chemical shifts. Nat. Methods 2015, 12, 747–750. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Bax, A. SPARTA+: A modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR 2010, 48, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, D.E.; Kulikowski, C.A.; Huang, Y.; Feng, W.; Tashiro, M.; Shimotakahara, S.; Chien, C.-Y.; Powers, R.; Montelione, G.T. Automated analysis of protein NMR assignments using methods from artificial intelligence11Edited by P. E. Wright. J. Mol. Biol. 1997, 269, 592–610. [Google Scholar] [CrossRef] [PubMed]
- Maciejewski, M.W.; Schuyler, A.D.; Gryk, M.R.; Moraru, I.I.; Romero, P.R.; Ulrich, E.L.; Eghbalnia, H.R.; Livny, M.; Delaglio, F.; Hoch, J.C. NMRbox: A Resource for Biomolecular NMR Computation. Biophys. J. 2017, 112, 1529–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.; Markley, J.L. PINE-SPARKY.2 for automated NMR-based protein structure research. Bioinformatics 2018, 34, 1586–1588. [Google Scholar] [CrossRef]
- Lemke, C.T.; Goudreau, N.; Zhao, S.; Hucke, O.; Thibeault, D.; Llinas-Brunet, M.; White, P.W. Combined X-ray, NMR, and kinetic analyses reveal uncommon binding characteristics of the hepatitis C virus NS3-NS4A protease inhibitor BI 201335. J. Biol. Chem. 2011, 286, 11434–11443. [Google Scholar] [CrossRef]
- Scott, D.E.; Coyne, A.G.; Hudson, S.A.; Abell, C. Fragment-Based Approaches in Drug Discovery and Chemical Biology. Biochemistry 2012, 51, 4990–5003. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, S.; Ng, E.Y.; Li, R.; Poulsen, A.; Hill, J.; Pobbati, A.V.; Hung, A.W.; Hong, W.; Keller, T.H.; et al. Structural and ligand-binding analysis of the YAP-binding domain of transcription factor TEAD4. Biochem. J. 2018, 475, 2043–2055. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Z.; Phoo, W.W.; Loh, Y.R.; Wang, W.; Liu, S.; Chen, M.W.; Hung, A.W.; Keller, T.H.; Luo, D.; et al. Structural Dynamics of Zika Virus NS2B-NS3 Protease Binding to Dipeptide Inhibitors. Structure 2017, 25, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- Ziarek, J.J.; Baptista, D.; Wagner, G. Recent developments in solution nuclear magnetic resonance (NMR)-based molecular biology. J. Mol. Med. 2018, 96, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.S.; Leung, E.W.; Chandrashekaran, I.R.; MacRaild, C.A. Applications of (19)F-NMR in Fragment-Based Drug Discovery. Molecules 2016, 21, 860. [Google Scholar] [CrossRef] [PubMed]
- Gee, C.T.; Arntson, K.E.; Urick, A.K.; Mishra, N.K.; Hawk, L.M.L.; Wisniewski, A.J.; Pomerantz, W.C.K. Protein-observed 19F-NMR for fragment screening, affinity quantification and druggability assessment. Nat. Protoc. 2016, 11, 1414. [Google Scholar] [CrossRef]
- Erlanson, D.A.; Fesik, S.W.; Hubbard, R.E.; Jahnke, W.; Jhoti, H. Twenty years on: The impact of fragments on drug discovery. Nat. Rev. Drug Discov. 2016, 15, 605–619. [Google Scholar] [CrossRef]
- Arntson, K.E.; Pomerantz, W.C.K. Protein-Observed Fluorine NMR: A Bioorthogonal Approach for Small Molecule Discovery. J. Med. Chem. 2016, 59, 5158–5171. [Google Scholar] [CrossRef] [PubMed]
- Yanamala, N.; Dutta, A.; Beck, B.; van Fleet, B.; Hay, K.; Yazbak, A.; Ishima, R.; Doemling, A.; Klein-Seetharaman, J. NMR-based screening of membrane protein ligands. Chem. Biol. Drug Des. 2010, 75, 237–256. [Google Scholar] [CrossRef]
- Hanzawa, H.; Takizawa, T. [NMR screening in fragment-based drug discovery]. Yakugaku Zasshi 2010, 130, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Shuker, S.B.; Hajduk, P.J.; Meadows, R.P.; Fesik, S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science 1996, 274, 1531–1534. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, C.; Otting, G. NMR studies of ligand binding. Curr. Opin. Struct. Biol. 2018, 48, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Skora, L.; Jahnke, W. 19F-NMR-Based Dual-Site Reporter Assay for the Discovery and Distinction of Catalytic and Allosteric Kinase Inhibitors. ACS Med. Chem. Lett. 2017, 8, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Mello, J.; Gomes, R.A.; Vital-Fujii, D.G.; Ferreira, G.M.; Trossini, G.H.G. Fragment-based drug discovery as alternative strategy to the drug development for neglected diseases. Chem. Biol. Drug Des. 2017, 90, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Curtis-Marof, R.; Doko, D.; Rowe, M.L.; Richards, K.L.; Williamson, R.A.; Howard, M.J. 19 F NMR spectroscopy monitors ligand binding to recombinantly fluorine-labelled b′x from human protein disulphide isomerase (hPDI). Org. Biomol. Chem. 2014, 12, 3808–3812. [Google Scholar] [CrossRef] [PubMed]
- Luchinat, E.; Banci, L. In-Cell NMR in Human Cells: Direct Protein Expression Allows Structural Studies of Protein Folding and Maturation. Acc. Chem. Res. 2018, 51, 1550–1557. [Google Scholar] [CrossRef] [PubMed]
- Serber, Z.; Dotsch, V. In-cell NMR spectroscopy. Biochemistry 2001, 40, 14317–14323. [Google Scholar] [CrossRef]
- Lippens, G.; Cahoreau, E.; Millard, P.; Charlier, C.; Lopez, J.; Hanoulle, X.; Portais, J.C. In-cell NMR: From metabolites to macromolecules. Analyst 2018, 143, 620–629. [Google Scholar] [CrossRef]
- Li, C.; Zhao, J.; Cheng, K.; Ge, Y.; Wu, Q.; Ye, Y.; Xu, G.; Zhang, Z.; Zheng, W.; Zhang, X.; et al. Magnetic Resonance Spectroscopy as a Tool for Assessing Macromolecular Structure and Function in Living Cells. Annu. Rev. Anal. Chem. 2017, 10, 157–182. [Google Scholar] [CrossRef]
- Kaplan, M.; Narasimhan, S.; de Heus, C.; Mance, D.; van Doorn, S.; Houben, K.; Popov-Čeleketić, D.; Damman, R.; Katrukha, E.A.; Jain, P.; et al. EGFR Dynamics Change during Activation in Native Membranes as Revealed by NMR. Cell 2016, 167, 1241–1251.e11. [Google Scholar] [CrossRef] [Green Version]
- Renault, M.; Tommassen-van Boxtel, R.; Bos, M.P.; Post, J.A.; Tommassen, J.; Baldus, M. Cellular solid-state nuclear magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 4863–4868. [Google Scholar] [CrossRef] [PubMed]
- Stadmiller, S.S.; Pielak, G.J. The Expanding Zoo of In-Cell Protein NMR. Biophys. J. 2018, 115, 1628–1629. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Charlton, L.M.; Lakkavaram, A.; Seagle, C.; Wang, G.; Young, G.B.; Macdonald, J.M.; Pielak, G.J. Differential dynamical effects of macromolecular crowding on an intrinsically disordered protein and a globular protein: Implications for in-cell NMR spectroscopy. J. Am. Chem. Soc. 2008, 130, 6310–6311. [Google Scholar] [CrossRef] [PubMed]
- Warnet, X.L.; Arnold, A.A.; Marcotte, I.; Warschawski, D.E. In-Cell Solid-State NMR: An Emerging Technique for the Study of Biological Membranes. Biophys. J. 2015, 109, 2461–2466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pielak, G.J.; Tian, F. Membrane proteins, magic-angle spinning, and in-cell NMR. Proc. Natl. Acad. Sci. USA 2012, 109, 4715–4716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serber, Z.; Keatinge-Clay, A.T.; Ledwidge, R.; Kelly, A.E.; Miller, S.M.; Dotsch, V. High-resolution macromolecular NMR spectroscopy inside living cells. J. Am. Chem. Soc. 2001, 123, 2446–2447. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Cefaro, C.; Ciofi-Baffoni, S.; Gallo, A.; Martinelli, M.; Sideris, D.P.; Katrakili, N.; Tokatlidis, K. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 2009, 16, 198–206. [Google Scholar] [CrossRef]
- Sakakibara, D.; Sasaki, A.; Ikeya, T.; Hamatsu, J.; Hanashima, T.; Mishima, M.; Yoshimasu, M.; Hayashi, N.; Mikawa, T.; Wälchli, M.; et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature 2009, 458, 102. [Google Scholar] [CrossRef]
- Müntener, T.; Häussinger, D.; Selenko, P.; Theillet, F.-X. In-Cell Protein Structures from 2D NMR Experiments. J. Phys. Chem. Lett. 2016, 7, 2821–2825. [Google Scholar] [CrossRef]
- Pan, B.-B.; Yang, F.; Ye, Y.; Wu, Q.; Li, C.; Huber, T.; Su, X.-C. 3D structure determination of a protein in living cells using paramagnetic NMR spectroscopy. Chem. Commun. 2016, 52, 10237–10240. [Google Scholar] [CrossRef]
- Hamatsu, J.; O’Donovan, D.; Tanaka, T.; Shirai, T.; Hourai, Y.; Mikawa, T.; Ikeya, T.; Mishima, M.; Boucher, W.; Smith, B.O.; et al. High-resolution heteronuclear multidimensional NMR of proteins in living insect cells using a baculovirus protein expression system. J. Am. Chem. Soc. 2013, 135, 1688–1691. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Barbieri, L.; Bertini, I.; Cantini, F.; Luchinat, E. In-cell NMR in E. coli to Monitor Maturation Steps of hSOD1. PLoS ONE 2011, 6, e23561. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, G.-F.; Wang, Y.; Creager-Allen, R.; Lutz, E.A.; Scronce, H.; Slade, K.M.; Ruf, R.A.S.; Mehl, R.A.; Pielak, G.J. Protein 19F NMR in Escherichia coli. J. Am. Chem. Soc. 2010, 132, 321–327. [Google Scholar] [CrossRef]
- Hough, L.E.; Dutta, K.; Sparks, S.; Temel, D.B.; Kamal, A.; Tetenbaum-Novatt, J.; Rout, M.P.; Cowburn, D. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife 2015, 4, e10027. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Barbieri, L.; Bertini, I.; Luchinat, E.; Secci, E.; Zhao, Y.; Aricescu, A.R. Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat. Chem. Biol. 2013, 9, 297. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; McLeod, S.; MacCormack, K.; Sriram, S.; Gao, N.; Breeze, A.L.; Hu, J. Real-Time Monitoring of New Delhi Metallo-β-Lactamase Activity in Living Bacterial Cells by 1H NMR Spectroscopy. Angew. Chem. Int. Ed. 2014, 53, 2130–2133. [Google Scholar] [CrossRef] [PubMed]
- Veronesi, M.; Giacomina, F.; Romeo, E.; Castellani, B.; Ottonello, G.; Lambruschini, C.; Garau, G.; Scarpelli, R.; Bandiera, T.; Piomelli, D.; et al. Fluorine nuclear magnetic resonance-based assay in living mammalian cells. Anal. Biochem. 2016, 495, 52–59. [Google Scholar] [CrossRef] [Green Version]
- Serber, Z.; Straub, W.; Corsini, L.; Nomura, A.M.; Shimba, N.; Craik, C.S.; Ortiz de Montellano, P.; Dotsch, V. Methyl groups as probes for proteins and complexes in in-cell NMR experiments. J. Am. Chem. Soc. 2004, 126, 7119–7125. [Google Scholar] [CrossRef]
- Barnes, C.O.; Pielak, G.J. In-cell protein NMR and protein leakage. Proteins 2011, 79, 347–351. [Google Scholar] [CrossRef]
- Burz, D.S.; Dutta, K.; Cowburn, D.; Shekhtman, A. Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR). Nat. Methods 2006, 3, 91–93. [Google Scholar] [CrossRef] [Green Version]
- Majumder, S.; Xue, J.; DeMott, C.M.; Reverdatto, S.; Burz, D.S.; Shekhtman, A. Probing Protein Quinary Interactions by In-Cell Nuclear Magnetic Resonance Spectroscopy. Biochemistry 2015, 54, 2727–2738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbieri, L.; Luchinat, E.; Banci, L. Protein interaction patterns in different cellular environments are revealed by in-cell NMR. Sci. Rep. 2015, 5, 14456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeMott, C.M.; Girardin, R.; Cobbert, J.; Reverdatto, S.; Burz, D.S.; McDonough, K.; Shekhtman, A. Potent Inhibitors of Mycobacterium tuberculosis Growth Identified by Using in-Cell NMR-based Screening. ACS Chem. Biol. 2018, 13, 733–741. [Google Scholar] [CrossRef]
- Xie, J.; Thapa, R.; Reverdatto, S.; Burz, D.S.; Shekhtman, A. Screening of Small Molecule Interactor Library by Using In-Cell NMR Spectroscopy (SMILI-NMR). J. Med. Chem. 2009, 52, 3516–3522. [Google Scholar] [CrossRef] [Green Version]
- Banci, L.; Bertini, I.; Cefaro, C.; Cenacchi, L.; Ciofi-Baffoni, S.; Felli, I.C.; Gallo, A.; Gonnelli, L.; Luchinat, E.; Sideris, D.; et al. Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc. Natl. Acad. Sci. USA 2010, 107, 20190–20195. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Tochio, H.; Tenno, T.; Ito, Y.; Kokubo, T.; Hiroaki, H.; Shirakawa, M. In-cell NMR spectroscopy of proteins inside Xenopus laevis oocytes. J. Biomol. NMR 2006, 36, 179–188. [Google Scholar] [CrossRef]
- Selenko, P.; Serber, Z.; Gadea, B.; Ruderman, J.; Wagner, G. Quantitative NMR analysis of the protein G B1 domain in Xenopus laevis egg extracts and intact oocytes. Proc. Natl. Acad. Sci. USA 2006, 103, 11904–11909. [Google Scholar] [CrossRef] [Green Version]
- Serber, Z.; Selenko, P.; Hansel, R.; Reckel, S.; Lohr, F.; Ferrell, J.E., Jr.; Wagner, G.; Dotsch, V. Investigating macromolecules inside cultured and injected cells by in-cell NMR spectroscopy. Nat. Protoc. 2006, 1, 2701–2709. [Google Scholar] [CrossRef]
- Selenko, P.; Frueh, D.P.; Elsaesser, S.J.; Haas, W.; Gygi, S.P.; Wagner, G. In situ observation of protein phosphorylation by high-resolution NMR spectroscopy. Nat. Struct. Mol. Biol. 2008, 15, 321–329. [Google Scholar] [CrossRef]
- Bertrand, K.; Reverdatto, S.; Burz, D.S.; Zitomer, R.; Shekhtman, A. Structure of proteins in eukaryotic compartments. J. Am. Chem. Soc. 2012, 134, 12798–12806. [Google Scholar] [CrossRef]
- Ogino, S.; Kubo, S.; Umemoto, R.; Huang, S.; Nishida, N.; Shimada, I. Observation of NMR signals from proteins introduced into living mammalian cells by reversible membrane permeabilization using a pore-forming toxin, streptolysin O. J. Am. Chem. Soc. 2009, 131, 10834–10835. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, A.; Saso, A.; Zhao, Q.; Kubo, S.; Nishida, N.; Shimada, I. Balanced Regulation of Redox Status of Intracellular Thioredoxin Revealed by in-Cell NMR. J. Am. Chem. Soc. 2018, 140, 3784–3790. [Google Scholar] [CrossRef] [PubMed]
- Inomata, K.; Ohno, A.; Tochio, H.; Isogai, S.; Tenno, T.; Nakase, I.; Takeuchi, T.; Futaki, S.; Ito, Y.; Hiroaki, H.; et al. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature 2009, 458, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Binolfi, A.; Limatola, A.; Verzini, S.; Kosten, J.; Theillet, F.X.; May Rose, H.; Bekei, B.; Stuiver, M.; van Rossum, M.; Selenko, P. Intracellular repair of oxidation-damaged alpha-synuclein fails to target C-terminal modification sites. Nat. Commun. 2016, 7, 10251. [Google Scholar] [CrossRef] [PubMed]
- Theillet, F.X.; Binolfi, A.; Bekei, B.; Martorana, A.; Rose, H.M.; Stuiver, M.; Verzini, S.; Lorenz, D.; van Rossum, M.; Goldfarb, D.; et al. Structural disorder of monomeric alpha-synuclein persists in mammalian cells. Nature 2016, 530, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Luchinat, E.; Barbieri, L.; Rubino, J.T.; Kozyreva, T.; Cantini, F.; Banci, L. In-cell NMR reveals potential precursor of toxic species from SOD1 fALS mutants. Nat. Commun. 2014, 5, 5502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luchinat, E.; Barbieri, L.; Banci, L. A molecular chaperone activity of CCS restores the maturation of SOD1 fALS mutants. Sci. Rep. 2017, 7, 17433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capper, M.J.; Wright, G.S.A.; Barbieri, L.; Luchinat, E.; Mercatelli, E.; McAlary, L.; Yerbury, J.J.; O’Neill, P.M.; Antonyuk, S.V.; Banci, L.; et al. The cysteine-reactive small molecule ebselen facilitates effective SOD1 maturation. Nat. Commun. 2018, 9, 1693. [Google Scholar] [CrossRef]
- Banci, L.; Barbieri, L.; Luchinat, E.; Secci, E. Visualization of Redox-Controlled Protein Fold in Living Cells. Chem. Biol. 2013, 20, 747–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercatelli, E.; Barbieri, L.; Luchinat, E.; Banci, L. Direct structural evidence of protein redox regulation obtained by in-cell NMR. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Dzatko, S.; Krafcikova, M.; Hänsel-Hertsch, R.; Fessl, T.; Fiala, R.; Loja, T.; Krafcik, D.; Mergny, J.-L.; Foldynova-Trantirkova, S.; Trantirek, L. Evaluation of the Stability of DNA i-Motifs in the Nuclei of Living Mammalian Cells. Angew. Chem. Int. Ed. 2018, 57, 2165–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luchinat, E.; Secci, E.; Cencetti, F.; Bruni, P. Sequential protein expression and selective labeling for in-cell NMR in human cells. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2016, 1860, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Luchinat, E.; Banci, L. Intracellular metal binding and redox behavior of human DJ-1. JBIC J. Biol. Inorg. Chem. 2018, 23, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Primikyri, A.; Sayyad, N.; Quilici, G.; Vrettos, E.I.; Lim, K.; Chi, S.-W.; Musco, G.; Gerothanassis, I.P.; Tzakos, A.G. Probing the interaction of a quercetin bioconjugate with Bcl-2 in living human cancer cells with in-cell NMR spectroscopy. FEBS Lett. 2018, 592, 3367–3379. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.; Sarkar, D.; Srivatsan, S.G. A Dual-App Nucleoside Probe Provides Structural Insights into the Human Telomeric Overhang in Live Cells. J. Am. Chem. Soc. 2018, 140, 12622–12633. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Zhiping, L.L.; Tan, S.-M.; Bhattacharjya, S. Interaction Analyses of the Integrin β2 Cytoplasmic Tail with the F3 FERM Domain of Talin and 14-3-3ζ Reveal a Ternary Complex with Phosphorylated Tail. J. Mol. Biol. 2016, 428, 4129–4142. [Google Scholar] [CrossRef] [PubMed]
- Luchinat, E.; Banci, L. In-cell NMR: A topical review. IUCrJ 2017, 4 Pt 2, 108–118. [Google Scholar] [CrossRef]
- Pervushin, K.; Riek, R.; Wider, G.; Wuthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 1997, 94, 12366–12371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riek, R.; Fiaux, J.; Bertelsen, E.B.; Horwich, A.L.; Wuthrich, K. Solution NMR techniques for large molecular and supramolecular structures. J. Am. Chem. Soc. 2002, 124, 12144–12153. [Google Scholar] [CrossRef] [PubMed]
- Cruzeiro-Silva, C.; Albernaz, F.P.; Valente, A.P.; Almeida, F.C. In-Cell NMR spectroscopy: Inhibition of autologous protein expression reduces Escherichia coli lysis. Cell Biochem. Biophys. 2006, 44, 497–502. [Google Scholar] [CrossRef]
- Kubo, S.; Nishida, N.; Udagawa, Y.; Takarada, O.; Ogino, S.; Shimada, I. A Gel-Encapsulated Bioreactor System for NMR Studies of Protein–Protein Interactions in Living Mammalian Cells. Angew. Chem. Int. Ed. 2013, 52, 1208–1211. [Google Scholar] [CrossRef] [PubMed]
- Breindel, L.; DeMott, C.; Burz, D.S.; Shekhtman, A. Real-Time In-Cell Nuclear Magnetic Resonance: Ribosome-Targeted Antibiotics Modulate Quinary Protein Interactions. Biochemistry 2018, 57, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Elowitz, M.B.; Surette, M.G.; Wolf, P.E.; Stock, J.B.; Leibler, S. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 1999, 181, 197–203. [Google Scholar] [PubMed]
- Robinson, K.E.; Reardon, P.N.; Spicer, L.D. In-cell NMR spectroscopy in Escherichia coli. Methods Mol. Biol. 2012, 831, 261–277. [Google Scholar] [PubMed]
- Keizers, P.H.J.; Ubbink, M. Paramagnetic tagging for protein structure and dynamics analysis. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 58, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Luchinat, E.; Banci, L. Characterization of proteins by in-cell NMR spectroscopy in cultured mammalian cells. Nat. Protoc. 2016, 11, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, G.; Simenel, C.; Jang, J.; Kalia, N.P.; Choi, I.; Nilges, M.; Pethe, K.; Izadi-Pruneyre, N. Target engagement and binding mode of an anti-tuberculosis drug to its bacterial target deciphered in whole living cells by NMR. Biochemistry 2018. [Google Scholar] [CrossRef]
- Tochio, H. Watching protein structure at work in living cells using NMR spectroscopy. Curr. Opin. Chem. Biol. 2012, 16, 609–613. [Google Scholar] [CrossRef]
- Serber, Z.; Ledwidge, R.; Miller, S.M.; Dotsch, V. Evaluation of parameters critical to observing proteins inside living Escherichia coli by in-cell NMR spectroscopy. J. Am. Chem. Soc. 2001, 123, 8895–8901. [Google Scholar] [CrossRef]
- Pastore, A.; Temussi, P.A. The Emperor’s new clothes: Myths and truths of in-cell NMR. Arch. Biochem. Biophys. 2017, 628, 114–122. [Google Scholar] [CrossRef]
- Ito, Y.; Mikawa, T.; Smith, B.O. In-cell NMR of intrinsically disordered proteins in prokaryotic cells. Methods Mol. Biol. 2012, 895, 19–31. [Google Scholar] [PubMed]
- Theillet, F.X.; Binolfi, A.; Frembgen-Kesner, T.; Hingorani, K.; Sarkar, M.; Kyne, C.; Li, C.; Crowley, P.B.; Gierasch, L.; Pielak, G.J.; et al. Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs). Chem. Rev. 2014, 114, 6661–6714. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, J.; Mu, X.; Lang, L.; Wang, H.; Binolfi, A.; Theillet, F.X.; Bekei, B.; Logan, D.T.; Selenko, P.; Wennerstrom, H.; et al. Thermodynamics of protein destabilization in live cells. Proc. Natl. Acad. Sci. USA 2015, 112, 12402–12407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbs, E.B.; Cook, E.C.; Showalter, S.A. Application of NMR to studies of intrinsically disordered proteins. Arch. Biochem. Biophys. 2017, 628, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; DeMott, C.M.; Reverdatto, S.; Burz, D.S.; Shekhtman, A. Total Cellular RNA Modulates Protein Activity. Biochemistry 2016, 55, 4568–4573. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.; Vanoye, C.G.; Welch, R.C.; Van Horn, W.D.; Sanders, C.R. Functional delivery of a membrane protein into oocyte membranes using bicelles. Biochemistry 2010, 49, 653–655. [Google Scholar] [CrossRef]
- Tian, C.; Vanoye, C.G.; Kang, C.; Welch, R.C.; Kim, H.J.; George, A.L., Jr.; Sanders, C.R. Preparation, functional characterization, and NMR studies of human KCNE1, a voltage-gated potassium channel accessory subunit associated with deafness and long QT syndrome. Biochemistry 2007, 46, 11459–11472. [Google Scholar] [CrossRef]
- Otting, G. Protein NMR Using Paramagnetic Ions. Annu. Rev. Biophys. 2010, 39, 387–405. [Google Scholar] [CrossRef]
- Barbieri, L.; Luchinat, E.; Banci, L. Structural insights of proteins in sub-cellular compartments: In-mitochondria NMR. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 2492–2496. [Google Scholar] [CrossRef] [Green Version]
- Breindel, L.; Burz, D.S.; Shekhtman, A. Interaction proteomics by using in-cell NMR spectroscopy. J. Proteom. 2018, 191, 202–211. [Google Scholar] [CrossRef]
- Burz, D.S.; Shekhtman, A. The STINT-NMR method for studying in-cell protein-protein interactions. Curr. Protoc. Protein Sci. 2010, 61. [Google Scholar]
- Trbovic, N.; Smirnov, S.; Zhang, F.; Bruschweiler, R. Covariance NMR spectroscopy by singular value decomposition. J. Magn. Reson. 2004, 171, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Selvaratnam, R.; Chowdhury, S.; VanSchouwen, B.; Melacini, G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc. Natl. Acad. Sci. USA 2011, 108, 6133–6138. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.; Ferreon, J.C.; Wright, P.E. Quantitative analysis of multisite protein-ligand interactions by NMR: Binding of intrinsically disordered p53 transactivation subdomains with the TAZ2 domain of CBP. J. Am. Chem. Soc. 2012, 134, 3792–3803. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; DeMott, C.M.; Burz, D.S.; Shekhtman, A. Using singular value decomposition to characterize protein-protein interactions by in-cell NMR spectroscopy. ChemBioChem 2014, 15, 929–933. [Google Scholar] [CrossRef] [PubMed]
- Lambruschini, C.; Veronesi, M.; Romeo, E.; Garau, G.; Bandiera, T.; Piomelli, D.; Scarpelli, R.; Dalvit, C. Development of Fragment-Based n-FABS NMR Screening Applied to the Membrane Enzyme FAAH. ChemBioChem 2013, 14, 1611–1619. [Google Scholar] [CrossRef]
- Rahman, S.; Byun, Y.; Hassan, M.I.; Kim, J.; Kumar, V. Towards understanding cellular structure biology: In-cell NMR. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2017, 1865, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Durham, T.B.; Blanco, M.-J. Target Engagement in Lead Generation. Bioorganic Med. Chem. Lett. 2015, 25, 998–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez Molina, D.; Jafari, R.; Ignatushchenko, M.; Seki, T.; Larsson, E.A.; Dan, C.; Sreekumar, L.; Cao, Y.; Nordlund, P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 2013, 341, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Jafari, R.; Almqvist, H.; Axelsson, H.; Ignatushchenko, M.; Lundback, T.; Nordlund, P.; Martinez Molina, D. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014, 9, 2100–2122. [Google Scholar] [CrossRef]
- Dubach, J.M.; Kim, E.; Yang, K.; Cuccarese, M.; Giedt, R.J.; Meimetis, L.G.; Vinegoni, C.; Weissleder, R. Quantitating drug-target engagement in single cells in vitro and in vivo. Nat. Chem. Biol. 2017, 13, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Reckel, S.; Lohr, F.; Dotsch, V. In-cell NMR spectroscopy. ChemBioChem 2005, 6, 1601–1606. [Google Scholar] [CrossRef] [PubMed]
- Luchinat, E.; Gianoncelli, A.; Mello, T.; Galli, A.; Banci, L. Combining in-cell NMR and X-ray fluorescence microscopy to reveal the intracellular maturation states of human superoxide dismutase 1. Chem. Commun. 2015, 51, 584–587. [Google Scholar] [CrossRef] [PubMed]
- Mitri, E.; Barbieri, L.; Vaccari, L.; Luchinat, E. 15N isotopic labelling for in-cell protein studies by NMR spectroscopy and single-cell IR synchrotron radiation FTIR microscopy: A correlative study. Analyst 2018, 143, 1171–1181. [Google Scholar] [CrossRef] [PubMed]


| Experiment | Remarks | Reference |
|---|---|---|
| 1H-15N-HSQC (heteronuclear single quantum coherence) | Protein–protein/ligand interactions | [66,67] |
| 3D experiments | Backbone assignment | [68] |
| PCS (pseudo-contact shift) | Protein structure determination using lanthanide tags | [69,70] |
| NOESY (Nuclear Overhauser effect spectroscopy) | Protein structure determination | [71] |
| SOFAST-HMQC (Band-Selective Optimized Flip Angle Short Transient- heteronuclear multiple quantum coherence) | Protein–protein/ligand interactions | [72] |
| 1H-13C HSQC | Protein structure analysis using selectively protonation and 13C labeling | [68] |
| 19F-NMR | In-cell protein-observed 19F can be obtained | [73] |
| Relaxation | Protein dynamics | [74] |
| Residue dipolar couplings | Lanthanide tags can also be used to generate RDCs | [69] |
| Protein-based-1H NMR | 1H-NMR at His residue regions | [75] |
| Ligand-based 1H NMR | Protein-ligand interactions | [76] |
| 19F-NMR | Ligand observed 19F-NMR was used in ligand binding studies | [77] |
| Cells | Targets | Studies | Reference |
|---|---|---|---|
| Bacteria | TTHA1718 | Structure was determined in the living cells | [68] |
| calmodulin, NmerA, and FKBP (FK506 binding protein) | Labeling methyl groups of protein was used in-cell NMR studies | [78] | |
| HdeA, alpha-synuclein, chymotrypsin inhibitor 2 (CI2) ubiquitin | Protein dynamics in cells, protein leakage, and protein–protein interactions were analyzed | [63,79,80] | |
| Thioredoxin | Quandary interactions of proteins in cells was addressed in the study | [81] | |
| ADK (adenosine kinase) | |||
| FKBP | |||
| Alpha-synuclein, ubiquitin, HDH (histidinol dehydrogensase), GFP (Green fluorescence protein) | Protein-based 19F-NMR study was carried out | [73] | |
| SOD1 SOD1 (human copper, zinc superoxide dismutase 1) | Protein folding in living cells was analyzed. | [72] | |
| PFN1 (protein profilin 1) | Protein–protein interaction was studied in living cells | [82] | |
| Pup (prokaryotic ubiquitin like protein) | In-cell NMR was used to screen compounds disrupting protein–protein interactions | [83] | |
| Mpa (mycobacterial protease ATPase) | |||
| FKBP12 | In-cell NMR was used to screen a library. | [84] | |
| Cox17 (cytochrome c oxidase copper chaperone) | In-cell NMR was used to probe protein folding in living cells | [85] | |
| oocyte | Ubiquitin, calmodulin | Protein–protein interactions were probed in oocyte | [86] |
| GB1 (the B domain of G protein) | Structural studies were performed using PRE restrains | [70,87,88] | |
| XT-GB1 (SV40 regulatory domain-GB1) | Protein phosphorylation was monitored in cells | [89] | |
| yeast | Ubiquitin | Structural studies were carried out in cell compartments | [90] |
| Insect | GB1, HB8 TTHA1718, rat calmodulin, and human HAH1 | 3D experiments were collected in living insect cells for structural studies. | [71] |
| Mammalian cells | Tβ4 (thymosin β4) | Introducing proteins into cells using toxin was used for in-cell NMR studies. | [91] |
| Thioredoxin | Redox status of intracellular thioredoxin was measured in living cells | [92] | |
| GB1 | Labeled protein was delivered into mammalian cells using peptides for in-cell NMR | [93] | |
| FKBP12 | |||
| Alpha-synuclein | Protein modification and folding were monitored | [94,95] | |
| hSOD1 and mutants | Folding in living cells and protein–protein interactions were analyzed | [96,97] | |
| SOD1 | Effect of ebselen and ebsulphur on protein structure was investigated | [98] | |
| Mia40 (mitochondrial intermembrane space import and assembly protein 40) | Protein folding in living cells was investigated | [99] | |
| Cox17 | Protein folding was investigated in living cells | [100] | |
| DNA i-motif | Stability of DNA i-motif was investigated. | [101] | |
| copper binding protein HAH1 | Sequential protein expression in mammalian cells and selective labeling proteins was used in-cell NMR studies | [102] | |
| DJ1 | Protein folding was investigated | [103] | |
| Bcl-2 (B-cell lymphoma 2) | Protein-ligand interactions. Saturation-Transfer Difference (STD) and TrNOE experiments were carried out | [104] | |
| PFN1 | Specific and unspecific interactions in cells was explored using in-cell NMR | [82] |
| System | Experimental Outcome | Reference |
|---|---|---|
| E. coli | Heteronuclear spectra of proteins were collected in living cells | [66] |
| E. coli | Protein structure was determined in living cells | [68] |
| Mammalian cells | In-cell NMR study of proteins that were delivered into cells was performed | [93] |
| Oocyte | Lanthanide tag was used in generating distance restraints in living cells | [115] |
| HEK293T | Protein was overexpressed in mammalian cells for in-cell NMR studies | [116] |
| E. coli | In-cell NMR was used to screening a library | [84] |
| M. smegmatis | The first application of in-cell NMR in target engagement | [117] |
| Hela | In-cell NMR study on DNA was carried out | [101] |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, C. Applications of In-Cell NMR in Structural Biology and Drug Discovery. Int. J. Mol. Sci. 2019, 20, 139. https://doi.org/10.3390/ijms20010139
Kang C. Applications of In-Cell NMR in Structural Biology and Drug Discovery. International Journal of Molecular Sciences. 2019; 20(1):139. https://doi.org/10.3390/ijms20010139
Chicago/Turabian StyleKang, CongBao. 2019. "Applications of In-Cell NMR in Structural Biology and Drug Discovery" International Journal of Molecular Sciences 20, no. 1: 139. https://doi.org/10.3390/ijms20010139
APA StyleKang, C. (2019). Applications of In-Cell NMR in Structural Biology and Drug Discovery. International Journal of Molecular Sciences, 20(1), 139. https://doi.org/10.3390/ijms20010139