Functional Polymorphisms in DNA Repair Genes Are Associated with Sporadic Colorectal Cancer Susceptibility and Clinical Outcome
Abstract
1. Introduction
2. Results
2.1. SNP Selection
2.2. Case-Control Study
2.3. Survival Analyses
2.4. Survival and Therapy
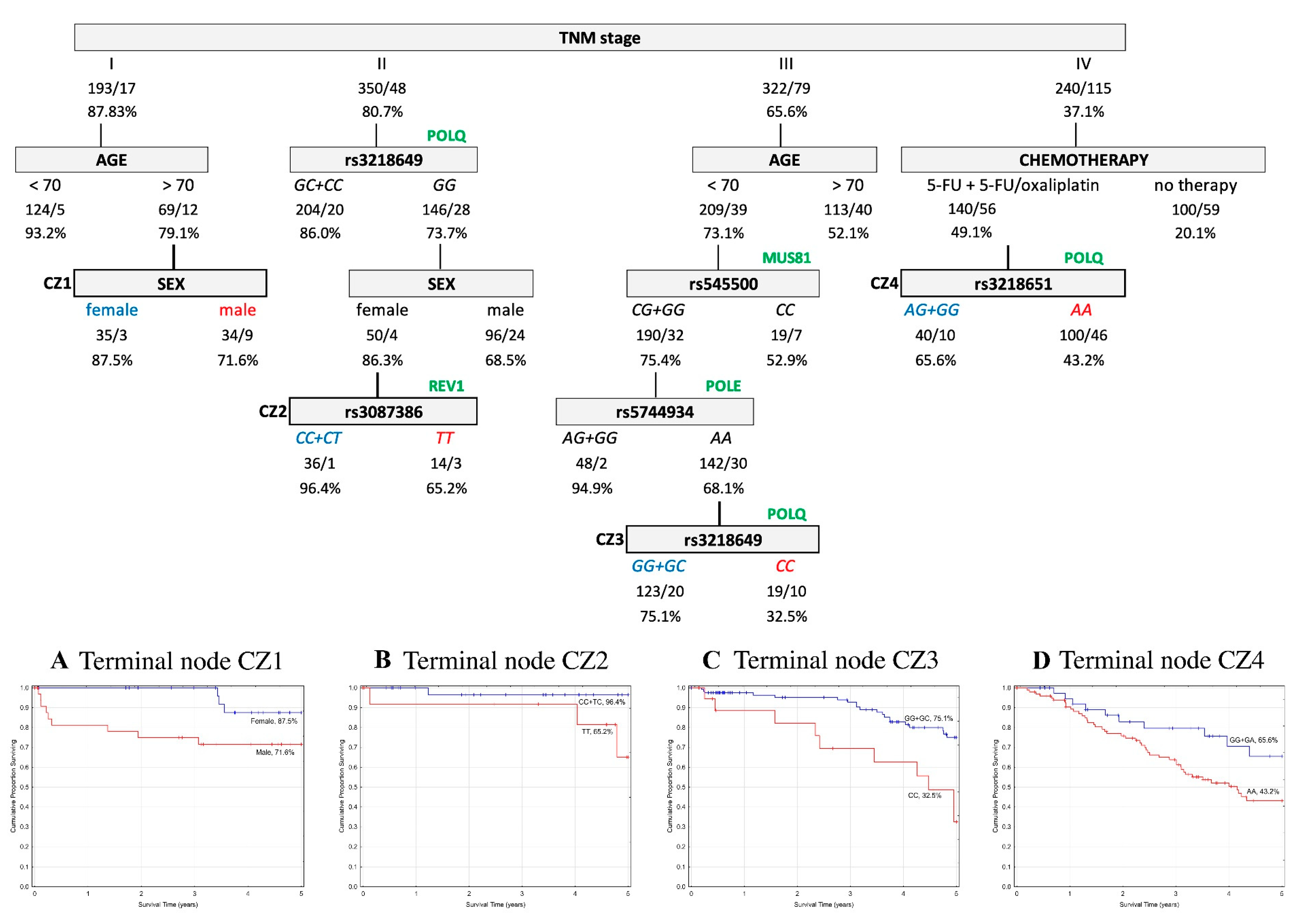
2.5. Classification and Regression Tree Survival Analysis
2.5.1. Overall Survival
2.5.2. Event-Free Survival
3. Discussion
4. Material and Methods
4.1. SNP Selection and In Silico Analysis of Functional Relevance and Conservation
4.2. Study Populations and Data Collection
4.2.1. Discovery Set—Czech Republic
4.2.2. Replication Set—Austria
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-FU | 5-Fluorouracil |
| BER | Base excision repair |
| BMI | Body mass index |
| CART | Classification and regression tree analysis |
| CI | Confidence intervals |
| CRC | Colorectal cancer |
| DSB | Double strand break repair |
| EFS | Event-free survival |
| FA | Fanconi anemia |
| FDR | False discovery rate |
| GERP | Genomic evolutionary rate profiling |
| GWAS | Genome-wide association study |
| HRs | Hazard ratios |
| ICL | Interstrand cross-links repair |
| LD | Linkage disequilibrium |
| MAF | Minor allele frequency |
| NER | Nucleotide excision repair |
| nsSNP | Non-synonymous single nucleotide polymorphism |
| ORs | Odds ratios |
| OS | Overall survival |
| RS | Rejected substitutions |
| TLS | Translesion synthesis |
| TNM | Tumor–node–metastasis stage system |
| UICC | International union against cancer |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Steliarova-Foucher, E.; Lortet-Tieulent, J.; Rosso, S.; Coebergh, J.W.; Comber, H.; Forman, D.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer 2013, 49, 1374–1403. [Google Scholar] [CrossRef] [PubMed]
- Aran, V.; Victorino, A.P.; Thuler, L.C.; Ferreira, C.G. Colorectal Cancer: Epidemiology, Disease Mechanisms and Interventions to Reduce Onset and Mortality. Clin. Colorectal Cancer 2016, 15, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Binefa, G.; Rodriguez-Moranta, F.; Teule, A.; Medina-Hayas, M. Colorectal cancer: From prevention to personalized medicine. World J. Gastroenterol. 2014, 20, 6786–6808. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, B.; Carlsson, G.; Machover, D.; Petrelli, N.; Roth, A.; Schmoll, H.J.; Tveit, K.M.; Gibson, F. A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin. Colorectal Cancer 2015, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Vodicka, P.; Stetina, R.; Polakova, V.; Tulupova, E.; Naccarati, A.; Vodickova, L.; Kumar, R.; Hanova, M.; Pardini, B.; Slyskova, J.; et al. Association of DNA repair polymorphisms with DNA repair functional outcomes in healthy human subjects. Carcinogenesis 2007, 28, 657–664. [Google Scholar] [CrossRef]
- He, J.; Shi, T.Y.; Zhu, M.L.; Wang, M.Y.; Li, Q.X.; Wei, Q.Y. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: A meta-analysis. Int. J. Cancer 2013, 133, 1765–1775. [Google Scholar] [CrossRef] [PubMed]
- Slyskova, J.; Naccarati, A.; Pardini, B.; Polakova, V.; Vodickova, L.; Smerhovsky, Z.; Levy, M.; Lipska, L.; Liska, V.; Vodicka, P. Differences in nucleotide excision repair capacity between newly diagnosed colorectal cancer patients and healthy controls. Mutagenesis 2012, 27, 225–232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peters, U.; Jiao, S.; Schumacher, F.R.; Hutter, C.M.; Aragaki, A.K.; Baron, J.A.; Berndt, S.I.; Bezieau, S.; Brenner, H.; Butterbach, K.; et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology 2013, 144, 799–807.e24. [Google Scholar] [CrossRef]
- Whiffin, N.; Hosking, F.J.; Farrington, S.M.; Palles, C.; Dobbins, S.E.; Zgaga, L.; Lloyd, A.; Kinnersley, B.; Gorman, M.; Tenesa, A.; et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum. Mol. Genet. 2014, 23, 4729–4737. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Yan, T.; Wang, Z.; Su, H.C.; Zhu, X.; Tian, X.; Fang, J.Y.; Chen, H.; Hong, J. Variant of SNP rs1317082 at CCSlnc362 (RP11-362K14.5) creates a binding site for miR-4658 and diminishes the susceptibility to CRC. Cell Death Dis. 2018, 9, 1177. [Google Scholar] [CrossRef] [PubMed]
- Tanskanen, T.; van den Berg, L.; Valimaki, N.; Aavikko, M.; Ness-Jensen, E.; Hveem, K.; Wettergren, Y.; Bexe Lindskog, E.; Tonisson, N.; Metspalu, A.; et al. Genome-wide association study and meta-analysis in Northern European populations replicate multiple colorectal cancer risk loci. Int. J. Cancer 2018, 142, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Huyghe, J.R.; Bien, S.A.; Harrison, T.A.; Kang, H.M.; Chen, S.; Schmit, S.L.; Conti, D.V.; Qu, C.; Jeon, J.; Edlund, C.K.; et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 2018. [Google Scholar] [CrossRef] [PubMed]
- Slyskova, J.; Cordero, F.; Pardini, B.; Korenkova, V.; Vymetalkova, V.; Bielik, L.; Vodickova, L.; Pitule, P.; Liska, V.; Matejka, V.M.; et al. Post-treatment recovery of suboptimal DNA repair capacity and gene expression levels in colorectal cancer patients. Mol. Carcinog. 2015, 54, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, M.D.; Wilson, D.M., 3rd. Participation of DNA repair in the response to 5-fluorouracil. Cell. Mol. Life Sci. 2009, 66, 788–799. [Google Scholar] [CrossRef]
- Martin, L.P.; Hamilton, T.C.; Schilder, R.J. Platinum resistance: The role of DNA repair pathways. Clin. Cancer Res. 2008, 14, 1291–1295. [Google Scholar] [CrossRef]
- De Mattia, E.; Cecchin, E.; Toffoli, G. Pharmacogenomics of intrinsic and acquired pharmacoresistance in colorectal cancer: Toward targeted personalized therapy. Drug Resist. Updat. 2015, 20, 39–70. [Google Scholar] [CrossRef]
- Vaisman, A.; Woodgate, R. Translesion DNA polymerases in eukaryotes: What makes them tick? Crit. Rev. Biochem. Mol. Biol. 2017, 52, 274–303. [Google Scholar] [CrossRef]
- Wittschieben, J.P.; Patil, V.; Glushets, V.; Robinson, L.J.; Kusewitt, D.F.; Wood, R.D. Loss of DNA polymerase zeta enhances spontaneous tumorigenesis. Cancer Res. 2010, 70, 2770–2778. [Google Scholar] [CrossRef]
- Aparicio, T.; Baer, R.; Gautier, J. DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst.) 2014, 19, 169–175. [Google Scholar] [CrossRef]
- Knobel, P.A.; Marti, T.M. Translesion DNA synthesis in the context of cancer research. Cancer Cell Int. 2011, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, S.Y.; Wang, S.; Lu, J.; Wu, W.; Weng, L.; Chen, D.; Zhang, Y.; Lu, Z.; Yang, J.; et al. REV3L confers chemoresistance to cisplatin in human gliomas: The potential of its RNAi for synergistic therapy. Neuro Oncol. 2009, 11, 790–802. [Google Scholar] [CrossRef]
- Roos, W.P.; Tsaalbi-Shtylik, A.; Tsaryk, R.; Guvercin, F.; de Wind, N.; Kaina, B. The translesion polymerase Rev3L in the tolerance of alkylating anticancer drugs. Mol. Pharmacol. 2009, 76, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Stallons, L.J.; McGregor, W.G. Translesion synthesis polymerases in the prevention and promotion of carcinogenesis. J. Nucleic Acids 2010, 2010, 643857. [Google Scholar] [CrossRef]
- Brondello, J.M.; Pillaire, M.J.; Rodriguez, C.; Gourraud, P.A.; Selves, J.; Cazaux, C.; Piette, J. Novel evidences for a tumor suppressor role of Rev3, the catalytic subunit of Pol zeta. Oncogene 2008, 27, 6093–6101. [Google Scholar] [CrossRef] [PubMed]
- Varadi, V.; Bevier, M.; Grzybowska, E.; Johansson, R.; Enquist, K.; Henriksson, R.; Butkiewicz, D.; Pamula-Pilat, J.; Tecza, K.; Hemminki, K.; et al. Genetic variation in genes encoding for polymerase zeta subunits associates with breast cancer risk, tumour characteristics and survival. Breast Cancer Res. Treat. 2011, 129, 235–245. [Google Scholar] [CrossRef]
- Pan, J.; Chi, P.; Lu, X.; Xu, Z. Genetic polymorphisms in translesion synthesis genes are associated with colorectal cancer risk and metastasis in Han Chinese. Gene 2012, 504, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.K.; Mu, L.N.; Cai, L.; Chang, S.C.; Park, S.L.; Oh, S.S.; Wang, Y.; Goldstein, B.Y.; Ding, B.G.; Jiang, Q.; et al. Genetic variation in immune regulation and DNA repair pathways and stomach cancer in China. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2304–2309. [Google Scholar] [CrossRef] [PubMed]
- Yousefzadeh, M.J.; Wood, R.D. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair (Amst.) 2013, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Beagan, K.; McVey, M. Linking DNA polymerase theta structure and function in health and disease. Cell. Mol. Life Sci. 2016, 73, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Bahar, R.; Seimiya, M.; Chiyo, M.; Wada, A.; Okada, S.; Hatano, M.; Tokuhisa, T.; Kimura, H.; Watanabe, S.; et al. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer 2004, 109, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Lemee, F.; Bergoglio, V.; Fernandez-Vidal, A.; Machado-Silva, A.; Pillaire, M.J.; Bieth, A.; Gentil, C.; Baker, L.; Martin, A.L.; Leduc, C.; et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl. Acad. Sci. USA 2010, 107, 13390–13395. [Google Scholar] [CrossRef] [PubMed]
- Allera-Moreau, C.; Rouquette, I.; Lepage, B.; Oumouhou, N.; Walschaerts, M.; Leconte, E.; Schilling, V.; Gordien, K.; Brouchet, L.; Delisle, M.B.; et al. DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis 2012, 1, e30. [Google Scholar] [CrossRef] [PubMed]
- Pillaire, M.J.; Selves, J.; Gordien, K.; Gourraud, P.A.; Gentil, C.; Danjoux, M.; Do, C.; Negre, V.; Bieth, A.; Guimbaud, R.; et al. A ‘DNA replication’ signature of progression and negative outcome in colorectal cancer. Oncogene 2010, 29, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Q.; Hu, N.; Hyland, P.L.; Gao, Y.; Wang, Z.M.; Yu, K.; Su, H.; Wang, C.Y.; Wang, L.M.; Chanock, S.J.; et al. Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis 2013, 34, 1536–1542. [Google Scholar] [CrossRef]
- Brandalize, A.P.; Schuler-Faccini, L.; Hoffmann, J.S.; Caleffi, M.; Cazaux, C.; Ashton-Prolla, P. A DNA repair variant in POLQ (c.-1060A > G) is associated to hereditary breast cancer patients: A case-control study. BMC Cancer 2014, 14, 850. [Google Scholar] [CrossRef]
- Rendleman, J.; Antipin, Y.; Reva, B.; Adaniel, C.; Przybylo, J.A.; Dutra-Clarke, A.; Hansen, N.; Heguy, A.; Huberman, K.; Borsu, L.; et al. Genetic variation in DNA repair pathways and risk of non-Hodgkin’s lymphoma. PLoS ONE 2014, 9, e101685. [Google Scholar] [CrossRef]
- Family, L.; Bensen, J.T.; Troester, M.A.; Wu, M.C.; Anders, C.K.; Olshan, A.F. Single-nucleotide polymorphisms in DNA bypass polymerase genes and association with breast cancer and breast cancer subtypes among African Americans and Whites. Breast Cancer Res. Treat. 2015, 149, 181–190. [Google Scholar] [CrossRef][Green Version]
- Krokeide, S.Z.; Laerdahl, J.K.; Salah, M.; Luna, L.; Cederkvist, F.H.; Fleming, A.M.; Burrows, C.J.; Dalhus, B.; Bjoras, M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst.) 2013, 12, 1159–1164. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S.; Murphy, D.L.; Sweasy, J.B. Base excision repair and cancer. Cancer Lett. 2012, 327, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Hildrestrand, G.A.; Neurauter, C.G.; Diep, D.B.; Castellanos, C.G.; Krauss, S.; Bjoras, M.; Luna, L. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci. 2009, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Shinmura, K.; Kato, H.; Kawanishi, Y.; Igarashi, H.; Goto, M.; Tao, H.; Inoue, Y.; Nakamura, S.; Misawa, K.; Mineta, H.; et al. Abnormal Expressions of DNA Glycosylase Genes NEIL1, NEIL2, and NEIL3 Are Associated with Somatic Mutation Loads in Human Cancer. Oxid. Med. Cell. Longev. 2016, 2016, 1546392. [Google Scholar] [CrossRef] [PubMed]
- Kauffmann, A.; Rosselli, F.; Lazar, V.; Winnepenninckx, V.; Mansuet-Lupo, A.; Dessen, P.; van den Oord, J.J.; Spatz, A.; Sarasin, A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene 2008, 27, 565–573. [Google Scholar] [CrossRef] [PubMed]
- D’Errico, M.; Parlanti, E.; Pascucci, B.; Fortini, P.; Baccarini, S.; Simonelli, V.; Dogliotti, E. Single nucleotide polymorphisms in DNA glycosylases: From function to disease. Free Radic. Biol. Med. 2017, 107, 278–291. [Google Scholar] [CrossRef]
- Barry, K.H.; Koutros, S.; Berndt, S.I.; Andreotti, G.; Hoppin, J.A.; Sandler, D.P.; Burdette, L.A.; Yeager, M.; Freeman, L.E.; Lubin, J.H.; et al. Genetic variation in base excision repair pathway genes, pesticide exposure, and prostate cancer risk. Environ. Health Perspect. 2011, 119, 1726–1732. [Google Scholar] [CrossRef]
- Bethke, L.; Webb, E.; Murray, A.; Schoemaker, M.; Johansen, C.; Christensen, H.C.; Muir, K.; McKinney, P.; Hepworth, S.; Dimitropoulou, P.; et al. Comprehensive analysis of the role of DNA repair gene polymorphisms on risk of glioma. Hum. Mol. Genet. 2008, 17, 800–805. [Google Scholar] [CrossRef]
- Cipollini, M.; Figlioli, G.; Maccari, G.; Garritano, S.; De Santi, C.; Melaiu, O.; Barone, E.; Bambi, F.; Ermini, S.; Pellegrini, G.; et al. Polymorphisms within base and nucleotide excision repair pathways and risk of differentiated thyroid carcinoma. DNA Repair (Amst.) 2016, 41, 27–31. [Google Scholar] [CrossRef][Green Version]
- Allione, A.; Pardini, B.; Viberti, C.; Oderda, M.; Allasia, M.; Gontero, P.; Vineis, P.; Sacerdote, C.; Matullo, G. The prognostic value of basal DNA damage level in peripheral blood lymphocytes of patients affected by bladder cancer. Urol. Oncol. 2018, 36, 241.e15–241.e23. [Google Scholar] [CrossRef]
- Lee, P.H.; Shatkay, H. F-SNP: Computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008, 36, D820–D824. [Google Scholar] [CrossRef] [PubMed]
- Vymetalkova, V.; Pardini, B.; Rosa, F.; Jiraskova, K.; Di Gaetano, C.; Bendova, P.; Levy, M.; Veskrnova, V.; Buchler, T.; Vodickova, L.; et al. Polymorphisms in microRNA binding sites of mucin genes as predictors of clinical outcome in colorectal cancer patients. Carcinogenesis 2017, 38, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Schneiderova, M.; Naccarati, A.; Pardini, B.; Rosa, F.; Gaetano, C.D.; Jiraskova, K.; Opattova, A.; Levy, M.; Veskrna, K.; Veskrnova, V.; et al. MicroRNA-binding site polymorphisms in genes involved in colorectal cancer etiopathogenesis and their impact on disease prognosis. Mutagenesis 2017, 32, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Hofer, P.; Hagmann, M.; Brezina, S.; Dolejsi, E.; Mach, K.; Leeb, G.; Baierl, A.; Buch, S.; Sutterluty-Fall, H.; Karner-Hanusch, J.; et al. Bayesian and frequentist analysis of an Austrian genome-wide association study of colorectal cancer and advanced adenomas. Oncotarget 2017, 8, 98623–98634. [Google Scholar] [CrossRef] [PubMed]
- Lemon, S.C.; Roy, J.; Clark, M.A.; Friedmann, P.D.; Rakowski, W. Classification and regression tree analysis in public health: Methodological review and comparison with logistic regression. Ann. Behav. Med. 2003, 26, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Statist. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]




| Genomic Annotation | Functional Genomics | Comparative Genomics | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | DNA Repair Pathway | UniProtKB | SNP ID | Base Change | Amino Acid Change | MAF in EUR a | LD with Other SNPs Associated with CRC | LD within the Same Gene | F-SNP Prediction Result (on Protein Coding) | ELASPIC (∆∆G) | DUET (∆∆G) | Element GERP RS Score >800 | SIPHYs |
| EME1 | DSB | Q96AY2 | rs12450550 | T > C | Ile350Thr | 0.24 | no | no | deleterious | Destabilizing (Core 1.646) | Destabilizing (−3.002 Kcal/mol) | ||
| FAAP24 | DSB | Q9BTP7 | rs3816032 b | T > C | Ile192Thr | 0.11 | no | no | deleterious | Destabilizing (Core 1.133) | Destabilizing (−1.653 Kcal/mol) | X | |
| FANCI | DSB | Q9NVI1 | rs2283432 | C > G | Cys742Ser | 0.38 | no | no | deleterious | NA | NA | X | X |
| MUS81 | DSB | Q96NY9 | rs545500 b | C > G | Arg180Pro | 0.33 | no | no | deleterious | NA | NA | X | |
| NEIL3 | BER | Q8TAT5 | rs7689099 | C > G | Pro117Arg | 0.12 | no | no | deleterious | NA | NA | X | X |
| POLE | BER, DSB, NER | Q07864 | rs5744934 | A > G | Asn1396Ser | 0.13 | no | no | deleterious | NA | NA | X | |
| POLN | DSB | Q7Z5Q5 | rs2353552 | C > A | Gln121His | 0.13 | no | no | deleterious | NA | NA | ||
| rs9328764 | A > G | Arg425Cys | 0.12 | no | no | deleterious | NA | Destabilizing (−1.765 Kcal/mol) | X | ||||
| POLQ | DSB | O75417 | rs1381057 | C > T | Gln2513Arg | 0.33 | no | no | deleterious | Destabilizing (Core 1.648) | NA | X | |
| rs3218649 | C > G | Thr982Arg | 0.39 | no | no | deleterious | NA | NA | X | ||||
| rs3218651 | T > C | His1201Arg | 0.15 | no | no | damaging | NA | NA | X | ||||
| RAD51D | DSB | O75771 | rs4796033 | C > T | Arg165Gln | 0.13 | no | no | deleterious | Destabilizing (Core 1.843) | NA | X | |
| REV1 | DSB | Q9UBZ9 | rs3087386 | G > A | Phe257Ser | 0.43 | no | no | deleterious | NA | NA | X | |
| rs3087399 | A > G | Asn373Ser | 0.12 | no | no | deleterious | NA | Destabilizing (−0.596 Kcal/mol) | X | X | |||
| REV3L | DSB | O60673 | rs3204953 | G > A | Val2986Ile | 0.17 | no | no | deleterious | Destabilizing (Core 1.965) | NA | X | X |
| RPA1 | BER, DSB, NER | P27694 | rs5030755 | A > G | Thr351Ala | 0.10 | no | no | deleterious | NA | Destabilizing (−1.037 Kcal/mol) | X | |
| Czech Republic | Austria | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Controls | Cases | OR | 95% CI | p-Value | Controls | Cases | OR | 95% CI | p-Value | |
| No. (%) | No. (%) | No. (%) | No. (%) | ||||||||
| Sex | Female | 478 (40.8) | 696 (38.1) | Ref | 353 (43.1) | 389 (40.9) | Ref | ||||
| Male | 694 (59.2) | 1133 (61.9) | 1.20 | 1.02–1.40 | 0.03 | 467 (56.9) | 561 (59.1) | 1.39 | 1.09–1.78 | 0.01 | |
| Age (years) | <50 | 269 (22.9) | 140 (9.2) | Ref | 92 (11.2) | 129 (13.5) | Ref | ||||
| (50, 60] | 546 (46.6) | 433 (28.4) | 1.52 | 1.20–1.94 | 0.0006 | 147 (17.9) | 208 (21.9) | 2.18 | 0.72–1.43 | 0.92 | |
| (60, 70] | 183 (15.6) | 639 (42.0) | 6.71 | 5.16–8.72 | <0.0001 | 282 (34.4) | 323 (34.0) | 0.82 | 0.60–1.13 | 0.22 | |
| >70 | 174 (14.9) | 310 (20.4) | 3.42 | 2.60–4.51 | <0.0001 | 299 (36.5) | 291 (30.6) | 0.70 | 0.51–0.96 | 0.03 | |
| BMI | (18.5, 25] | 93 (8.0) | 358 (23.5) | Ref | 189 (23.7) | 296 (35.7) | Ref | ||||
| <18.5 | 334 (28.5) | 370 (24.3) | 3.22 | 2.45–4.24 | <0.0001 | 2 (0.3) | 17 (2.0) | 5.43 | 1.24–23.76 | 0.02 | |
| (25, 30] | 529 (45.1) | 508 (33.4) | 0.84 | 0.69–1.02 | 0.08 | 364 (45.6) | 336 (40.5) | 0.59 | 0.47–0.75 | <0.0001 | |
| >30 | 213 (18.4) | 286 (18.8) | 1.13 | 0.89–1.43 | 0.31 | 243 (30.4) | 181 (21.8) | 0.48 | 0.37–0.62 | <0.0001 | |
| Smoking habit | No | 638 (57.6) | 769 (53.1) | Ref | 447 (55.7) | 251 (48.8) | Ref | ||||
| Yes a | 470 (42.4) | 679 (46.9) | 1.33 | 1.13–1.56 | <0.001 | 356 (44.3) | 263 (51.2) | 1.30 | 0.97–1.73 | 0.08 | |
| DM | No | 555 (85.5) | 1076 (80.4) | Ref | 370 (82.8) | 817 (86.0) | Ref | ||||
| Yes | 94 (14.5) | 263 (19.6) | 1.41 | 1.09–1.84 | 0.01 | 77 (17.2) | 133 (14.0) | 0.62 | 0.42–0.92 | 0.02 | |
| Family history of CRC | No | 942 (89.3) | 1103 (84.1) | Ref | NDA | NDA | |||||
| Yes | 113 (10.7) | 209 (15.9) | 1.65 | 1.28–2.12 | <0.001 | NDA | NDA | ||||
| Diagnosis | Colon | 1192 (65.8) | 586 (62.6) | ||||||||
| Rektum | 621 (34.2) | 350 (37.4) | |||||||||
| tnm stage | I | 277 (16.8) | 188 (21.2) | ||||||||
| II | 498 (30.2) | 227 (25.5) | |||||||||
| III | 491 (29.8) | 354 (39.8) | |||||||||
| IV | 384 (23.3) | 120 (13.5) | |||||||||
| Chemotherapy | None | 795 (49.9) | 389 (43.0) | ||||||||
| 5-FU | 494 (31.0) | 253 (28.0) | |||||||||
| 5-FU combined with oxaliplatin | 303 (19.1) | 262 (29.0) | |||||||||
| All CRC Patients | Colon Cancer Patients | Rectal Cancer Patients | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Genotype | Controls a | Cases a | OR b | 95% CI | p-Value | Cases a | OR b | 95% CI | p-Value | Cases a | OR b | 95% CI | p-Value | HWE c |
| SNP | Χ2, p-Value | ||||||||||||||
| Czech Republic | |||||||||||||||
| EME1 rs12450550 | TT | 678 | 815 | Ref | 526 | Ref | 284 | Ref | 1.07, 0.58 | ||||||
| TC | 410 | 570 | 1.19 | 1.00–1.40 | 0.05 | 363 | 1.17 | 0.97–1.40 | 0.11 | 198 | 1.20 | 0.96–1.50 | 0.11 | ||
| CC | 73 | 108 | 1.24 | 0.90–1.70 | 0.20 | 64 | 1.15 | 0.80–1.65 | 0.46 | 41 | 1.38 | 0.91–2.09 | 0.13 | ||
| TC+CC | 483 | 678 | 1.19 | 1.02–1.40 | 0.03 | 427 | 1.16 | 0.97–1.39 | 0.10 | 239 | 1.23 | 0.99–1.52 | 0.06 | ||
| TT+TC | 1088 | 1385 | Ref | 889 | Ref | 482 | Ref | ||||||||
| CC | 73 | 108 | 1.16 | 0.84–1.59 | 0.36 | 64 | 1.08 | 0.76–1.55 | 0.66 | 41 | 1.28 | 0.86–1.93 | 0.23 | ||
| REV3L rs3204953 | GG | 839 | 1049 | Ref | 666 | Ref | 371 | Ref | 4.68, 0.10 | ||||||
| GA | 304 | 405 | 1.09 | 0.91–1.30 | 0.37 | 261 | 1.12 | 0.91–1.37 | 0.27 | 139 | 1.06 | 0.83–1.34 | 0.66 | ||
| AA | 15 | 43 | 2.32 | 1.27–4.25 | 0.006 * | 30 | 2.59 | 1.36–4.91 | 0.004 * | 13 | 1.97 | 0.92–4.22 | 0.08 | ||
| GA+AA | 319 | 448 | 1.14 | 0.96–1.36 | 0.13 | 291 | 1.19 | 0.98–1.44 | 0.08 | 152 | 1.10 | 0.87–1.39 | 0.42 | ||
| GG+GA | 1143 | 1454 | Ref | 927 | Ref | 510 | Ref | ||||||||
| AA | 15 | 43 | 2.28 | 1.24–4.17 | 0.008 * | 30 | 2.52 | 1.33–4.77 | 0.005 * | 13 | 1.95 | 0.91–4.18 | 0.09 | ||
| Austria | |||||||||||||||
| POLQ rs1381057 | CC | 372 | 413 | Ref | 267 | Ref | 134 | Ref | 1.49, 0.47 | ||||||
| CT | 349 | 423 | 1.09 | 0.90–1.34 | 0.38 | 250 | 1.00 | 0.80–1.25 | 1.00 | 166 | 1.32 | 1.01–1.74 | 0.04 | ||
| TT | 99 | 114 | 1.05 | 0.77–1.42 | 0.76 | 65 | 0.93 | 0.65–1.32 | 0.68 | 49 | 1.40 | 0.94–2.08 | 0.10 | ||
| CT+TT | 448 | 537 | 1.08 | 0.90–1.31 | 0.40 | 315 | 0.98 | 0.80–1.22 | 0.89 | 215 | 1.34 | 1.04–1.74 | 0.03 | ||
| CC+CT | 721 | 836 | Ref | 517 | Ref | 300 | Ref | ||||||||
| TT | 99 | 114 | 1.00 | 0.75–1.34 | 0.98 | 65 | 0.93 | 0.66–1.29 | 0.66 | 49 | 1.21 | 0.84–1.75 | 0.32 | ||
| REV1 rs3087399 | AA | 593 | 673 | Ref | 414 | Ref | 243 | Ref | 0.02, 0.99 | ||||||
| AG | 208 | 259 | 1.10 | 0.89–1.36 | 0.39 | 151 | 1.04 | 0.81–1.32 | 0.78 | 105 | 1.25 | 0.95–1.66 | 0.11 | ||
| GG | 19 | 18 | 0.83 | 0.43–1.60 | 0.58 | 17 | 1.27 | 0.65–2.48 | 0.48 | 1 | 0.13 | 0.02–0.96 | 0.05 | ||
| AG+GG | 227 | 277 | 1.08 | 0.87–1.32 | 0.50 | 168 | 1.06 | 0.83–1.34 | 0.65 | 106 | 1.16 | 0.88–1.53 | 0.30 | ||
| AA+AG | 801 | 932 | Ref | 565 | Ref | 348 | Ref | ||||||||
| GG | 19 | 18 | 0.81 | 0.42–1.56 | 0.53 | 17 | 1.26 | 0.65–2.44 | 0.50 | 1 | 0.12 | 0.02–090 | 0.04 | ||
| Czech Republic | Austria | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | N a | OS | EFS | N a | OS | EFS | |||||
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | HR (95% CI) | p-Value | HR (95% CI) | p-Value | ||||
| Sex | Female | 696 | Ref | Ref | 389 | Ref | Ref | ||||
| Male | 1133 | 1.47 (1.20–1.80) | 0.0002 | 1.29 (1.09–1.52) | 0.003 | 561 | 1.37 (1.03–1.83) | 0.03 | 1.43 (1.11–1.83) | 0.005 | |
| Age (years) | <50 | 149 | Ref | Ref | 129 | Ref | Ref | ||||
| (50, 60] | 433 | 0.96 (0.62–1.50) | 0.87 | 1.06 (0.76–1.49) | 0.72 | 208 | 1.44 (0.77–2.69) | 0.26 | 1.62 (0.99–2.65) | 0.05 | |
| (60, 70] | 639 | 1.08 (0.71–1.65) | 0.72 | 0.90 (0.65–1.25) | 0.54 | 323 | 2.14 (1.21–3.79) | 0.01 | 1.68 (1.05–2.68) | 0.03 | |
| >70 | 610 | 1.47 (0.97–2.24) | 0.07 | 1.05 (0.76–1.46) | 0.77 | 291 | 3.11 (1.77–5.47) | <0.0001 | 2.53 (1.60–4.00) | <0.0001 | |
| BMI | (18.5, 25] | 434 | Ref | Ref | 296 | Ref | Ref | ||||
| <18.5 | 456 | 0.99 (0.77–1.27) | 0.92 | 1.06 (0.86–1.32) | 0.58 | 17 | 1.12 (0.41–3.07) | 0.83 | 1.31 (0.57–3.00) | 0.52 | |
| (25, 30] | 626 | 0.83 (066–1.06) | 0.13 | 0.94 (0.77–1.15) | 0.54 | 336 | 0.79 (0.55–1.12) | 0.18 | 0.85 (0.63–1.15) | 0.29 | |
| >30 | 315 | 0.58 (0.43–0.80) | 0.0008 | 0.83 (0.65–1.06) | 0.13 | 181 | 1.13 (0.77–1.65) | 0.54 | 1.04 (0.74–1.46) | 0.83 | |
| Smoking habit | No | 967 | Ref | Ref | 251 | Ref | Ref | ||||
| Yes b | 777 | 1.266 (1.049–1.529) | 0.01 | 1.27 (1.08–1.48) | 0.003 | 263 | 0.93 (0.65–1.32) | 0.67 | 1.02 (0.75–1.39) | 0.91 | |
| Stage | I | 277 | Ref | Ref | 188 | Ref | Ref | ||||
| II | 498 | 1.75 (1.10–2.80) | 0.02 | 1.99 (1.41–2.82) | 0.0001 | 227 | 1.00 (0.57–1.76) | 1.00 | 0.90 (0.56–1.45) | 0.67 | |
| III | 491 | 3.46 (2.22–5.39) | <0.0001 | 3.45 (2.47–4.83) | <0.0001 | 354 | 1.51 (0.92–2.45) | 0.10 | 1.55 (1.03–2.32) | 0.03 | |
| IV | 384 | 8.91 (5.78–13.74) | <0.0001 | 6.00 (4.30–8.38) | <0.0001 | 120 | 7.98 (4.95–12.88) | <0.0001 | 9.33 (6.21–14.02) | <0.0001 | |
| 5FU-based chemotherapy | No | 765 | Ref | Ref | 389 | Ref | Ref | ||||
| Yes | 797 | 1.022 (0.84–1.24) | 0.82 | 1.387 (1.18–1.63) | <0.0001 | 515 | 1.33 (0.99–1.78) | 0.06 | 1.79 (1.38–2.32) | <0.0001 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiraskova, K.; Hughes, D.J.; Brezina, S.; Gumpenberger, T.; Veskrnova, V.; Buchler, T.; Schneiderova, M.; Levy, M.; Liska, V.; Vodenkova, S.; et al. Functional Polymorphisms in DNA Repair Genes Are Associated with Sporadic Colorectal Cancer Susceptibility and Clinical Outcome. Int. J. Mol. Sci. 2019, 20, 97. https://doi.org/10.3390/ijms20010097
Jiraskova K, Hughes DJ, Brezina S, Gumpenberger T, Veskrnova V, Buchler T, Schneiderova M, Levy M, Liska V, Vodenkova S, et al. Functional Polymorphisms in DNA Repair Genes Are Associated with Sporadic Colorectal Cancer Susceptibility and Clinical Outcome. International Journal of Molecular Sciences. 2019; 20(1):97. https://doi.org/10.3390/ijms20010097
Chicago/Turabian StyleJiraskova, Katerina, David J. Hughes, Stefanie Brezina, Tanja Gumpenberger, Veronika Veskrnova, Tomas Buchler, Michaela Schneiderova, Miroslav Levy, Vaclav Liska, Sona Vodenkova, and et al. 2019. "Functional Polymorphisms in DNA Repair Genes Are Associated with Sporadic Colorectal Cancer Susceptibility and Clinical Outcome" International Journal of Molecular Sciences 20, no. 1: 97. https://doi.org/10.3390/ijms20010097
APA StyleJiraskova, K., Hughes, D. J., Brezina, S., Gumpenberger, T., Veskrnova, V., Buchler, T., Schneiderova, M., Levy, M., Liska, V., Vodenkova, S., Di Gaetano, C., Naccarati, A., Pardini, B., Vymetalkova, V., Gsur, A., & Vodicka, P. (2019). Functional Polymorphisms in DNA Repair Genes Are Associated with Sporadic Colorectal Cancer Susceptibility and Clinical Outcome. International Journal of Molecular Sciences, 20(1), 97. https://doi.org/10.3390/ijms20010097