Conserved miR396b-GRF Regulation Is Involved in Abiotic Stress Responses in Pitaya (Hylocereus polyrhizus)
Abstract
1. Introduction
2. Results
2.1. Cloning and Sequence Analysis of miR396b in Pitaya
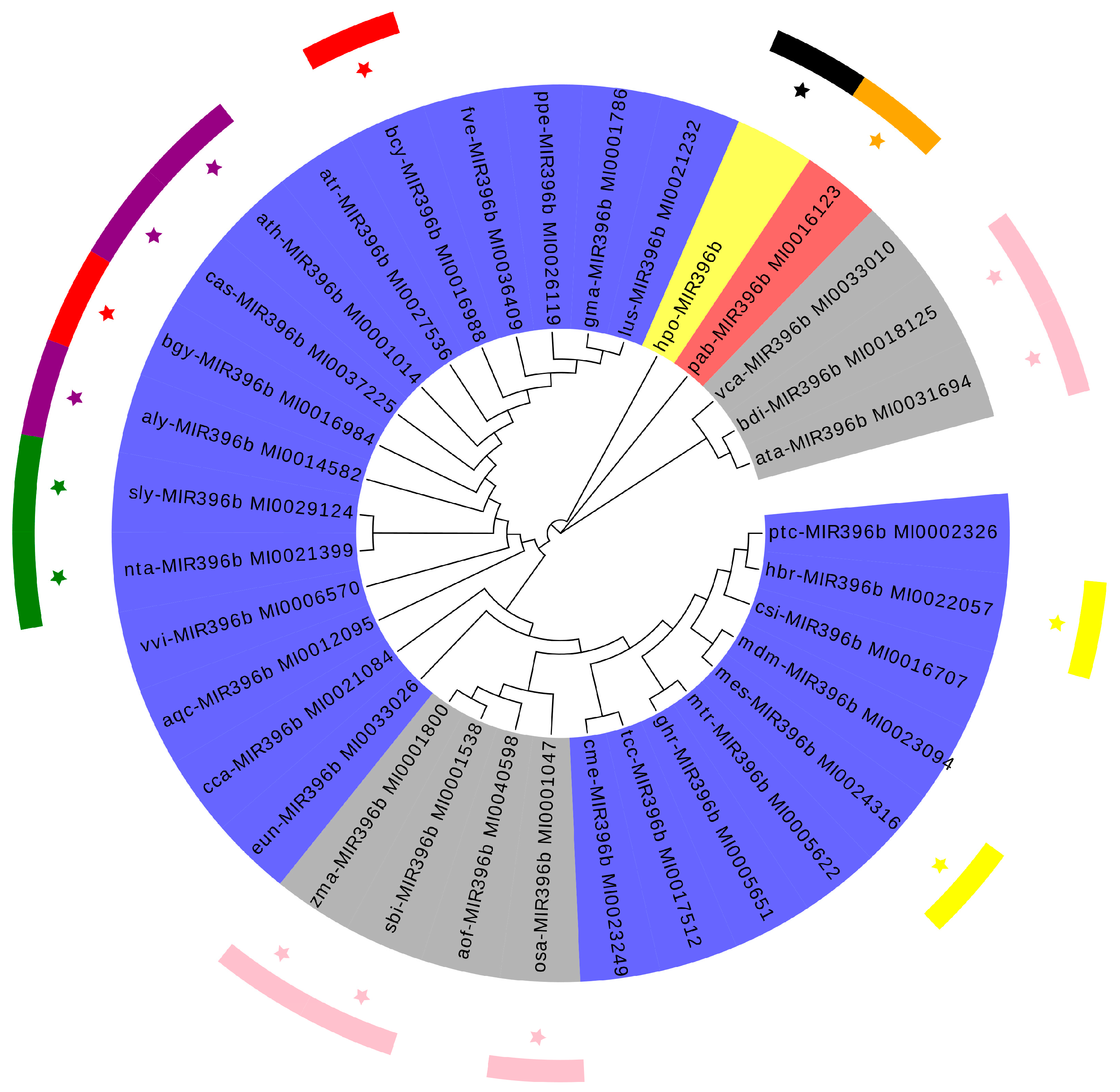
2.2. Phylogenetic Analysis of Plants MIR396b
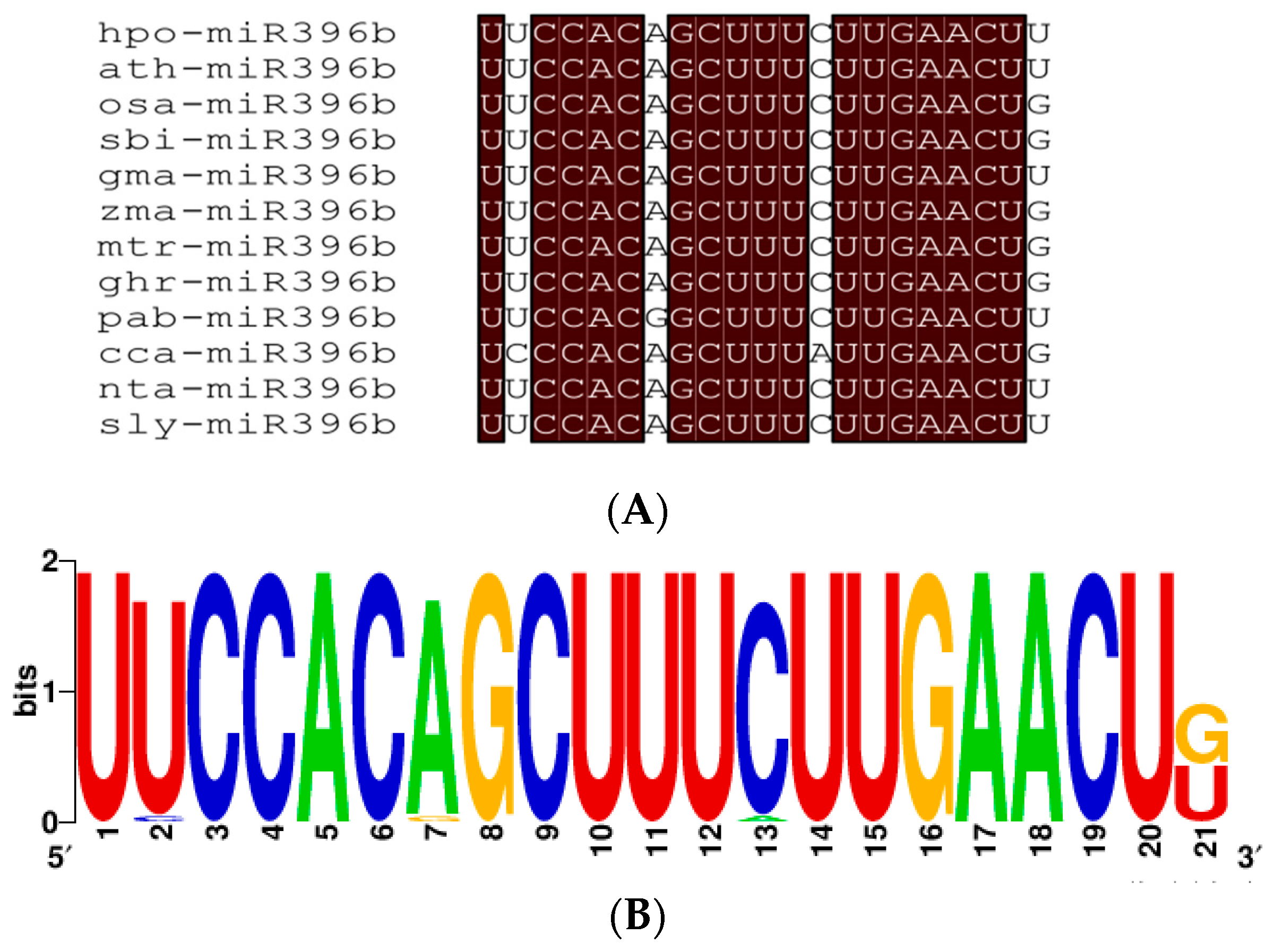
2.3. Conservation Analysis of Plants miR396b
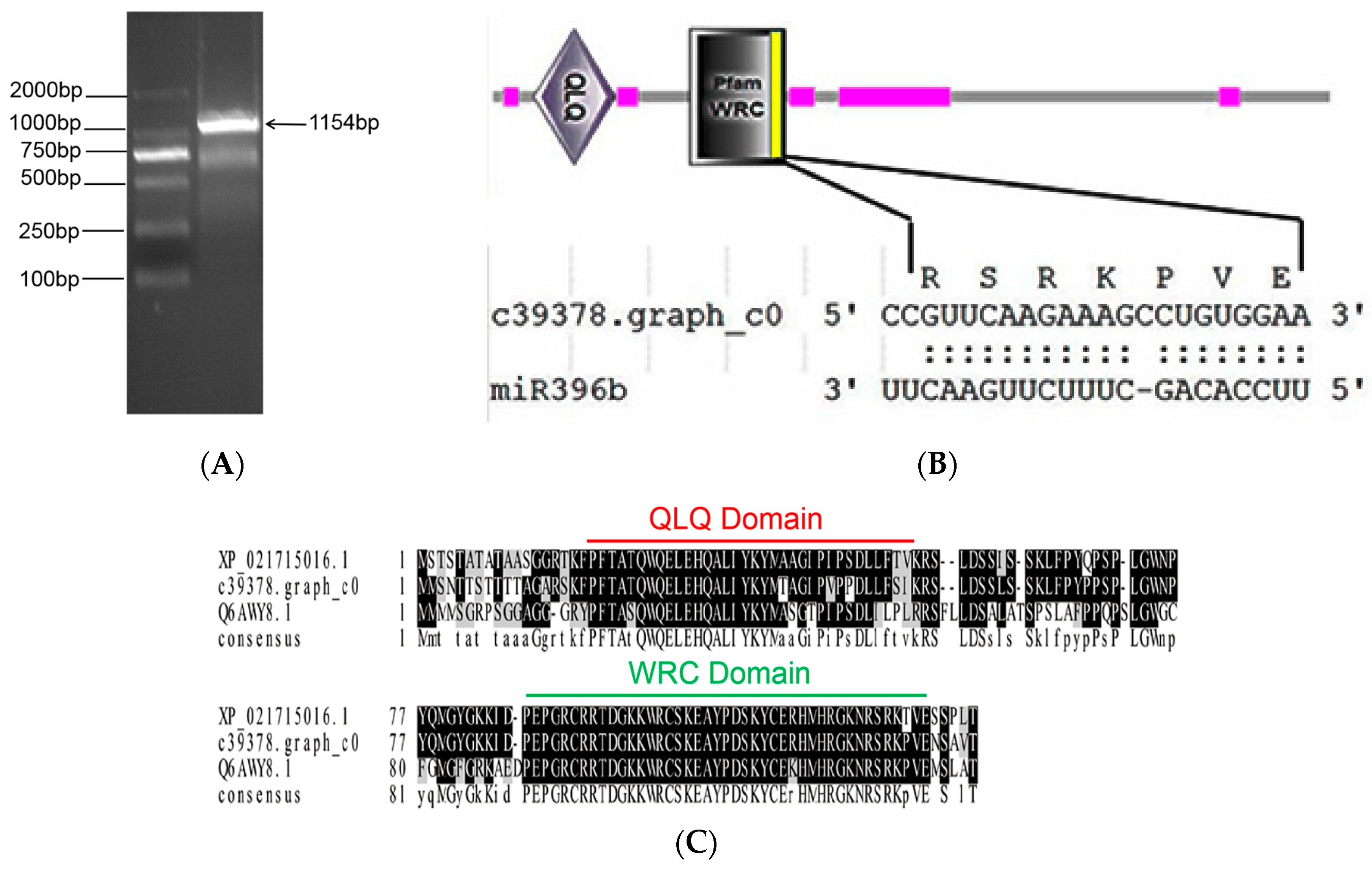
2.4. Prediction of Hpo-miR396b Target Genes
2.5. Cloning and Structure Analysis of HpGRF6
2.6. Subcellular Localization of HpGRF6 Protein
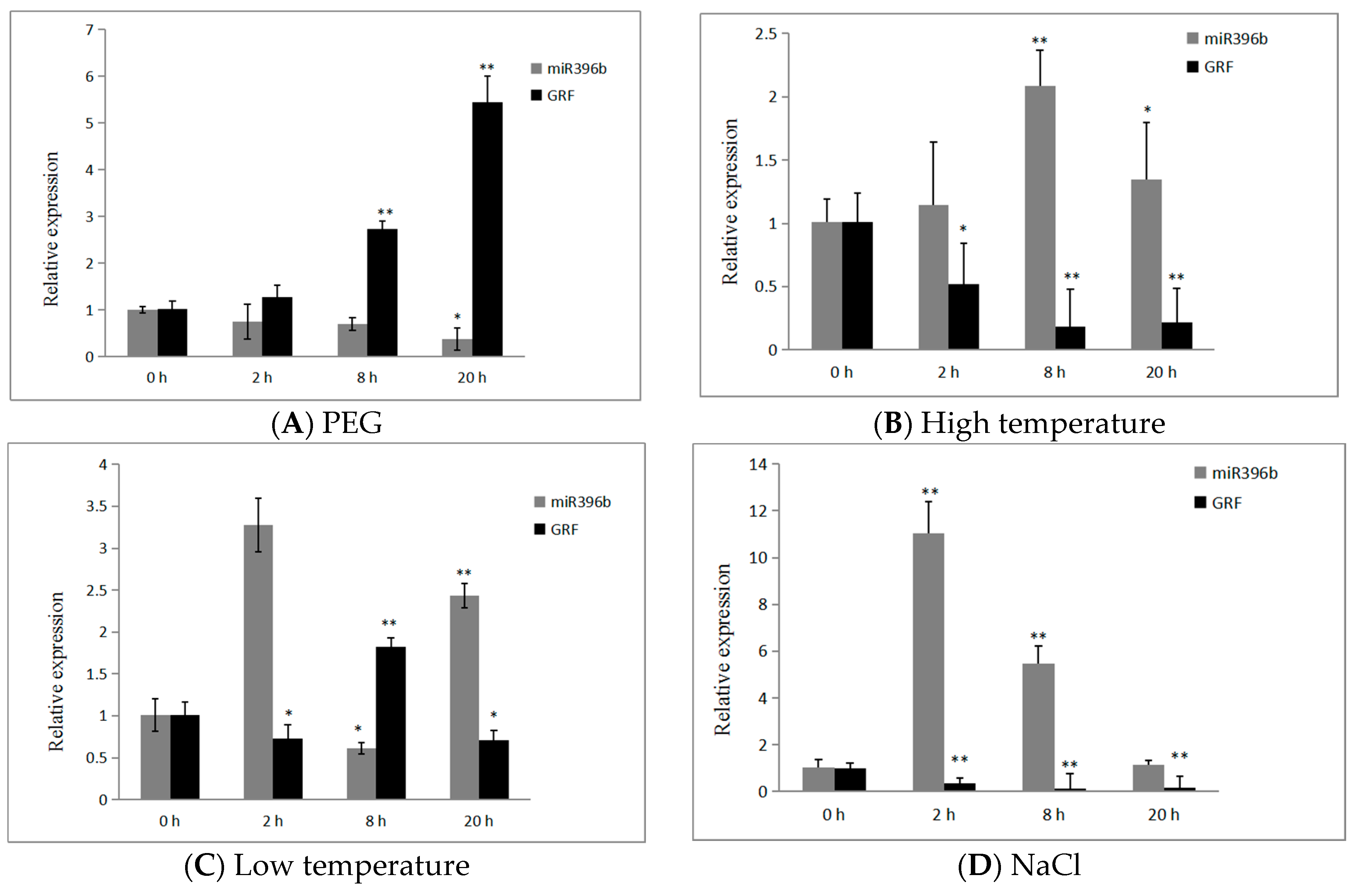
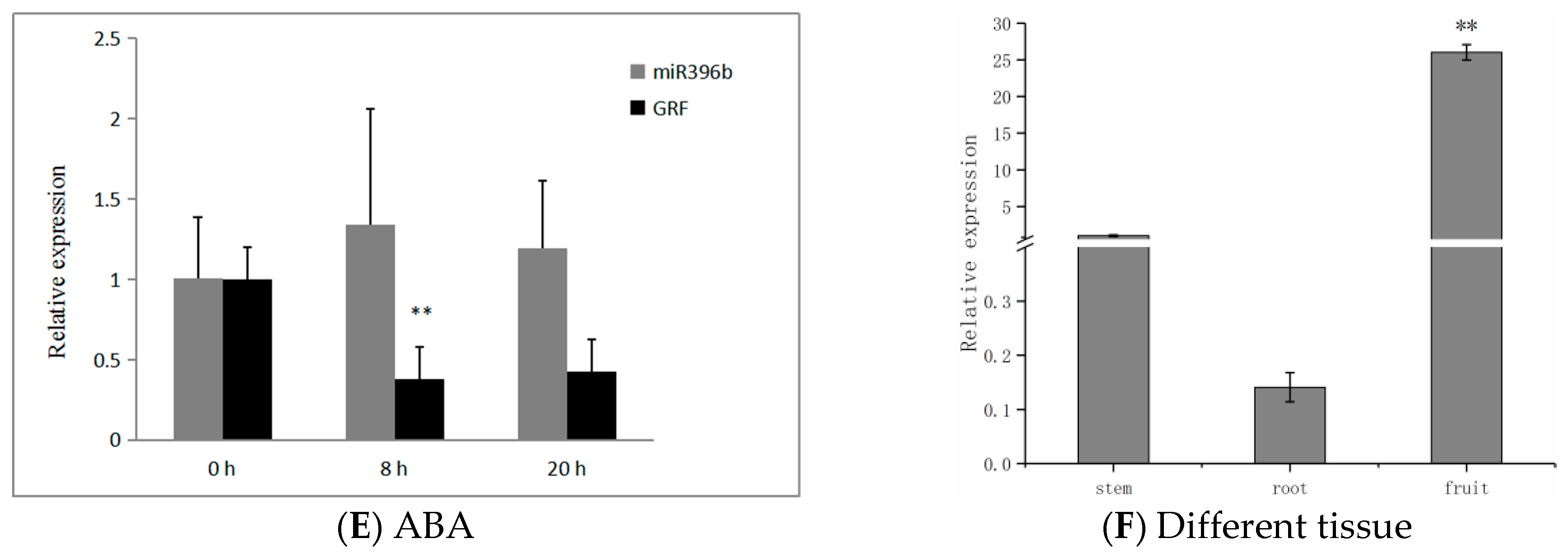
2.7. Expression of hpo-miR396b and HpGRF6 during Exposure to Abiotic Stresses
3. Discussion
3.1. Evolution and Conservation Analysis of Plants miR396b
3.2. Target Genes of miR396b
3.3. Tissue-Specific and Abiotic Stress Response of miR396 in Plant
4. Materials and Methods
4.1. Plant Material and Stress Treatment
4.2. Prediction of Hpo-miR396b Target Genes in Pitaya
4.3. Gene Cloning and Sequence Analysis
4.4. Bioinformatics Analysis
4.5. HpGRF6 Protein Subcellular Localization
4.6. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
4.7. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| GFP | Green fluorescent protein |
| GRF | Growth regulating factor |
| HpGRF | Hylocereus polyrhizu growth regulating factor |
| miRNA | Mature microRNA |
| MIRNA | Precursor microRNA |
| PCR | Polymerase chain reaction |
| PEG | Polyethylene glycol |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| RACE | Rapid amplification of cDNA ends |
| RT | Reverse transcription |
Appendix A
| Target Gene ID | Homologous Gene in A. thaliana | Encoding Protein | Annotation of the Target Protein |
|---|---|---|---|
| c56908.graph_c0 | AT2G22840 | AtGRF1, GRF1 | transcription factor |
| c43469.graph_c0 | AT4G37740 | AtGRF2, GRF2 | transcription factor |
| c52862.graph_c0 | AT3G52910 | GRF4, AtGRF4 | transcription factor |
| c39378.graph_c0 | AT2G06200 | GRF6, AtGRF6 | transcription factor |
| c21696.graph_c0 | AT4G24150 | AtGRF8, GRF8 | transcription factor |
| c54658.graph_c0 | AT2G45480 | AtGRF9, GRF9 | transcription factor |
| c56441.graph_c0 | AT2G20180 | PIF1, PIL5 | transcription factor |
| c57103.graph_c0 | AT4G30080 | ARF16 | Auxin response factor |
| c60026.graph_c0 | AT5G13220 | JAZ10, TIFY9, JAS1 | protein binding |
| c53553.graph_c0 | AT1G73960 | TFIID | transcription initiation factor |
| c49149.graph_c0 | AT5G15020 | SNL2 | - |
| c80735.graph_c0 | AT1G11870 | SRS, OVA7, ATSRS | Nucleotide binding |
| c60533.graph_c0 | AT3G14110 | FLU | - |
| c63676.graph_c0 | AT5G03960 | IQD12 | calmodulin binding |
| c62238.graph_c0 | AT3G12580 | HSP70 ATHSP70 | stress response protein |
| c67010.graph_c0 | AT4G09020 | ATISA3, ISA3 | Material metabolism |
| c56337.graph_c0 | AT1G04050 | CPR30 | Stress response protein |
| c81492.graph_c0 | AT2G31830 | 5PTase14 | Nuclear protein |
| c44847.graph_c0 | AT5G52460 | protein TONSOKU-like | Nuclear protein |
| Gene Name | Primer | Primer Sequence (5′→3′) | Use |
|---|---|---|---|
| hpo-MIR396b | Forward | ACCTTTCTCTCTCTCGTCTTCT | Gene Clone |
| Reversal | TGCGAGATGGAGAGGCAATT | ||
| HpGRF6 | Forward | TCTTGAAATGATGAGTAATACTACTTCTACTACAAC | Gene Clone |
| Reversal | TCCCACCTTCTCCCTTCTCTTGAAC | ||
| HpGRF6 | Forward | CAGTGGTCTCACAACATGATGAGTAATACTACTTC | Subcellular Localization |
| Reversal | CAGTGGTCTCATACAACCTCGTGATGATGAGGCC | ||
| hpo-miR396b | Stem-loop RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTCAAGA | cDNA Synthesis |
| Forward | CGGCGGTTCCACAGCTTTC | qRT-PCR | |
| Reversal | CCAGTGCAGGGTCCGAGGT | ||
| HpGRF6 | Forward | ACTGCTGGTATCCCTGTTCC | qRT-PCR |
| Reversal | TGCCTCTTTTGAACATCTCC | ||
| U6 | Forward | GGGGACATCCGATAAAATTGG | qRT-PCR |
| Reversal | GATTTGTGCGTGTCATCCTTG | ||
| RP40S | Forward | GACACTGATTCTCCTTTGCGTTAT | qRT-PCR |
| Reversal | CCTTTGGTCTCCTCTGGCTCT |
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Zhang, B.H.; Stellwag, E.J.; Pan, X.P. Large-scale genome analysis reveals unique features of microRNAs. Gene 2009, 443, 100–109. [Google Scholar] [CrossRef]
- Mallory, A.C.; Bouché, N. MicroRNA-directed regulation: To cleave or not to cleave. Trends Plant Sci. 2008, 13, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Bao, N.; Lye, K.W.; Barton, M.K. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 2004, 7, 653–662. [Google Scholar] [CrossRef]
- Khraiwesh, B.; Arif, M.A.; Seumel, G.I.; Ossowski, S.; Weigel, D.; Reski, R.; Frank, W. Transcriptional control of gene expression by microRNAs. Cell 2010, 140, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhou, H.Y.; Zhang, Q.Q.; Zhang, J.G.; Ni, F.R.; Liu, C.; Qi, Y.J. DNA methylation mediated by a microRNA pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar] [CrossRef]
- Xie, M.; Zhang, S.; Yu, B. MicroRNA biogenesis, degradation and activity in plants. Cell Mol. Life Sci. 2015, 72, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef]
- Elton, T.S.; Yalowich, J.C. Experimental procedures to identify and validate specific mRNA targets of miRNAS. EXCLI J. 2015, 14, 758–790. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309. [Google Scholar] [CrossRef]
- Peng, T.; Sun, H.; Du, Y.; Zhang, J.; Li, J.; Liu, Y.; Zhao, Y.; Zhao, Q. Characterization and expression patterns of microRNAs involved in rice grain filling. PLoS ONE 2013, 8, e54148. [Google Scholar] [CrossRef]
- Trindade, I.; Capitão, C.; Dalmay, T.; Fevereiro, M.P.; Santos, D.M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2009, 231, 705–716. [Google Scholar] [CrossRef]
- Zhao, M.; Ding, H.; Zhu, J.K.; Zhang, F.; Li, W.X. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011, 190, 906–915. [Google Scholar] [CrossRef]
- Rodriguez, R.E.; Mecchia, M.A.; Debernardi, J.M.; Schommer, C.; Weigel, D.; Palatnik, J.F. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 2010, 137, 103–112. [Google Scholar] [CrossRef]
- Vander Knaap, E.; Kim, J.H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant. J. 2003, 36, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Rice, J.H.; Chen, N.; Baum, T.J.; Hewezi, T. Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PLoS ONE 2014, 9, e98477. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef]
- Vaillant, F.; Perez, A.; Davila, I.; Dornier, M.; Reyne, M. Colorant and antioxidant properties of red-purple pitahaya (Hylocereus sp.). Fruits 2005, 60, 3–12. [Google Scholar] [CrossRef]
- Ramli, N.S.; Brown, L.; Ismail, P.; Rahmat, A. Effects of red pitaya juice supplementation on cardiovascular and hepatic changes in high-carbohydrate, high-fat diet-induced metabolic syndrome rats. BMC Complement. Altern. Med. 2014, 14, 189. [Google Scholar] [CrossRef]
- Fan, Q.J.; Yan, F.X.; Qiao, G.; Zhang, B.X.; Wen, X.P. Identification of differentially-expressed genes potentially implicated in drought response in pitaya (Hylocereus undatus) by suppression subtractive hybridization and cDNA microarray analysis. Gene 2014, 533, 322–331. [Google Scholar] [CrossRef]
- Nong, Q.; Zhang, M.; Chen, J.; Zhang, M.; Cheng, H.; Jian, S.; Lu, H.; Xia, K. RNA-Seq De Novo Assembly of Red Pitaya (Hylocereus polyrhizus) Roots and Differential Transcriptome Analysis in Response to Salt Stress. Tropical Plant. Biol. 2019, 1–12. [Google Scholar] [CrossRef]
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef]
- Tanzer, A.; Stadler, P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Lu, J.; Shen, Y.; Wu, Q.; Kumar, S.; He, B.; Shi, S.H.; Carthew, R.W.; Wang, S.M.; Wu, C.I. The birth and death of microRNA genes in Drosophila. Nat. Genet. 2008, 40, 351–355. [Google Scholar] [CrossRef]
- Felippes, F.F.; Schneeberger, K.; Dezulian, T.; Huson, D.H.; Weigel, D. Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA 2008, 14, 2455–2459. [Google Scholar] [CrossRef]
- Bentwich, I.; Avniel, A.; Karov, Y.; Aharonov, R.; Gilad, S.; Barad, O.; Barzilai, A.; Einat, P.; Einav, U.; Meiri, E.; et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005, 37, 766–770. [Google Scholar] [CrossRef]
- Alaba, S.; Piszczalka, P.; Pietrykowska, H.; Pacak, A.M.; Sierocka, I.; Nuc, P.W.; Singh, K.; Plewka, P.; Sulkowska, A.; Jarmolowski, A.; et al. The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytol. 2015, 206, 352–367. [Google Scholar] [CrossRef]
- Lin, P.C.; Lu, C.W.; Shen, B.N.; Lee, G.Z.; Bowman, J.L.; Arteaga-Vazquez, M.A.; Liu, L.-Y.D.; Hong, S.F.; Lo, C.F.; Su, G.M.; et al. Identification of miRNAs and their targets in the liverwort Marchantia polymorpha by integrating RNA-Seq and degradome analyses. Plant. Cell Physiol. 2016, 57, 339–358. [Google Scholar] [CrossRef]
- Thomson, D.W.; Bracken, C.P.; Goodall, G.J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011, 39, 6845–6853. [Google Scholar] [CrossRef]
- German, M.A.; Pillay, M.; Jeong, D.H.; Hetawal, A.; Luo, S.; Janardhanan, P.; Kannan, V.; Rymarquis, L.A.; Nobuta, K.; German, R. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008, 26, 941–946. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, Q.Q.; Zhou, H.Y.; Ni, F.R.; Wu, X.Y.; Qi, Y.J. Rice microRNA effector complexes and targets. Plant Cell 2009, 21, 3421–3435. [Google Scholar] [CrossRef]
- Pantaleo, V.; Szittya, G.; Moxon, S.; Miozzi, L.; Moulton, V.; Dalmay, T.; Burgyan, J. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 2010, 62, 960–976. [Google Scholar] [CrossRef]
- Addo-Quaye, C.; Eshoo, T.W.; Bartel, D.P.; Axtell, M.J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008, 18, 758–762. [Google Scholar] [CrossRef]
- Zhao, Y.P.; Xu, Z.H.; Mo, Q.C.; Zou, C.; Li, W.X.; Xu, Y.B.; Xie, C.X. Combined small RNA and degradome sequencing reveals novel miRNAs and their targets in response to low nitrate availability in maize. Ann. Bot. 2013, 112, 633–642. [Google Scholar] [CrossRef]
- Yang, X.Y.; Wang, L.C.; Yuan, D.J.; Lindsey, K.; Zhang, X.L. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J. Exp. Bot. 2013, 64, 1521–1536. [Google Scholar] [CrossRef]
- Shamimuzzaman, M.; Vodkin, L. Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genom. 2012, 13, 310. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, X.; Zhao, K.; Li, F.; Li, K.; Ning, L.; He, J.; Xin, Z.; Yin, D. Small RNA and degradome deep sequencing reveals the roles of microRNAs in seed expansion in peanut (Arachis hypogaea L.). Front. Plant Sci. 2018, 9, 349. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, Y.; Li, X.; Wang, X.; Dong, Y.; Wang, N.; Liu, X.; Chen, H.; Yao, N.; Cui, X. Tissue-Specific regulation of Gma-miR396 family on coordinating development and low water availability responses. Front. Plant Sci. 2017, 8, 1112. [Google Scholar] [CrossRef]
- Hewezi, T.; Maier, T.R.; Nettleton, D.; Baum, T.J. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 2012, 159, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Szécsi, J.; Joly, C.; Bordji, K.; Varaud, E.; Cock, J.M.; Dumas, C.; Bendahmane, M. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006, 25, 3912–3920. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.L.; Bian, H.W.; Zha, Y.L.; Li, F.Y.; Sun, Y.Z.; Bai, B.; Che, Z.H.; Wang, J.H.; Zhu, M.Y.; Han, N. miR396a-mediated basic helix-loop-helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol. 2014, 55, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, J.M.; Rodriguez, R.E.; Mecchia, M.A.; Palatnik, J.F. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 2012, 8, e1002419. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Cheng, C.; Lin, Y.; Zhu, Q.; Lin, J.; Lai, Z. Combined small RNA and degradome sequencing reveals complex microRNA regulation of catechin biosynthesis in tea (Camellia sinensis). PLoS ONE 2017, 12, e0171173. [Google Scholar] [CrossRef]
- Baucher, M.; Moussawi, J.; Vandeputte, O.M.; Monteyne, D.; Mol, A.; Pérez-Morga, D.; El-Jaziri, M. A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biol. 2012, 15, 892–898. [Google Scholar] [CrossRef]
- Bazin, J.; Khan, G.A.; Combier, J.P.; Bustos-Sanmamed, P.; Debernardi, J.M.; Rodriguez, R.; Sorin, C.; Palatnik, J.; Hartmann, C.; Crespi, M. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant. J. 2013, 74, 920–934. [Google Scholar] [CrossRef]
- Heidel, A.J.; Clarke, J.D.; Antonovics, J.; Dong, X.N. Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 2004, 168, 2197–2206. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant. Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef]
- Liu, D.M.; Song, Y.; Chen, Z.X.; Yu, D.Q. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 2009, 136, 223–236. [Google Scholar] [CrossRef]
- Beltramino, M.; Ercoli, M.F.; Debernardi, J.M.; Goldy, C.; Rojas, A.M.L.; Nota, F.; Alvarez, M.E.; Vercruyssen, L.; Inzé, D.; Palatnik, J.F.; et al. Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci. Rep. 2018, 8, 13447. [Google Scholar] [CrossRef]
- Chen, L.; Luan, Y.S.; Zhai, J.M. Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant. Cell Rep. 2015, 34, 2013–2025. [Google Scholar] [CrossRef]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.K.; Li, Y.; Cai, H.; Ji, W.; Guo, D.J.; Zhu, Y.M. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 2010, 231, 991–1001. [Google Scholar] [CrossRef]
- Noon, J.B.; Hewezi, T.; Baum, T.J. Homeostasis in the soybean miRNA396-GRF network is essential for productive soybean cyst nematode infections. J. Exp. Bot. 2019, 70, 1653–1668. [Google Scholar] [CrossRef]
- Casadevall, R.; Rodriguez, R.E.; Debernardi, J.M.; Palatnik, J.F.; Casati, P. Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell 2013, 25, 3570–3583. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef]
- Kiełbasa, S.M.; Blüthgen, N.; Fähling, M.; Mrowka, R. Targetfinder.org: A resource for systematic discovery of transcription factor target genes. Nucleic Acids Res. 2010, 38, W233–W238. [Google Scholar] [CrossRef]
- Li, X. Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio-Protocol 2011, 1, e95. [Google Scholar] [CrossRef]
- Chen, C.F.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.H.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Pant, B.D.; Musialak-Lange, M.; Nuc, P.; May, P.; Buhtz, A.; Kehr, J.; Walther, D.; Scheible, W.R. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant. Physiol. 2009, 150, 1541–1555. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, A.-L.; Wen, Z.; Yang, K.; Wen, X.-P. Conserved miR396b-GRF Regulation Is Involved in Abiotic Stress Responses in Pitaya (Hylocereus polyrhizus). Int. J. Mol. Sci. 2019, 20, 2501. https://doi.org/10.3390/ijms20102501
Li A-L, Wen Z, Yang K, Wen X-P. Conserved miR396b-GRF Regulation Is Involved in Abiotic Stress Responses in Pitaya (Hylocereus polyrhizus). International Journal of Molecular Sciences. 2019; 20(10):2501. https://doi.org/10.3390/ijms20102501
Chicago/Turabian StyleLi, A-Li, Zhuang Wen, Kun Yang, and Xiao-Peng Wen. 2019. "Conserved miR396b-GRF Regulation Is Involved in Abiotic Stress Responses in Pitaya (Hylocereus polyrhizus)" International Journal of Molecular Sciences 20, no. 10: 2501. https://doi.org/10.3390/ijms20102501
APA StyleLi, A.-L., Wen, Z., Yang, K., & Wen, X.-P. (2019). Conserved miR396b-GRF Regulation Is Involved in Abiotic Stress Responses in Pitaya (Hylocereus polyrhizus). International Journal of Molecular Sciences, 20(10), 2501. https://doi.org/10.3390/ijms20102501





