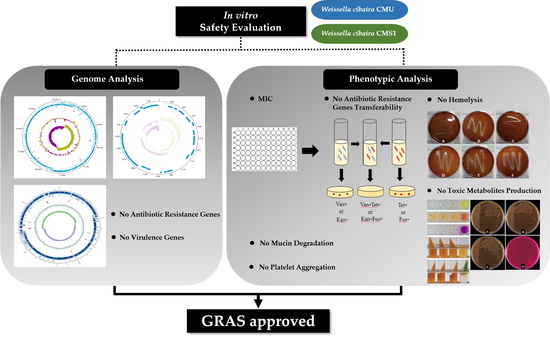
Safety Evaluation of Oral Care Probiotics Weissella cibaria CMU and CMS1 by Phenotypic and Genotypic Analysis
Abstract
:1. Introduction
2. Results
2.1. Antibiotic Resistance
2.2. PCR Detection for Antibiotic Resistance Genes (ARGs)
2.3. Transferability of ARGs
2.4. Genome Analysis for ARGs
2.5. Virulence Genes
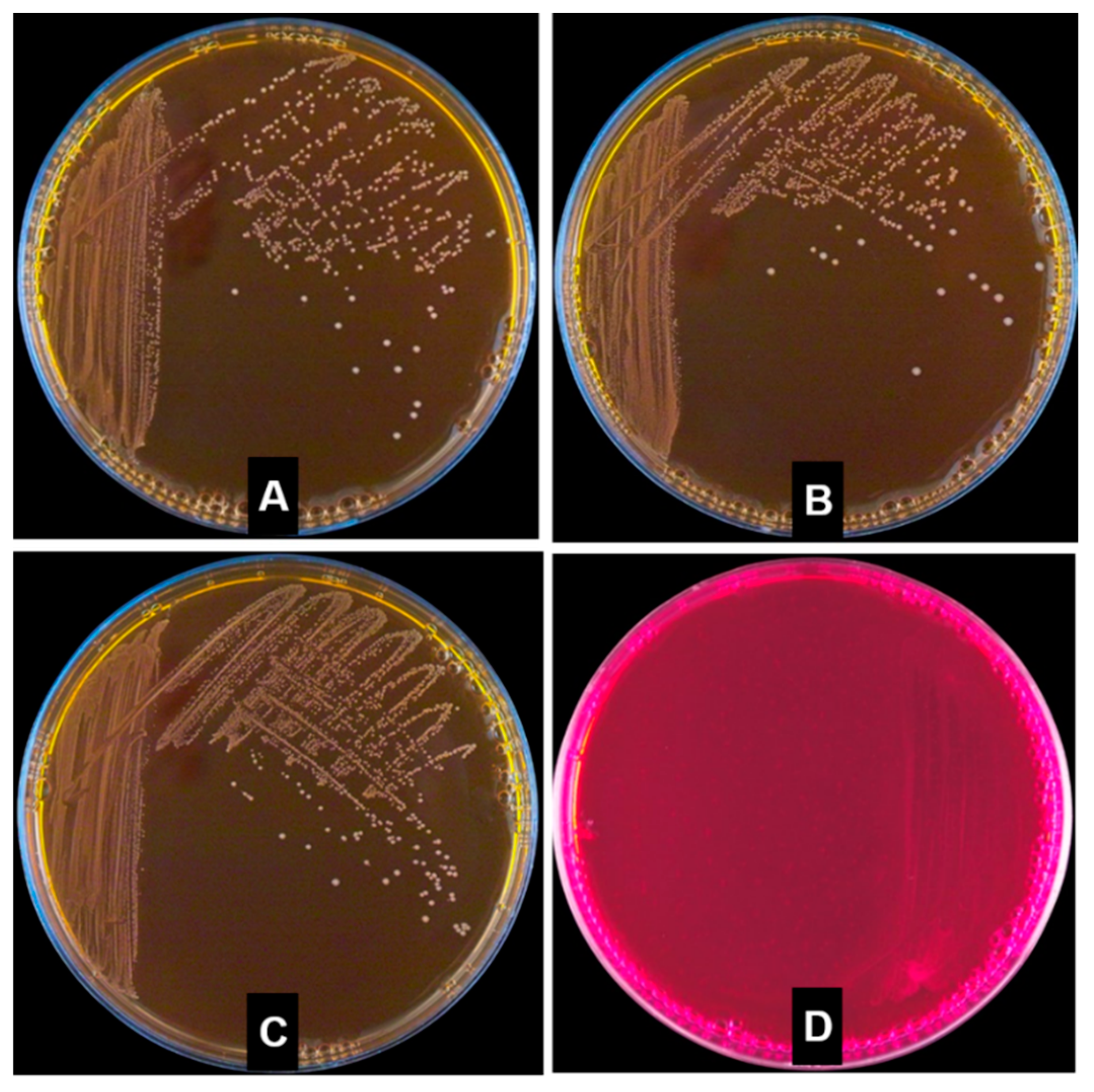
2.6. Hemolytic Properties
2.7. Mucin Degradation
2.8. Toxic Metabolite Production
2.9. Platelet Aggregation
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Antibiotic Resistance
4.2.1. Preparation of Antibiotics
4.2.2. Determination of Minimum Inhibitory Concentrations
4.2.3. ARG Detection by PCR
4.2.4. Antibiotic Resistance Transferability Test
4.2.5. Genome Analysis for ARGs
4.2.6. Detection of Virulence Genes
4.3. Hemolytic Activity Test
4.4. Mucin Degradation Test
4.5. Toxic Metabolite Production Test
4.5.1. d-Lactic Acid Production Test
4.5.2. Bile Salt Deconjugation Test
4.5.3. Urease Activity Test
4.5.4. β-Glucuronidase Activity Test
4.5.5. Indole Production Test
4.5.6. Nitroreductase Activity Test
4.5.7. Gelatin Liquefaction Test
4.5.8. Phenylalanine Degradation Test
4.6. Platelet Aggregation Test
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lilly, D.M.; Stillwell, R.H. Probiotics: Growth-promoting factors produced by microorganisms. Science 1965, 147, 747–748. [Google Scholar] [CrossRef] [PubMed]
- Behnsen, J.; Deriu, E.; Sassone-Corsi, M.; Raffatellu, M. Probiotics: Properties, examples, and specific applications. Cold Spring Harb. Perspect. Med. 2013, 3, a010074. [Google Scholar] [CrossRef] [PubMed]
- Global Market Insights Inc. Probiotics Market Size to Exceed USD 64 Billion by 2023: Global Market Insights Inc. Available online: https://www.prnewswire.com/news-releases/the-global-probiotics-market-size-is-expected-to-reach-usd-66-03-billion-by-2024--300726946.html (accessed on 4 April 2019).
- Ku, S.; Park, M.S.; Ji, G.E.; You, H.J. Review on bifidobacterium bifidum BGN4: Functionality and nutraceutical applications as a probiotic microorganism. Int. J. Mol. Sci. 2016, 17, 1544. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization-World Health Organization (FAO/WHO). Report on Joint FAO/WHO Guidelines for the Evaluation of Probiotics in Food. 2002. Available online: http://www.who.int/foodsafety/fs management/en/probiotic_guidelines.pdf (accessed on 4 April 2019).
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Romani Vestman, N.; Chen, T.; Lif Holgerson, P.; Öhman, C.; Johansson, I. Oral microbiota shift after 12-week supplementation with lactobacillus reuteri DSM 17938 and PTA 5289; a randomized control trial. PLoS ONE 2015, 10, e0125812. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, E.; Tecco, S.; Santonico, M.; Vernile, C.; Ciciarelli, D.; Tarantino, E.; Marzo, G.; Pennazza, G. Multi-sensor approach for the monitoring of halitosis treatment via lactobacillus brevis (CD2)-containing lozenges—A randomized, double-blind placebo-controlled clinical trial. Sensors 2015, 15, 19583–19596. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Yoneda, M.; Tanabe, K.; Fujimoto, A.; Iha, K.; Seno, K.; Yamada, K.; Iwamoto, T.; Masuo, Y.; Hirofuji, T. Lactobacillus salivarius WB21—Containing tablets for the treatment of oral malodor: A double-blind, randomized, placebo-controlled crossover trial. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2014, 117, 462–470. [Google Scholar] [CrossRef]
- Mahasneh, S.A.; Mahasneh, A.M. Probiotics: A promising role in dental health. Dent. J. 2017, 5, 26. [Google Scholar] [CrossRef]
- Allaker, R.P.; Stephen, A.S. Use of probiotics and oral health. Curr. Oral Health Rep. 2017, 4, 309–318. [Google Scholar] [CrossRef]
- Kang, M.S.; Chung, J.; Kim, S.M.; Yang, K.H.; Oh, J.S. Effect of Weissella cibaria isolates on the formation of Streptococcus mutans biofilm. Caries Res. 2006, 40, 418–425. [Google Scholar] [CrossRef]
- Kang, M.S.; Kim, B.G.; Chung, J.; Lee, H.C.; Oh, J.S. Inhibitory effect of Weissella cibaria isolates on the production of volatile sulphur compounds. J. Clin. Periodontol. 2006, 33, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Do, K.H.; Park, H.E.; Kang, M.S.; Kim, J.T.; Yeu, J.E.; Lee, W.K. Effects of Weissella cibaria CMU on Halitosis and Calculus, Plaque, and Gingivitis Indices in Beagles. J. Vet. Dent. 2019. accepted. [Google Scholar]
- Lim, H.S.; Yeu, J.E.; Hong, S.P.; Kang, M.S. Characterization of antibacterial cell-free supernatant from oral care probiotic Weissella cibaria, CMU. Molecules 2018, 23, 1984. [Google Scholar] [CrossRef] [PubMed]
- List of Raw Materials Available for Food. Available online: https://www.foodsafetykorea.go.kr/foodcode/01_03.jsp?idx=12135 (accessed on 4 April 2019).
- EFSA Panel on Additives and Products of Substances used in Animal Feed (FEEDAP). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- Woodford, N.; Ellington, M.J. The emergence of antibiotic resistance by mutation. Clin. Microbiol. Infect. 2007, 13, 5–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, S.; Singh, R. Antibiotic resistance in food lactic acid bacteria—A review. Int. J. Food Microbiol. 2005, 105, 281–295. [Google Scholar] [CrossRef]
- Choi, A.R.; Patra, J.K.; Kim, W.; Kang, S.S. Antagonistic activities and probiotic potential of lactic acid bacteria derived from a plant-based fermented food. Front. Microbiol. 2018, 9, 1963. [Google Scholar] [CrossRef]
- Wang, J.; Wei, X.; Fan, M. Assessment of antibiotic susceptibility within lactic acid bacteria and coagulase-negative staphylococci isolated from hunan smoked pork, a naturally fermented meat product in China. J. Food Sci. 2018, 83, 1707–1715. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Mardassi, B.B.; Aissani, N.; Moalla, I.; Dhahri, D.; Dridi, A.; Mlik, B. Evidence for the predominance of a single tet(M) gene sequence type in tetracycline-resistant Ureaplasma parvum and Mycoplasma hominis isolates from tunisian patients. J. Med. Microbiol. 2012, 61, 1254–1261. [Google Scholar] [CrossRef]
- Bahl, M.I.; Sørensen, S.J.; Hansen, L.H.; Licht, T.R. Effect of tetracycline on transfer and establishment of the tetracycline-inducible conjugative transposon Tn916 in the guts of gnotobiotic rats. Appl. Environ. Microbiol. 2004, 70, 758–764. [Google Scholar] [CrossRef]
- Scornec, H.; Bellanger, X.; Guilloteau, H.; Groshenry, G.; Merlin, C. Inducibility of Tn916 conjugative transfer in Enterococcus faecalis by subinhibitory concentrations of ribosome-targeting antibiotics. J. Antimicrob. Chemother. 2017, 72, 2722–2728. [Google Scholar] [CrossRef] [PubMed]
- Giraffa, G. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 2003, 88, 215–222. [Google Scholar] [CrossRef]
- Olawale, K.O.; Fadiora, S.O.; Taiwo, S.S. Prevalence of hospital-acquired enterococci infections in two primary-care hospitals in Osogbo, Southwestern Nigeria. Afr. J. Infect. Dis. 2011, 5, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.M.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottschalk, M.G.; Lacouture, S.; Dubreuil, J.D. Characterization of Streptococcus suis capsular type 2 haemolysin. Microbiology 1995, 141, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Prakash, R.; Bharathi Raja, S.; Devaraj, H.; Devaraj, S.N. Up-Regulation of MUC2 and IL-1β expression in human colonic epithelial cells by Shigella and its interaction with mucins. PLoS ONE 2011, 6, e27046. [Google Scholar] [CrossRef] [PubMed]
- Domellöf, L.; Reddy, B.S.; Weisburger, J.H. Microflora and deconjugation of bile acids in alkaline reflux after partial gastrectomy. Am. J. Surg. 1980, 140, 291–295. [Google Scholar] [CrossRef]
- Begley, M.; Hill, C.; Gahan, C.G. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef]
- Muñoz-Atienza, E.; Gómez-Sala, B.; Araújo, C.; Campanero, C.; del Campo, R.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Antimicrobial activity, antibiotic susceptibility and virulence factors of lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 2013, 13, 15. [Google Scholar] [CrossRef]
- Adeva, M.; González-Lucán, M.; Seco, M.; Donapetry, C. Enzymes involved in l-lactate metabolism in humans. Mitochondrion 2013, 13, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Vitetta, L.; Coulson, S.; Thomsen, M.; Nguyen, T.; Hall, S. Probiotics, D-Lactic acidosis, oxidative stress and strain specificity. Gut Microbes 2017, 8, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Isenberg, H.D. Urease. In Clinical Microbiology Procedures Handbook; American Society of Microbiology: Washington, DC, USA, 1992; Volume 1, p. 2.6.8. [Google Scholar]
- Heavey, P.M.; Rowland, I.R. Microbial-gut interactions in health and disease. Gastrointestinal cancer. Best Pract. Res. Clin. Gastroenterol. 2004, 18, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Snell, E.E. Reversibility of the Tryptophanase reaction: Synthesis of tryptophan from indole, pyruvate, and ammonia. Proc. Natl. Acad. Sci. USA 1972, 69, 1086–1090. [Google Scholar] [CrossRef] [PubMed]
- Rafil, F.; Franklin, W.; Heflich, R.H.; Cerniglia, C.E. Reduction of nitroaromatic compounds by anaerobic bacteria isolated from the human gastrointestinal tract. Appl. Environ. Microbiol. 1991, 57, 962–968. [Google Scholar] [PubMed]
- Zemelman, R.; Longeri, L. Characterization of staphylococci isolated from raw milk. Appl. Microbiol. 1965, 13, 167–170. [Google Scholar] [PubMed]
- Korpela, R.; Moilanen, E.; Saxelin, M.; Vapaatalo, H. Lactobacillus rhamnosus GG (ATCC 53103) and platelet aggregation in vitro. Int. J. Food Microbiol. 1997, 37, 83–86. [Google Scholar] [CrossRef]
- Favaloro, E.J. Clinical utility of the PFA-100. Semin. Thromb. Hemost. 2008, 34, 709–733. [Google Scholar] [CrossRef]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef]
- Björkroth, K.J.; Schillinger, U.; Geisen, R.; Weiss, N.; Hoste, B.; Holzapfel, W.H.; Korkeala, H.J.; Vandamme, P. Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov., detected in food and clinical samples. Int. J. Syst. Evol. Microbiol. 2002, 52, 141–148. [Google Scholar]
- Kwak, S.H.; Cho, Y.M.; Noh, G.M.; Om, A.S. Cancer preventive potential of kimchi lactic acid bacteria (Weissella cibaria, Lactobacillus plantarum). J. Cancer Prev. 2014, 19, 253–258. [Google Scholar] [CrossRef]
- Lee, H.A.; Song, B.R.; Kim, H.R.; Kim, J.E.; Yun, W.B.; Park, J.J.; Lee, M.L.; Choi, J.Y.; Lee, H.S.; Hwang, D.Y. Butanol extracts of Asparagus cochinchinensis fermented with Weissella cibaria inhibit iNOS-mediated COX-2 induction pathway and inflammatory cytokines in LPS-stimulated RAW264.7 macrophage cells. Exp. Ther. Med. 2017, 14, 4986–4994. [Google Scholar] [CrossRef]
- Hong, Y.F.; Lee, Y.D.; Park, J.Y.; Kim, S.; Lee, Y.W.; Jeon, B.; Jagdish, D.; Kim, H.; Chung, D.K. Lipoteichoic acid isolated from Weissella cibaria increases cytokine production in human monocyte-like THP-1 cells and mouse splenocytes. J. Microbiol. Biotechnol. 2016, 26, 1198–1205. [Google Scholar] [CrossRef]
- Baruah, R.; Maina, N.H.; Katina, K.; Juvonen, R.; Goyal, A. Functional food applications of dextran from Weissella cibaria RBA12 from pummelo (Citrus maxima). Int. J. Food Microbiol. 2017, 242, 124–131. [Google Scholar] [CrossRef]
- Abriouel, H.; Lerma, L.L.; Casado Muñoz Mdel, C.; Montoro, B.P.; Kabisch, J.; Pichner, R.; Cho, G.S.; Neve, H.; Fusco, V.; Franz, C.M.; et al. The controversial nature of the Weissella genus: Technological and functional aspects versus whole genome analysis-based pathogenic potential for their application in food and health. Front. Microbiol. 2015, 6, 1197. [Google Scholar] [CrossRef]
- International Organization for Standardization (ISO). Milk and Milk Products-Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB); ISO 10932:2010 (IDF 223:2010); ISO: Geneve, Switzerland, 2010. [Google Scholar]
- Rojo-Bezares, B.; Sáenz, Y.; Poeta, P.; Zarazaga, M.; Ruiz-Larrea, F.; Torres, C. Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. Int. J. Food Microbiol. 2006, 111, 234–240. [Google Scholar] [CrossRef]
- Bujnakova, D.; Strakova, E.; Kmet, V. In vitro evaluation of the safety and probiotic properties of lactobacilli isolated from chicken and calves. Anaerobe 2014, 29, 118–127. [Google Scholar] [CrossRef]
- Hummel, A.S.; Hertel, C.; Holzapfel, W.H.; Franz, C.M. Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl. Environ. Microbiol. 2007, 73, 730–739. [Google Scholar] [CrossRef]
- Ouoba, L.I.; Lei, V.; Jensen, L.B. Resistance of potential probiotic lactic acid bacteria and bifidobacteria of african and european origin to antimicrobials: Determination and transferability of the resistance genes to other bacteria. Int. J. Food Microbiol. 2008, 121, 217–224. [Google Scholar] [CrossRef]
- Zhou, N.; Zhang, J.X.; Fan, M.T.; Wang, J.; Guo, G.; Wei, X.Y. Antibiotic resistance of lactic acid bacteria isolated from chinese yogurts. J. Dairy Sci. 2012, 95, 4775–4783. [Google Scholar] [CrossRef]
- Kastner, S.; Perreten, V.; Bleuler, H.; Hugenschmidt, G.; Lacroix, C.; Meile, L. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst. Appl. Microbiol. 2006, 29, 145–155. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Z.Y.; Dong, K.; Yuan, J.P.; Guo, X.K. Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomed. Environ. Sci. 2009, 22, 401–412. [Google Scholar] [CrossRef]
- Aquilanti, L.; Garofalo, C.; Osimani, A.; Silvestri, G.; Vignaroli, C.; Clementi, F. Isolation and molecular characterization of antibiotic-resistant lactic acid bacteria from poultry and swine meat products. J. Food Prot. 2007, 70, 557–565. [Google Scholar] [CrossRef]
- Gad, G.F.; Abdel-Hamid, A.M.; Farag, Z.S. Antibiotic resistance in lactic acid bacteria isolated from some pharmaceutical and dairy products. Braz. J. Microbiol. 2014, 45, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Pan, L.; Li, L.; Lu, J.; Kwok, L.; Menghe, B.; Zhang, H.; Zhang, W. characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J. Food Sci. 2017, 82, 724–730. [Google Scholar] [CrossRef]
- Morales, G.; Picazo, J.J.; Baos, E.; Candel, F.J.; Arribi, A.; Peláez, B.; Andrade, R.; de la Torre, M.A.; Fereres, J.; Sánchez-García, M. Resistance to linezolid is mediated by the cfr Gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 2010, 50, 821–825. [Google Scholar] [CrossRef]
- Macovei, L.; Zurek, L. Ecology of antibiotic resistance genes: Characterization of enterococci from houseflies collected in food settings. Appl. Environ. Microbiol. 2006, 72, 4028–4035. [Google Scholar] [CrossRef]
- Tannock, G.W. Conjugal transfer of plasmid pAM β1 in Lactobacillus reuteri and between lactobacilli and Enterococcus faecalis. Appl. Environ. Microbiol. 1987, 53, 2693–2695. [Google Scholar]
- Liu, B.; Pop, M. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Micobiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]







| Antibiotics | EFSA Cut-Off (mg/L) a | CMU | CMS1 | Lr | Ls | Lrh | Ef | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lactobacillus Obligate Heterofermentative | L. reuteri | L. salivarius | L. rhamnosus | Enterococcus | |||||||
| AMP | 4 | 2 | 4 | 4 | 4 | 0.5 | 0.5 | 0.5 | 0.5 | 1 | 1 |
| VAN | N/R | N/R | N/R | N/R | 4 | >256 | >256 | 256 | >256 | >256 | 2 |
| GEN | 16 | 8 | 16 | 16 | 32 | 16 | 4 | 8 | 8 | 8 | 256 |
| KAN | 32 | 64 | 64 | 64 | 512 | 128 | 32 | 128 | 128 | 32 | 256 |
| STR | 64 | 64 | 64 | 32 | 128 | 64 | 16 | 32 | 32 | 8 | >256 |
| ERY | 1 | 1 | 1 | 1 | 4 | 0.03 | 0.03 | 0.25 | 0.03 | 0.03 | 2 |
| CLIN | 1 | 1 | 1 | 1 | 4 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 16 |
| TET | 8 | 16 | 8 | 8 | 2 | 8 | 4 | 4 | 1 | 0.25 | 32 |
| CHL | 4 | 4 | 4 | 4 | 8 | 4 | 4 | 4 | 2 | 1 | 4 |
| FUS | 32 | 16 | 8 | 2 | 256 | 4 | |||||
| OXY | 8 | 4 | 4 | 0.5 | 0.1 | 8 | |||||
| RIF | 16 | 8 | 8 | 0.5 | 0.1 | 2 | |||||
| CIP | 2 | 2 | 4 | 1 | 0.25 | 1 | |||||
| LIN | 2 | 1 | 2 | 0.5 | 0.5 | 2 | |||||
| Antibiotics | Target Genes | CMU | CMS1 | Lr | Ls | Lrh | Ef |
|---|---|---|---|---|---|---|---|
| GEN | aac(6′)-aph(2”) | - | - | - | - | - | - |
| aac(6′)Ie-aph(2”)La | - | - | - | - | - | - | |
| CHL | catA | - | - | - | - | - | - |
| cat | - | - | - | - | - | - | |
| STR | aadA | - | - | - | - | - | - |
| aadE | - | - | - | - | - | - | |
| ant(6) | - | - | - | - | - | - | |
| dfrD | - | - | - | - | - | - | |
| VAN | vanE | - | - | - | - | - | - |
| vanX | - | - | - | - | - | - | |
| AMP | blaZ | - | - | - | - | - | - |
| bla | - | - | - | - | - | - | |
| mecA | - | - | - | - | - | - | |
| TET | tet(M) | - | - | - | - | - | + |
| tet(K) | - | - | - | - | - | - | |
| tet(W) | - | - | - | - | - | - | |
| RIF | rpoB | - | - | - | - | - | - |
| KAN | aph(3”)-III | - | - | - | - | - | - |
| ant(2”)-I | - | - | - | - | - | - | |
| aph(3”)-I | - | - | - | - | - | - | |
| CIP | gyrA | - | - | - | - | - | - |
| parC | - | - | - | - | - | - | |
| CLIN | lnu(A) | - | - | - | - | - | - |
| lnu(B) | - | - | - | - | - | - | |
| ERY | erm(B) | - | - | - | - | - | - |
| erm(B)-1 | - | - | - | - | - | - | |
| erm(C) | - | - | - | - | - | - | |
| LIN | cfr | - | - | - | - | - | - |
| Conjugative transposon integrons | Tn916/Tn1545 | - | - | - | - | - | + |
| Antibiotics | Donor | Recipients | Transconjugants | Transfer Frequency | ||
|---|---|---|---|---|---|---|
| CMU | Ef | Lrh | CMU + Ef | CMU + Lrh | ||
| Van 64 | 1.03 × 108 ± 8.41 × 107 | 0 | - | - | - | - |
| Tet 16 | 0 | 3.70 × 108 ± 3.68 × 108 | - | - | - | - |
| Van 64 + Tet 16 | 0 | 0 | - | 0 | - | 0 |
| Kan 64 | 2.50 × 106 ± 7.07 × 105 | - | 0 | - | - | - |
| Fus 64 | 0 | - | 2.15 × 109 ± 7.07 × 106 | - | - | - |
| Kan 64 + Fus 64 | 0 | - | 0 | - | 0 | 0 |
| Name | Accession No. | RGI Criteria | Perfect Hits | Strict Hits | Loose Hits |
|---|---|---|---|---|---|
| W. cibaria CMU, complete genome | CP013936 | Perfect, Strict, complete genes only | 0 | 0 | 0 |
| W. cibaria CMS1, complete genome | CP022606 | 0 | 0 | 0 | |
| L. rhamnosus GG, complete genome | 013198 | 0 | 0 | 0 | |
| E. faecalis ATCC 29212, complete genome | CP008816 | 0 | 2 | 0 | |
| RGI Criteria * | Strict | Strict | |||
| ARO | dfrE | tet(W/N/W) | |||
| Detection Criteria | Protein homologous model | Protein homologous model | |||
| AMR Gene family | Trimethoprim resistant dihydrofolate reductase dfr | Tetracycline-ribosomal protection protein | |||
| Drug Class | Diaminopyrimidine antibiotic | Tetracycline antibiotic | |||
| Resistance mechanism | Antibiotic target replacement | Antibiotic target protection | |||
| % Identity of Matching Region | 98.78 | 68.65 | |||
| % Length of Reference Sequence | 100.00 | 100.00 | |||
| Virulence Genes | CMU | CMS1 | Lrh | Ef | |
| Shiga-toxin genes for E. coli | - | - | - | - | |
| Virulence genes for E. coli | - | - | - | - | |
| Virulence genes for Listeria | - | - | - | - | |
| Exoenzyme genes for S. aureus | - | - | - | - | |
| Toxin genes for S. aureus | - | - | - | - | |
| Hostimm genes for S. aureus | - | - | - | - | |
| Virulence genes for Enterococcus | - | - | - | + | |
| Virulence Factor * | Identity | Query/Template Length | Position in Contig | Protein Function | Accession Number |
| ElrA | 99.68 | 2172/2172 | 1,447,067.. 1,449,238 | CP003726.1 | |
| SrtA | 100 | 735/735 | 1,813,853.. 1,814,587 | CP003726.1 | |
| ace | 97.76 | 1743/1743 | 157,931.. 159,673 | Collagen adhesin precursor | AF260879.1 |
| cCF10 | 99.88 | 828/828 | 2,112,855.. 2,113,662 | CP002491.1 | |
| cOB1 | 100 | 819/819 | 1,298,659.. 1,299,477 | CP002621.1 | |
| cad | 100 | 930/930 | 2,022,216.. 2,023,145 | CP003726.1 | |
| camE | 99.6 | 501/501 | 406,237.. 406,737 | Sex pheromone cAM373 precursor | AF435437.1 |
| ebpA | 99.97 | 3312/3312 | 147,276.. 150,587 | CP002491.1 | |
| ebpB | 99.86 | 1431/1431 | 150,591.. 152,021 | CP002491.1 | |
| efaAfs | 100 | 927/927 | 995,578.. 996,504 | FP929058.1 | |
| gelE | 99.87 | 1530/1530 | 824,819.. 826,348 | CP002491.1 | |
| hylA | 99.42 | 3266/3264 | 1,790,825.. 1,794,090 | CP002491.1 | |
| tpx | 100 | 510/510 | 1,704,855.. 1,705,364 | CP002621.1 | |
| Enzyme | CMU | CMS1 | Lr | Ls | Lrh | Ef | Li |
|---|---|---|---|---|---|---|---|
| Alkaline phosphatase | - | - | - | - | - | - | - |
| Esterase (C4) | - | - | + | - | - | + | + |
| Esterase lipase (C8) | - | - | - | - | + | - | - |
| Lipase (C14) | - | - | - | - | - | - | - |
| Leucine arylamidase | - | - | + | + | + | + | - |
| Valine arylamidase | - | - | - | - | + | - | - |
| Cystine arylamidase | - | - | - | - | - | - | - |
| Trypsin | - | - | - | - | - | - | - |
| α-chymotrypsin | - | - | - | - | + | + | - |
| Acid phosphatase | + | + | + | + | + | + | + |
| Naphthol-AS-BI-phosphohydrolase | + | + | + | + | + | + | + |
| α-galactosidase | - | - | + | + | - | - | - |
| β-galactosidase | - | - | + | + | + | - | - |
| β-glucuronidase | - | - | - | - | - | - | - |
| α-glucosidase | - | - | + | - | + | - | - |
| β-glucosidase | - | - | - | - | + | - | + |
| N-acetyl-β-glucosaminidase | - | - | - | - | - | - | + |
| α-mannosidase | - | - | - | - | - | - | - |
| α-fucosidase | - | - | - | - | + | - | - |
| Antibiotics | Target Genes | Oligo Sequences (5′–3′) | Annealing Temperature (°C) | Amplicon Size (bp) | References |
|---|---|---|---|---|---|
| GEN | aac(6′)-aph(2”) | CCAAGAGCAATAAGGGCATA | 60 | 220 | [51] |
| CACTATCATAACCACTACCG | |||||
| aac(6′)Ie-aph(2”)La | CAGAGCCTTGGGAAGATGAAG | 58 | 348 | [52] | |
| CCTCGTGTAATTCATGTTCTGGC | |||||
| CHL | catA | GGATATGAAATTTATCCCTC | 50 | 486 | [51] |
| CAATCATCTACCCTATGAAT | |||||
| cat | TTAGGTTATTGGGATAAGTTA | 48 | 300 | [53] | |
| GCATGRTAACCATCACAWAC | |||||
| STR | aadA | ATCCTTCGGCGCGATTTTG | 56 | 282 | [54] |
| GCAGCGCAATGACATTCTTG | |||||
| aadE | ATGGAATTATTCCCACCTGA | 50 | 565 | [54] | |
| TCAAAACCCCTATTAAAGCC | |||||
| ant(6) | ACTGGCTTAATCAATTTGGG | 53 | 597 | [55] | |
| GCCTTTCCGCCACCTCACCG | |||||
| VAN | vanE | TGTGGTATCGGAGCTGCAG | 52 | 513 | [56] |
| GTCGATTCTCGCTAATCC | |||||
| vanX | TCGCGGTAGTCCCACCATTCGTT | 55 | 454 | [57] | |
| AAATCATCGTTGACCTGCGTTAT | |||||
| AMP | blaZ | ACTTCAACACCTGCTGCTTTC | 58 | 240 | [58] |
| TAGGTTCAGATTGGCCCTTAG | |||||
| bla | CATARTTCCGATAATASMGCC | 51 | 297 | [53] | |
| CGTSTTTAACTAAGTATSGY | |||||
| mecA | GGGATCATAGCGTCATTATTC | 58 | 1429 | [58] | |
| AGTTCTGCAGTACCGGATTTGC | |||||
| TET | tet(M) | GGTGAACATCATAGACACGC | 55 | 401 | [59] |
| CTTGTTCGAGTTCCAATGC | |||||
| tet(K) | TCGATAGGAACAGCAGTA | 55 | 169 | [58] | |
| CAGCAGATCCTACTCCTT | |||||
| tet(W) | GAGAGCCTGCTATATGCCAGC | 64 | 168 | [56] | |
| GGGCGTATCCACAATGTTAAC | |||||
| RIF | rpoB | TAACCGTGGTGCTTGGCTDGAATWYGAAAC | 59 | 1100 | [60] |
| ATCAAACCAATGTTAGGNCCTTCWGGDGTTTC | |||||
| KAN | aph (3”)-III | GCCGATGTGGATTGCGAAAA | 52 | 292 | [54] |
| GCTTGATCCCCAGTAAGTCA | |||||
| ant(2”)-I | GGGCGCGTCATGGAGGAGTT | 67 | 329 | [54] | |
| TATCGCGACCTGAAAGCGGC | |||||
| aph(3”)-I | AACGTCTTGCTCGAGGCCGCG | 68 | 670 | [54] | |
| GGCAAGATCCTGGTATCGGTCTGCG | |||||
| CIP | gyrA | GAYTATGCWATGTCAGTTATTGT | 45 | 286 | [54] |
| GGAATRTTRGAYGTCATACCAAC | |||||
| parC | TATTCYAAATAYATCATTCARGA | 50 | 286 | [53] | |
| GCYTCNGTATAACGCATMGCCG | |||||
| CLIN | lnu(A) | GGTGGCTGGGGGGTAGATGTATTAACTGG | 55 | 323 | [56] |
| GCTTCTTTTGAAATACATGGTATTTTTCGATC | |||||
| lnu(B) | CCTACCTATTGTTTGTGGAA | 54 | 925 | [57] | |
| ATAACGTTACTCTCCTATTTC | |||||
| ERY | erm(B) | GAAAAGGTACTCAACCAAATA | 54 | 639 | [58] |
| AGTAACGGTACTTAAATTGTTTAC | |||||
| erm(B)-1 | CATTTAACGACGAAACTGGC | 54 | 405 | [55,59] | |
| GGAACATCTGTGGTATGGCG | |||||
| erm(C) | TCAAAACATAATATAGATAAA | 50 | 642 | [58] | |
| GCTAATATTGTTTAAATCGTCAAT | |||||
| LIN | cfr | TGAAGTATAAAGCAGGTTGGGAGTCA | 55 | 746 | [61] |
| ACCATATAATTGACCACAAGCAGC | |||||
| Conjugative transposon | int (Tn916/Tn1545) | GCGTGATTGTATCTCACT | 55 | 1046 | [62] |
| GACGCTCCTGTTGCTTCT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, M.-S.; Yeu, J.-E.; Hong, S.-P. Safety Evaluation of Oral Care Probiotics Weissella cibaria CMU and CMS1 by Phenotypic and Genotypic Analysis. Int. J. Mol. Sci. 2019, 20, 2693. https://doi.org/10.3390/ijms20112693
Kang M-S, Yeu J-E, Hong S-P. Safety Evaluation of Oral Care Probiotics Weissella cibaria CMU and CMS1 by Phenotypic and Genotypic Analysis. International Journal of Molecular Sciences. 2019; 20(11):2693. https://doi.org/10.3390/ijms20112693
Chicago/Turabian StyleKang, Mi-Sun, Ji-Eun Yeu, and Sang-Phil Hong. 2019. "Safety Evaluation of Oral Care Probiotics Weissella cibaria CMU and CMS1 by Phenotypic and Genotypic Analysis" International Journal of Molecular Sciences 20, no. 11: 2693. https://doi.org/10.3390/ijms20112693
APA StyleKang, M.-S., Yeu, J.-E., & Hong, S.-P. (2019). Safety Evaluation of Oral Care Probiotics Weissella cibaria CMU and CMS1 by Phenotypic and Genotypic Analysis. International Journal of Molecular Sciences, 20(11), 2693. https://doi.org/10.3390/ijms20112693