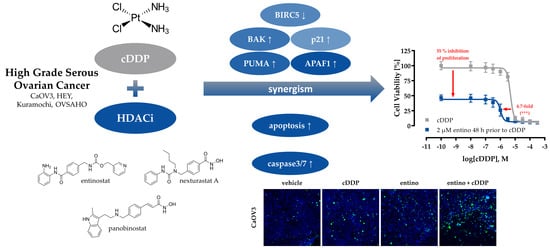
Class I-Histone Deacetylase (HDAC) Inhibition is Superior to pan-HDAC Inhibition in Modulating Cisplatin Potency in High Grade Serous Ovarian Cancer Cell Lines
Abstract
1. Introduction
2. Results
2.1. Characterization of the Human Ovarian Cancer Cell Lines
2.2. Cytotoxic and HDAC-Inhibitory Effects of Entinostat, Panobinostat, and Nexturastat A
2.3. Enhancement of Cisplatin-Induced Cytotoxicity
2.4. Enhancement of Cisplatin-Induced Cytotoxicity is Mediated via Apoptosis-Induction
2.5. Apoptosis-Induction of the Combination Treatment is Caspase3/7-Driven
2.6. Alterations in Apoptosis-Related Gene Expression
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Lines and Cell Culture
4.3. MTT Cell Viability Assay
4.4. Whole-Cell HDAC Inhibition Assay
4.5. Combination Experiments
4.6. Enzyme HDAC Inhibition Assay
4.7. Measurement of Apoptotic Nuclei
4.8. Caspase 3/7 Activation Assay
4.9. Immunoblotting
4.10. RT-PCR
4.11. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HDAC | histone deacetylase |
| HDACi | histone deacetylase inhibitor |
| entino | entinostat |
| cDDP | cis-diamminedichloridoplatinum(II) (cisplatin) |
| pano | panobinostat |
| next A | nexturastat A |
| BIRC5, survivin | baculoviral inhibitor of apoptosis repeat-containing 5 |
| PUMA | p53 upregulated modulator of apoptosis |
| BAK/Bak | Bcl-2 homologous antagonist killer |
| CDNK1A, p21 | cyclin-dependent kinase inhibitor 1 |
| APAF1 | apoptotic protease activating factor 1 |
| IAP | inhibitors of apoptosis |
| HPRT1 | hypoxanthine-guanine phosphoribosyltransferase |
| GUSB | beta-glucuronidase |
| TBP | TATA binding protein |
| HGSOC | high grade serous ovarian cancer |
| MTT | 3-(4,5-Dimethylthiazole-2-yl)-2,5-diphenyltetrazoliumbromide |
| CI | combination index |
| RT-PCR | reverse transcriptase polymerase chain reaction |
References
- Cancer of the Ovary - Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/ovary.html (accessed on 24 April 2019).
- Cancer of the Cervix Uteri - Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/cervix.html (accessed on 24 April 2019).
- Cancer of the Breast (Female) - Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/breast.html (accessed on 24 April 2019).
- Cancer of the Vulva - Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/vulva.html (accessed on 24 April 2019).
- Cancer of the Endometrium - Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/corp.html (accessed on 24 April 2019).
- Krebs – Datenbankabfrage. Available online: https://www.krebsdaten.de/Krebs/SiteGlobals/Forms/Datenbankabfrage/datenbankabfrage_stufe2_form.html (accessed on 14 May 2019).
- Singh, N.; McCluggage, W.G.; Gilks, C.B. High-grade serous carcinoma of tubo-ovarian origin: recent developments. Histopathology 2017, 71, 339–356. [Google Scholar] [CrossRef] [PubMed]
- Köbel, M.; Kalloger, S.E.; Huntsman, D.G.; Santos, J.L.; Swenerton, K.D.; Seidman, J.D.; Gilks, C.B. Cheryl Brown Ovarian Cancer Outcomes Unit of the British Columbia Cancer Agency, Vancouver BC Differences in tumor type in low-stage versus high-stage ovarian carcinomas. Int. J. Gynecol. Pathol. Off. J. Int. Soc. Gynecol. Pathol. 2010, 29, 203–211. [Google Scholar]
- Kohn, E.C.; Ivy, S.P. Whence High-Grade Serous Ovarian Cancer. Am. Soc. Clin. Oncol. Educ. Book Am. Soc. Clin. Oncol. Annu. Meet. 2017, 37, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Koshiyama, M.; Matsumura, N.; Konishi, I. Subtypes of Ovarian Cancer and Ovarian Cancer Screening. Diagnostics 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A. Real-world evidence in the treatment of ovarian cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, viii61–viii65. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorganic Chem. 2019, 88, 102925. [Google Scholar] [CrossRef] [PubMed]
- Ohmichi, M.; Hayakawa, J.; Tasaka, K.; Kurachi, H.; Murata, Y. Mechanisms of platinum drug resistance. Trends Pharmacol. Sci. 2005, 26, 113–116. [Google Scholar] [CrossRef]
- Jain, A.; Jahagirdar, D.; Nilendu, P.; Sharma, N.K. Molecular approaches to potentiate cisplatin responsiveness in carcinoma therapeutics. Expert Rev. Anticancer Ther. 2017, 17, 815–825. [Google Scholar] [CrossRef]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef]
- Kim, H.-J.; Bae, S.-C. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. Am. J. Transl. Res. 2011, 3, 166–179. [Google Scholar]
- Gregoretti, I.; Lee, Y.-M.; Goodson, H.V. Molecular Evolution of the Histone Deacetylase Family: Functional Implications of Phylogenetic Analysis. J. Mol. Biol. 2004, 338, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yang, Y.; Zhou, N.; Tang, K.; Lau, W.B.; Lau, B.; Wang, W.; Xu, L.; Yang, Z.; Huang, S.; et al. Epigenetics in ovarian cancer: premise, properties, and perspectives. Mol. Cancer 2018, 17, 109. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Yasuda, M.; Sakaki, M.; Nagata, K.; Fujino, T.; Arai, E.; Hasebe, T.; Miyazawa, M.; Miyazawa, M.; Ogane, N.; et al. Association of histone deacetylase expression with histology and prognosis of ovarian cancer. Oncol. Lett. 2018, 15, 3524–3531. [Google Scholar] [CrossRef] [PubMed]
- Marek, L.; Hamacher, A.; Hansen, F.K.; Kuna, K.; Gohlke, H.; Kassack, M.U.; Kurz, T. Histone deacetylase (HDAC) inhibitors with a novel connecting unit linker region reveal a selectivity profile for HDAC4 and HDAC5 with improved activity against chemoresistant cancer cells. J. Med. Chem. 2013, 56, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Stenzel, K.; Hamacher, A.; Hansen, F.K.; Gertzen, C.G.W.; Senger, J.; Marquardt, V.; Marek, L.; Marek, M.; Romier, C.; Remke, M.; et al. Alkoxyurea-Based Histone Deacetylase Inhibitors Increase Cisplatin Potency in Chemoresistant Cancer Cell Lines. J. Med. Chem. 2017, 60, 5334–5348. [Google Scholar] [CrossRef] [PubMed]
- Krieger, V.; Hamacher, A.; Gertzen, C.G.W.; Senger, J.; Zwinderman, M.R.H.; Marek, M.; Romier, C.; Dekker, F.J.; Kurz, T.; Jung, M.; et al. Design, Multicomponent Synthesis, and Anticancer Activity of a Focused Histone Deacetylase (HDAC) Inhibitor Library with Peptoid-Based Cap Groups. J. Med. Chem. 2017, 60, 5493–5506. [Google Scholar] [CrossRef] [PubMed]
- Cellosaurus Cell Line Caov-3 (CVCL_0201). Available online: https://web.expasy.org/cellosaurus/CVCL_0201 (accessed on 17 May 2019).
- Cellosaurus Cell Line HEY (CVCL_0297). Available online: https://web.expasy.org/cellosaurus/CVCL_0297 (accessed on 17 May 2019).
- Cellosaurus Cell Line Kuramochi (CVCL_1345). Available online: https://web.expasy.org/cellosaurus/CVCL_1345 (accessed on 17 May 2019).
- Cellosaurus Cell Line OVSAHO (CVCL_3114). Available online: https://web.expasy.org/cellosaurus/CVCL_3114 (accessed on 17 May 2019).
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef]
- Cellosaurus Cell Line A2780 (CVCL_0134). Available online: https://web.expasy.org/cellosaurus/CVCL_0134 (accessed on 17 May 2019).
- Beaufort, C.M.; Helmijr, J.C.A.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Besselink, N.; Murtaza, M.; van IJcken, W.F.J.; Heine, A.A.J.; Smid, M.; et al. Ovarian Cancer Cell Line Panel (OCCP): Clinical Importance of In Vitro Morphological Subtypes. PLoS ONE 2014, 9, e103988. [Google Scholar] [CrossRef]
- Panteix, G.; Beaujard, A.; Garbit, F.; Chaduiron-Faye, C.; Guillaumont, M.; Gilly, F.; Baltassat, P.; Bressolle, F. Population pharmacokinetics of cisplatin in patients with advanced ovarian cancer during intraperitoneal hyperthermia chemotherapy. Anticancer Res. 2002, 22, 1329–1336. [Google Scholar]
- Engelke, L.H.; Hamacher, A.; Proksch, P.; Kassack, M.U. Ellagic Acid and Resveratrol Prevent the Development of Cisplatin Resistance in the Epithelial Ovarian Cancer Cell Line A2780. J. Cancer 2016, 7, 353–363. [Google Scholar] [CrossRef]
- Chou, T.-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.-C.; Talalay, P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Waldman, T.; Kinzler, K.W.; Vogelstein, B. p21 Is Necessary for the p53-mediated G1 Arrest in Human Cancer Cells. Cancer Res. 1995, 55, 5187–5190. [Google Scholar] [PubMed]
- Pop, C.; Timmer, J.; Sperandio, S.; Salvesen, G.S. The Apoptosome Activates Caspase-9 by Dimerization. Mol. Cell 2006, 22, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Flemington, C.; Houghton, A.B.; Gu, Z.; Zambetti, G.P.; Lutz, R.J.; Zhu, L.; Chittenden, T. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc. Natl. Acad. Sci. 2001, 98, 11318–11323. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Vousden, K.H. PUMA, a Novel Proapoptotic Gene, Is Induced by p53. Mol. Cell 2001, 7, 683–694. [Google Scholar] [CrossRef]
- McArthur, K.; Whitehead, L.W.; Heddleston, J.M.; Li, L.; Padman, B.S.; Oorschot, V.; Geoghegan, N.D.; Chappaz, S.; Davidson, S.; Chin, H.S.; et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 2018, 359, eaao6047. [Google Scholar] [CrossRef]
- Shin, S.; Sung, B.-J.; Cho, Y.-S.; Kim, H.-J.; Ha, N.-C.; Hwang, J.-I.; Chung, C.-W.; Jung, Y.-K.; Oh, B.-H. An Anti-apoptotic Protein Human Survivin Is a Direct Inhibitor of Caspase-3 and -7. Biochemistry 2001, 40, 1117–1123. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Benton, C.B.; Fiskus, W.; Bhalla, K.N. Targeting Histone Acetylation: Readers and Writers in Leukemia and Cancer. Cancer J. Sudbury Mass 2017, 23, 286–291. [Google Scholar] [CrossRef]
- Pchejetski, D.; Alfraidi, A.; Sacco, K.; Alshaker, H.; Muhammad, A.; Monzon, L. Histone deacetylases as new therapy targets for platinum-resistant epithelial ovarian cancer. J. Cancer Res. Clin. Oncol. 2016, 142, 1659–1671. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, S.; Milstien, S.; Grant, S. Endogenous modulators and pharmacological inhibitors of histone deacetylases in cancer therapy. Oncogene 2012, 31, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, K.; Kishikawa, F.; Tanaka, M.; Sakamoto, T.; Tanimura, S.; Kohno, M. Histone deacetylase inhibitors enhance the chemosensitivity of tumor cells with cross-resistance to a wide range of DNA-damaging drugs. Cancer Sci. 2008, 99, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhao, Y.; Gou, W.; Zhao, S.; Takano, Y.; Zheng, H. The Anti-Tumor Effects and Molecular Mechanisms of Suberoylanilide Hydroxamic Acid (SAHA) on the Aggressive Phenotypes of Ovarian Carcinoma Cells. PLoS ONE 2013, 8, e79781. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-Y.; Lin, H.; Moh, J.-S.; Chen, K.-D.; Wang, I.-W.; Ou, Y.-C.; You, Y.-S.; Lung, C.-C. Low-dose LBH589 increases the sensitivity of cisplatin to cisplatin-resistant ovarian cancer cells. Taiwan. J. Obstet. Gynecol. 2011, 50, 165–171. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shah, R.R. Safety and Tolerability of Histone Deacetylase (HDAC) Inhibitors in Oncology. Drug Saf. 2019, 42, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Van Veggel, M.; Westerman, E.; Hamberg, P. Clinical Pharmacokinetics and Pharmacodynamics of Panobinostat. Clin. Pharmacokinet. 2018, 57, 21–29. [Google Scholar] [CrossRef]
- Tzogani, K.; van Hennik, P.; Walsh, I.; De Graeff, P.; Folin, A.; Sjöberg, J.; Salmonson, T.; Bergh, J.; Laane, E.; Ludwig, H.; et al. EMA Review of Panobinostat (Farydak) for the Treatment of Adult Patients with Relapsed and/or Refractory Multiple Myeloma. The Oncologist 2018, 23, 631–636. [Google Scholar] [CrossRef]
- Takai, N.; Narahara, H.; Narahara, T. Human Endometrial and Ovarian Cancer Cells: Histone Deacetylase Inhibitors Exhibit Antiproliferative Activity, Potently Induce Cell Cycle Arrest, and Stimulate Apoptosis. Curr. Med. Chem. 2007, 14, 2548–2553. [Google Scholar] [CrossRef]
- Buurman, R.; Sandbothe, M.; Schlegelberger, B.; Skawran, B. HDAC inhibition activates the apoptosome via Apaf1 upregulation in hepatocellular carcinoma. Eur. J. Med. Res. 2016, 21, 26. [Google Scholar] [CrossRef]
- Jaiswal, P.K.; Goel, A.; Mittal, R.D. Survivin: A molecular biomarker in cancer. Indian J. Med. Res. 2015, 141, 389–397. [Google Scholar] [PubMed]
- Tong, Q.-S.; Zheng, L.-D.; Wang, L.; Liu, J.; Qian, W. BAK overexpression mediates p53-independent apoptosis inducing effects on human gastric cancer cells. BMC Cancer 2004, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Rose, S.L.; Goodheart, M.J.; DeYoung, B.R.; Smith, B.J.; Buller, R.E. p21 expression predicts outcome in p53-null ovarian carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2003, 9, 1028–1032. [Google Scholar]
- Yu, J.; Zhang, L. The transcriptional targets of p53 in apoptosis control. Biochem. Biophys. Res. Commun. 2005, 331, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Sanders, Y.Y.; Hagood, J.S.; Liu, H.; Zhang, W.; Ambalavanan, N.; Thannickal, V.J. Histone deacetylase inhibition promotes fibroblast apoptosis and ameliorates pulmonary fibrosis in mice. Eur. Respir. J. 2014, 43, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- Glozak, M.A.; Seto, E. Histone deacetylases and cancer. Oncogene 2007, 26, 5420–5432. [Google Scholar] [CrossRef] [PubMed]
- Heltweg, B.; Jung, M. A Microplate Reader-Based Nonisotopic Histone Deacetylase Activity Assay. Anal. Biochem. 2002, 302, 175–183. [Google Scholar] [CrossRef]
- Ciossek, T.; Julius, H.; Wieland, H.; Maier, T.; Beckers, T. A homogeneous cellular histone deacetylase assay suitable for compound profiling and robotic screening. Anal. Biochem. 2008, 372, 72–81. [Google Scholar] [CrossRef]
- Bonfils, C.; Kalita, A.; Dubay, M.; Siu, L.L.; Carducci, M.A.; Reid, G.; Martell, R.E.; Besterman, J.M.; Li, Z. Evaluation of the pharmacodynamic effects of MGCD0103 from preclinical models to human using a novel HDAC enzyme assay. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 3441–3449. [Google Scholar] [CrossRef]
- Hoffmann, K.; Brosch, G.; Loidl, P.; Jung, M. A non-isotopic assay for histone deacetylase activity. Nucleic Acids Res. 1999, 27, 2057–2058. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]










| Cell Line | IC50 [µM] | pIC50 ± SEM |
|---|---|---|
| A2780 | 11.0 | 4.96 ± 0.01 |
| CaOV3 | 1.44 | 5.84 ± 0.01 |
| HEY | 5.25 | 5.28 ± 0.01 |
| Kuramochi | 4.68 | 5.33 ± 0.02 |
| OVSAHO | 4.83 | 5.32 ± 0.01 |
| Cell Line | HDACi | |||||
|---|---|---|---|---|---|---|
| Entinostat | Panobinostat | Nexturastat A | ||||
| IC50 [nM] | pIC50 ± SEM | IC50 [nM] | pIC50 ± SEM | IC50 [nM] | pIC50 ± SEM | |
| A2780 | 606 | 6.22 ± 0.03 | 15.3 | 7.82 ± 0.02 | 7778 | 5.11 ± 0.09 |
| CaOV3 | 1146 | 5.94 ± 0.03 | 7.62 | 8.12 ± 0.03 | 5291 | 5.28 ± 0.03 |
| HEY | 251 | 6.60 ± 0.02 | 2.68 | 8.57 ± 0.02 | 1724 | 5.76 ± 0.03 |
| Kuramochi | 485 | 6.31 ± 0.03 | 11.2 | 7.95 ± 0.02 | 5302 | 5.28 ± 0.02 |
| OVSAHO | 1828 | 5.74 ± 0.02 | 42.4 | 7.37 ± 0.05 | 16,218 | 4.79 ± 0.02 |
| Cell Line | HDACi | |||||
|---|---|---|---|---|---|---|
| Entinostat | Panobinostat | Nexturastat A | ||||
| IC50 [nM] | pIC50 ± SEM | IC50 [nM] | pIC50 ± SEM | IC50 [nM] | pIC50 ± SEM | |
| A2780 | 313 | 6.50 ± 0.04 | 12.1 | 7.91 ± 0.06 | 3633 | 5.44 ± 0.04 |
| CaOV3 | 333 | 6.48 ± 0.03 | 7.88 | 8.10 ± 0.05 | 3500 | 5.46 ± 0.03 |
| HEY | 219 | 6.60 ± 0.04 | 13.6 | 7.87 ± 0.03 | 3874 | 5.41 ± 0.03 |
| Kuramochi | 339 | 6.47 ± 0.04 | 9.87 | 8.01 ± 0.07 | 3733 | 5.43 ± 0.03 |
| OVSAHO | 326 | 6.49 ± 0.02 | 23.1 | 7.64 ± 0.04 | 5249 | 5.28 ± 0.02 |
| Cell Line | Entinostat [nM] | Panobinostat [nM] | Nexturastat A [nM] |
|---|---|---|---|
| A2780 | 750 | 40.0 | 7500 |
| CaOV3 | 750 | 15.0 | 7500 |
| HEY | 200 | 20.0 | 5000 |
| Kuramochi | 750 | 12.5 | 5000 |
| OVSAHO | 2000 | 70.0 | 12,500 |
| HDAC2 | HDAC4 | HDAC6 | HDAC8 | |||||
|---|---|---|---|---|---|---|---|---|
| Compd. | IC50 ± SD [µM] | Ki [µM] | IC50 ± SD [µM] | Ki [µM] | IC50 ± SD [µM] | Ki [µM] | IC50 ± SD [µM] | Ki [µM] |
| Nexturastat A | 1.99 ± 0.25 | 1.25 | 12.0 ± 1.69 | 7.57 | 0.05 ± 0.01 | 0.03 | 22.6 ± 2.55 | 12.8 |
| Cell Line | Cisplatin | + 48 h HDACi Pretreatment | |||||
|---|---|---|---|---|---|---|---|
| Entinostat | Panobinostat | Nexturastat A | |||||
| IC50 | IC50 | SF | IC50 | SF | IC50 | SF | |
| A2780 | 11.0 | 4.99 | 2.2 (***) | 2.58 | 5.6 (***) | 1.44 | 7.6 (***) |
| CaOV3 | 1.44 | 0.72 | 2.0 (*) | 0.74 | 2.0 (ns) | 1.33 | 1.1 (ns) |
| HEY | 5.25 | 1.39 | 3.8 (***) | 2.75 | 1.9 (*) | 1.28 | 4.1 (***) |
| Kuramochi | 4.68 | 3.28 | 1.4 (*) | 5.14 | < 1 (ns) | 3.22 | 1.5 (**) |
| OVSAHO | 4.83 | 1.02 | 4.7 (***) | 3.01 | 1.6 (**) | 1.43 | 3.4 (***) |
| Cisplatin [µM] | Entinostat [µM] | Panobinostat [nM] | Nexturastat A [µM] | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.10 | 0.25 | 0.50 | 0.75 | 10 | 20 | 30 | 40 | 1.25 | 2.50 | 5.00 | 7.50 | ||
| A2780 | 0.50 | * | 0.40 | 0.36 | 0.38 | 0.21 | 0.10 | 0.15 | 0.15 | * | 0.32 | 0.24 | 0.28 |
| 1.00 | 0.64 | 0.38 | 0.35 | 0.39 | 0.25 | 0.12 | 0.17 | 0.18 | * | 0.36 | 0.28 | 0.32 | |
| 2.00 | 0.83 | 0.43 | 0.34 | 0.40 | 0.35 | 0.16 | 0.22 | 0.21 | 0.97 | 0.47 | 0.30 | 0.33 | |
| 4.00 | 0.73 | 0.44 | 0.34 | 0.38 | 0.35 | 0.21 | 0.27 | 0.25 | >1.1 | 0.57 | 0.38 | 0.39 | |
| 6.00 | 0.59 | 0.35 | 0.32 | 0.37 | 0.29 | 0.23 | 0.26 | 0.26 | 0.83 | 0.52 | 0.37 | 0.40 | |
| 0.25 | 0.50 | 0.75 | 1.00 | - | - | - | - | - | - | - | - | ||
| CaOV3 | 0.20 | * | 0.66 | 0.71 | 0.42 | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ |
| 0.40 | * | 0.56 | 0.59 | 0.33 | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | |
| 0.60 | 0.56 | 0.41 | 0.49 | 0.28 | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | |
| 0.80 | 0.33 | 0.30 | 0.31 | 0.23 | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | |
| 1.00 | 0.21 | 0.20 | 0.21 | 0.16 | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | ◦ | |
| 0.10 | 0.15 | 0.20 | 0.25 | 5.0 | 10.0 | 15.0 | 20.0 | 1.25 | 2.50 | 3.75 | 5.00 | ||
| HEY | 0.32 | * | * | * | 0.27 | * | * | 0.19 | 0.14 | * | * | 0.32 | 0.27 |
| 0.50 | * | * | * | 0.26 | * | * | 0.21 | 0.15 | * | * | 0.33 | 0.29 | |
| 1.00 | * | * | 0.31 | 0.25 | * | * | 0.25 | 0.16 | * | * | 0.36 | 0.27 | |
| 2.00 | * | 0.37 | 0.29 | 0.24 | * | 0.41 | 0.25 | 0.17 | * | 0.63 | 0.33 | 0.26 | |
| 4.00 | 0.35 | 0.28 | 0.24 | 0.23 | 0.51 | 0.33 | 0.28 | 0.20 | 0.62 | 0.42 | 0.29 | 0.27 | |
| 0.25 | 0.50 | 0.75 | 1.00 | - | - | - | - | 1.25 | 2.50 | 3.75 | 5.00 | ||
| Kuramochi | 1.00 | 0.43 | 0.42 | 0.51 | 0.62 | ◦ | ◦ | ◦ | ◦ | * | 0.73 | 0.66 | 0.66 |
| 2.00 | 0.42 | 0.43 | 0.49 | 0.60 | ◦ | ◦ | ◦ | ◦ | * | 0.78 | 0.74 | 0.72 | |
| 3.00 | 0.46 | 0.44 | 0.48 | 0.57 | ◦ | ◦ | ◦ | ◦ | * | 0.80 | 0.77 | 0.75 | |
| 4.00 | 0.44 | 0.42 | 0.44 | 0.53 | ◦ | ◦ | ◦ | ◦ | 0.84 | 0.79 | 0.77 | 0.74 | |
| 5.00 | 0.45 | 0.43 | 0.47 | 0.52 | ◦ | ◦ | ◦ | ◦ | 0.78 | 0.78 | 0.74 | 0.70 | |
| 0.50 | 1.00 | 1.50 | 2.00 | 40 | 50 | 60 | 70 | 5.00 | 7.50 | 10.0 | 12.5 | ||
| OVSAHO | 1.00 | 0.50 | 0.39 | 0.41 | 0.44 | 0.36 | 0.33 | 0.27 | 0.26 | * | 0.82 | 0.84 | 0.78 |
| 2.00 | 0.41 | 0.29 | 0.34 | 0.37 | 0.25 | 0.23 | 0.21 | 0.21 | 0.87 | 0.77 | 0.75 | 0.67 | |
| 3.00 | 0.42 | 0.28 | 0.32 | 0.36 | 0.24 | 0.22 | 0.21 | 0.21 | 0.80 | 0.76 | 0.65 | 0.60 | |
| 4.00 | 0.39 | 0.28 | 0.31 | 0.35 | 0.23 | 0.22 | 0.21 | 0.21 | 0.72 | 0.69 | 0.61 | 0.57 | |
| 5.00 | 0.39 | 0.29 | 0.33 | 0.36 | 0.24 | 0.23 | 0.22 | 0.22 | 0.64 | 0.59 | 0.53 | 0.53 | |
| Gene | Primer Forward | Primer Reverse | Efficacy [%] |
|---|---|---|---|
| HPRT1 | CCTGGCGTCGTGATTAGTGA | CGAGCAAGACGTTCAGTCCT | 93.6 |
| TBP | GTGACCCAGCATCACTGTTTC | GAGCATCTCCAGCACACTCT | 86.9 |
| GUSB | ACCTCCAAGTATCCCAAGGGT | GTCTTGCTCCACGCTGGT | 83.1 |
| HDAC1 | TGCAAAGAAGTCCGAGGCAT | ACCCTCTGGTGATACTTTAGCA | 84.9 |
| HDAC2 | AATGGAAATATATAGGCCCC | GTTATCTGGTCTTATTGACCG | 96.4 |
| HDAC3 | GGCAACTTCCACTACGGAGC | GCATATTGGTGGGGCTGACT | 97.2 |
| HDAC4 | TTGGATGTCACAGACTCCGC | CCTTCTCGTGCCACAAGTCT | 80.8 |
| HDAC5 | GGAGAGCTCAAGAATGGATTTGC | CTGCTGTAGGAGTTTTGCG | 97.2 |
| HDAC6 | CTGGCGGAGTGGAAGAACC | TCTGCCTACTTCTTCGCTGC | 104 |
| HDAC7 | TCTCGTGAGCTAAAGAATGG | CTGTTGAATGATCTGCATGG | 96.5 |
| HDAC8 | CCACCTTCCACACTGATGCT | GCTGGGCAGTCATAACCTAGC | 97.7 |
| HDAC9 | TGTAGCTGGTGGAGTTCCCT | CTCTGAGGCAAAGGTGCAGA | 103 |
| HDAC10 | TGGCCTTTGAGTTTGACCCT | CCGATGGCTGAGTCAAATCCT | 97.3 |
| HDAC11 | CGGAAAATGGGGCAAAGTGA | CAACAGCAAAGGACCACTTG | 100 |
| CDNK1A (p21) | TGCCGAAGTCAGTTCCTTGT | GTTCTGACATGGCGCCTCC | 94.7 |
| BIRC5 (Survivin) | TGAGAACGAGCCAGACTTGG | TGTTCCTCTATGGGGTCGTCA | 108 |
| APAF1 | AGTGGAATAACTTCGTATGTAAGGA | AAACAACTGGCCTCTGTGGT | 98.7 |
| BAK1 | TCATCGGGGACGACATCAAC | CAAACAGGCTGGTGGCAATC | 111 |
| PUMA | GAGCGGCGGAGACAAGAG | TAAGGGCAGGAGTCCCATGA | 94.7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bandolik, J.J.; Hamacher, A.; Schrenk, C.; Weishaupt, R.; Kassack, M.U. Class I-Histone Deacetylase (HDAC) Inhibition is Superior to pan-HDAC Inhibition in Modulating Cisplatin Potency in High Grade Serous Ovarian Cancer Cell Lines. Int. J. Mol. Sci. 2019, 20, 3052. https://doi.org/10.3390/ijms20123052
Bandolik JJ, Hamacher A, Schrenk C, Weishaupt R, Kassack MU. Class I-Histone Deacetylase (HDAC) Inhibition is Superior to pan-HDAC Inhibition in Modulating Cisplatin Potency in High Grade Serous Ovarian Cancer Cell Lines. International Journal of Molecular Sciences. 2019; 20(12):3052. https://doi.org/10.3390/ijms20123052
Chicago/Turabian StyleBandolik, Jan J., Alexandra Hamacher, Christian Schrenk, Robin Weishaupt, and Matthias U. Kassack. 2019. "Class I-Histone Deacetylase (HDAC) Inhibition is Superior to pan-HDAC Inhibition in Modulating Cisplatin Potency in High Grade Serous Ovarian Cancer Cell Lines" International Journal of Molecular Sciences 20, no. 12: 3052. https://doi.org/10.3390/ijms20123052
APA StyleBandolik, J. J., Hamacher, A., Schrenk, C., Weishaupt, R., & Kassack, M. U. (2019). Class I-Histone Deacetylase (HDAC) Inhibition is Superior to pan-HDAC Inhibition in Modulating Cisplatin Potency in High Grade Serous Ovarian Cancer Cell Lines. International Journal of Molecular Sciences, 20(12), 3052. https://doi.org/10.3390/ijms20123052