Functional Dissection of pri-miR-290~295 in Dgcr8 Knockout Mouse Embryonic Stem Cells
Abstract
:1. Introduction
2. Results
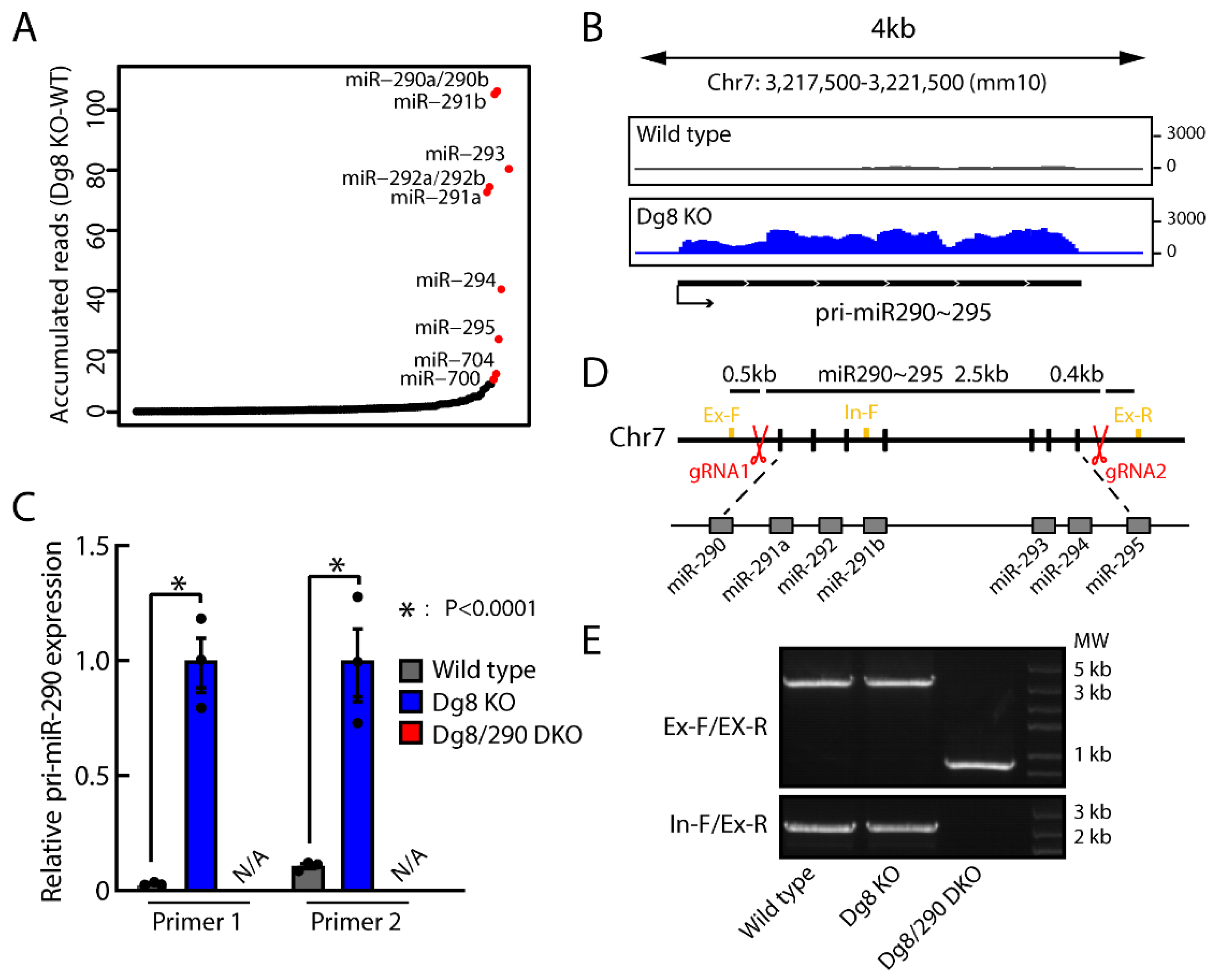
2.1. Establishment of Dgcr8/Pri-miR-290~295 Double Knockout ESCs
2.2. Pluripotency and Epithelial-to-Mesenchymal Transition (EMT)-Like Phenotype in Dgcr8/Pri-miR-290~295 Double Knockout ESCs
2.3. Proliferation and Cell Cycle Progression of Dgcr8/Pri-miR-290-295 Double Knockout ESCs
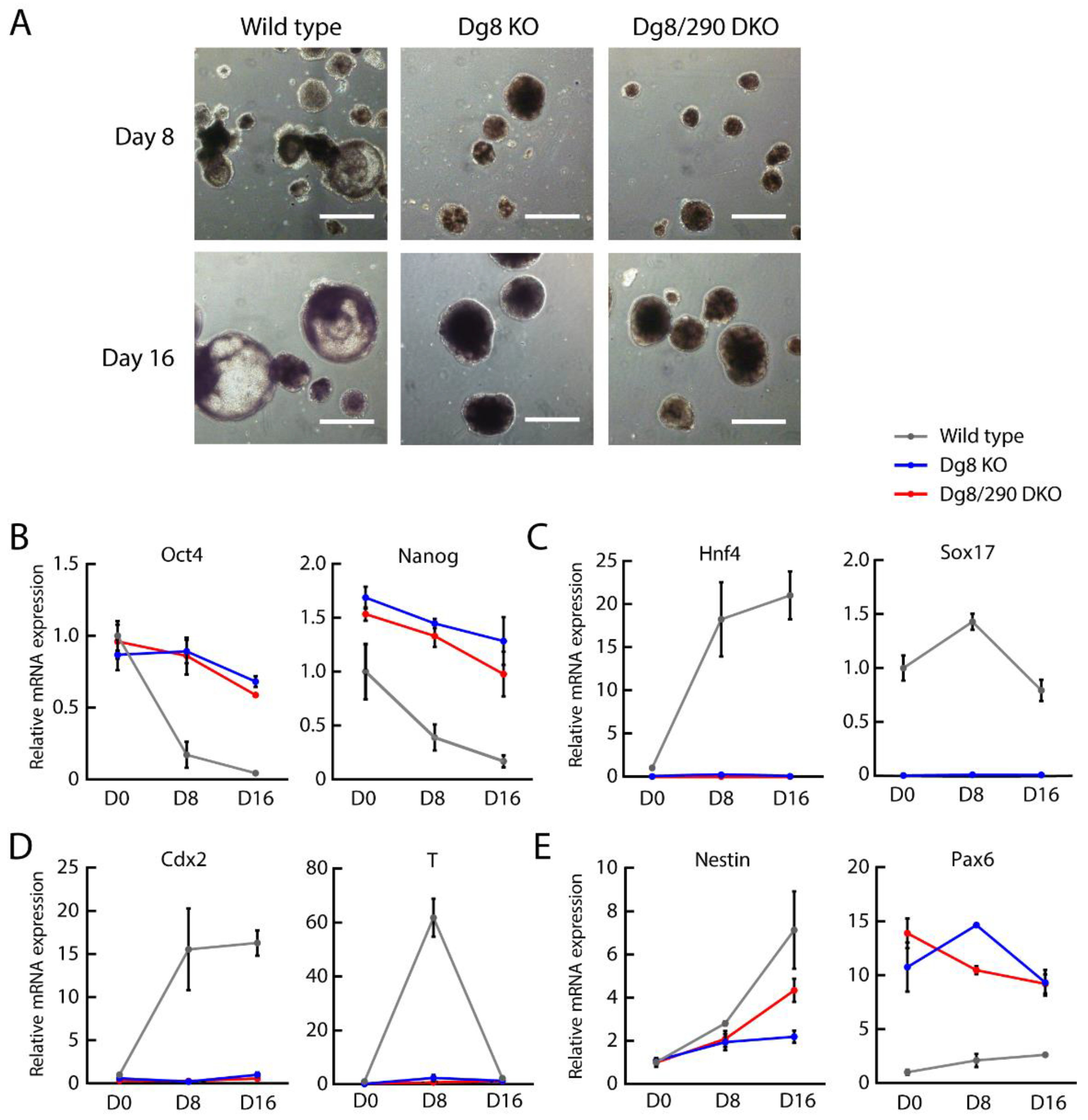
2.4. Embryoid Body Differentiation of Dgcr8/miR-290-295 Double Knockout ESCs
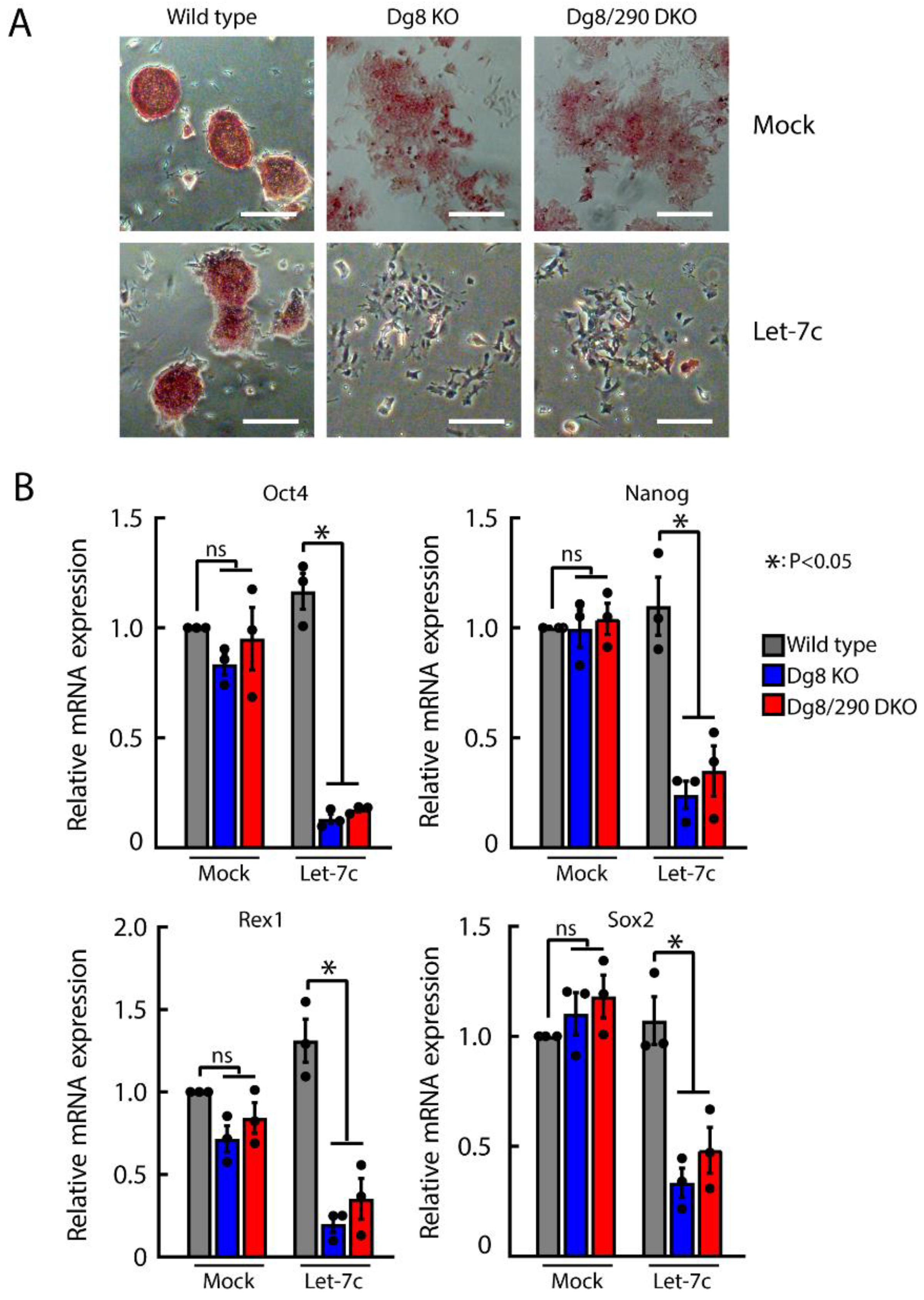
2.5. Let-7 Mediated Silencing of Self-Renewal in Dgcr8/Pri-miR-290 Double Knockout ESCs
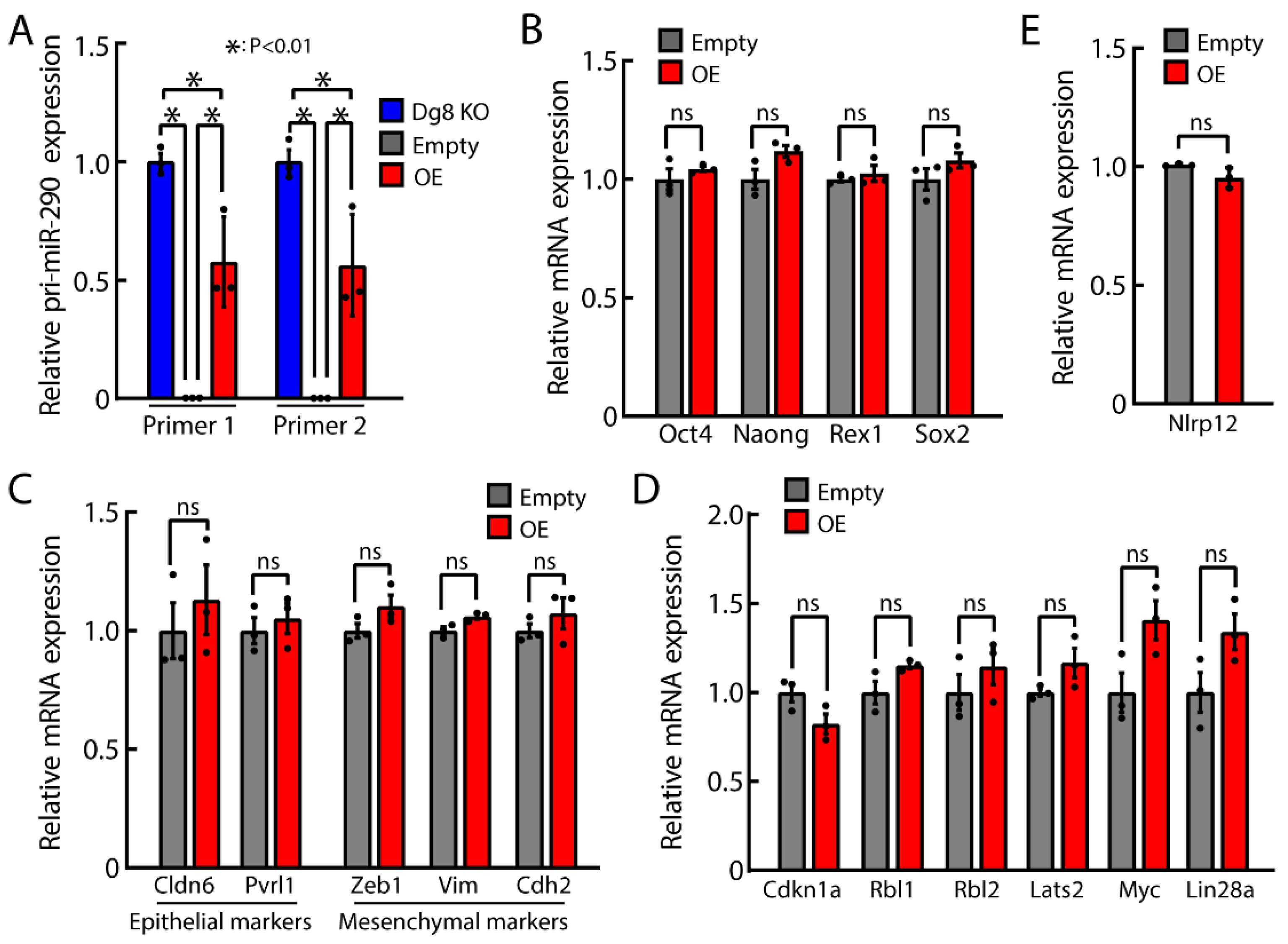
2.6. Impact of Pri-miR-290~295 Knockout on the Transcription of Neighboring Gene
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Pri-miR-290-295 Cluster Knockout
4.3. Transfection of miRNA Mimics
4.4. RNA Isolation and RT-qPCR
4.5. Alkaline Phosphatase Staining
4.6. Cell Cycle Analysis
4.7. RNA-Seq and Bioinformatics Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Handrick, R.; Aschrafi, A.; Otte, K. Unveiling the principle of microRNA-mediated redundancy in cellular pathway regulation. RNA Biol. 2015, 12, 238–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.T.; Wang, Y. Dgcr8 knockout approaches to understand microRNA functions in vitro and in vivo. Cell. Mol. Life Sci. 2019, 76, 1697–1711. [Google Scholar] [CrossRef] [PubMed]
- Murchison, E.P.; Partridge, J.F.; Tam, O.H.; Cheloufi, S.; Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 2005, 102, 12135–12140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005, 19, 489–501. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Medvid, R.; Melton, C.; Jaenisch, R.; Blelloch, R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007, 39, 380. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773. [Google Scholar] [CrossRef]
- Wang, Y.; Baskerville, S.; Shenoy, A.; Babiarz, J.E.; Baehner, L.; Blelloch, R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008, 40, 1478–1483. [Google Scholar] [CrossRef]
- Melton, C.; Judson, R.L.; Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010, 463, 621–626. [Google Scholar] [CrossRef]
- Wang, Y.; Melton, C.; Li, Y.P.; Shenoy, A.; Zhang, X.X.; Subramanyam, D.; Blelloch, R. miR-294/miR-302 Promotes Proliferation, Suppresses G1-S Restriction Point, and Inhibits ESC Differentiation through Separable Mechanisms. Cell Rep. 2013, 4, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Guo, W.T.; Tian, S.; He, X.; Wang, X.W.; Liu, X.; Gu, K.L.; Ma, X.; Huang, D.; Hu, L.; et al. miR-290/371-Mbd2-Myc circuit regulates glycolytic metabolism to promote pluripotency. EMBO J. 2015, 34, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.T.; Wang, X.W.; Yan, Y.L.; Li, Y.P.; Yin, X.; Zhang, Q.; Melton, C.; Shenoy, A.; Reyes, N.A.; Oakes, S.A.; et al. Suppression of epithelial-mesenchymal transition and apoptotic pathways by miR-294/302 family synergistically blocks let-7-induced silencing of self-renewal in embryonic stem cells. Cell Death Differ. 2015, 22, 1158–1169. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yao, N.; Liu, G.; Dong, L.; Liu, Y.; Zhang, M.; Wang, F.; Wang, B.; Wei, X.; Dong, H.; et al. Functional screen reveals essential roles of miR-27a/24 in differentiation of embryonic stem cells. EMBO J. 2015, 34, 361–378. [Google Scholar] [CrossRef] [PubMed]
- Gu, K.L.; Zhang, Q.; Yan, Y.; Li, T.T.; Duan, F.F.; Hao, J.; Wang, X.W.; Shi, M.; Wu, D.R.; Guo, W.T. Pluripotency-associated miR-290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Res. 2016, 26, 350–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Yang, X.; Li, T.T.; Gu, K.L.; Hao, J.; Zhang, Q.; Wang, Y. Significant differences of function and expression of microRNAs between ground state and serum-cultured pluripotent stem cells. J. Genet. Genom. 2017, 44, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Hao, J.; Guo, W.T.; Liao, L.Q.; Huang, S.Y.; Guo, X.; Bao, X.; Esteban, M.A.; Wang, Y. A DGCR8-Independent Stable MicroRNA Expression Strategy Reveals Important Functions of miR-290 and miR-183–182 Families in Mouse Embryonic Stem Cells. Stem Cell Rep. 2017, 9, 1618–1629. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [Green Version]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Liu, G.; Min, H.; Yue, S.B.; Chen, C.Z. Pre-miRNA Loop Nucleotides Control the Distinct Activities of mir-181a-1 and mir-181c in Early T Cell Development. PLoS ONE 2008, 3, e3592. [Google Scholar] [CrossRef]
- Trujillo, R.D.; Yue, S.B.; Tang, Y.; O’Gorman, W.E.; Chen, C.Z. The potential functions of primary microRNAs in target recognition and repression. EMBO J. 2014, 29, 3272–3285. [Google Scholar] [CrossRef] [PubMed]
- Mathiyalagan, P.; Okabe, J.; Chang, L.; Su, Y.; Du, X.J.; El-Osta, A. The primary microRNA-208b interacts with Polycomb-group protein, Ezh2, to regulate gene expression in the heart. Nucleic Acids Res. 2014, 42, 790–803. [Google Scholar] [CrossRef]
- Donayo, A.O.; Johnson, R.M.; Tseng, H.W.; Izreig, S.; Gariepy, A.; Mayya, V.K.; Wu, E.; Alam, R.; Lussier, C.; Jones, R.G. Oncogenic Biogenesis of pri-miR-17∼ 92 Reveals Hierarchy and Competition among Polycistronic MicroRNAs. Mol. Cell 2019, 75, 340–356. [Google Scholar] [CrossRef]
- Hao, J.; Duan, F.F.; Wang, Y. MicroRNAs and RNA binding protein regulators of microRNAs in the control of pluripotency and reprogramming. Curr. Opin. Genet. Dev. 2017, 46, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. eLife 2013, 2, e00471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–2586. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Houbaviy, H.B.; Dennis, L.; Jaenisch, R.; Sharp, P.A. Characterization of a highly variable eutherian microRNA gene. RNA 2005, 11, 1245–1257. [Google Scholar] [CrossRef] [Green Version]
- Zeisberg, M.; Neilson, E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Investig. 2009, 119, 1429–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Kaspi, H.; Chapnik, E.; Levy, M.; Beck, G.; Hornstein, E.; Soen, Y. Brief report: miR-290-295 regulate embryonic stem cell differentiation propensities by repressing Pax6. Stem Cells 2013, 31, 2266–2272. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, L.A.; Dennis, L.M.; Gill, M.E.; Houbaviy, H.; Markoulaki, S.; Fu, D.; White, A.C.; Kirak, O.; Sharp, P.A.; Page, D.C.; et al. Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc. Natl. Acad. Sci. USA 2011, 108, 14163–14168. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 2013, 10, 973–976. [Google Scholar] [CrossRef]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.W.; Wang, H.; Yang, H.; Shi, L.; Katz, Y.; Theunissen, T.W.; Rangarajan, S.; Shivalila, C.S.; Dadon, D.B.; Jaenisch, R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013, 23, 1163–1171. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Bio. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, M.; Hao, J.; Wang, X.-W.; Liao, L.-Q.; Cao, H.; Wang, Y. Functional Dissection of pri-miR-290~295 in Dgcr8 Knockout Mouse Embryonic Stem Cells. Int. J. Mol. Sci. 2019, 20, 4345. https://doi.org/10.3390/ijms20184345
Shi M, Hao J, Wang X-W, Liao L-Q, Cao H, Wang Y. Functional Dissection of pri-miR-290~295 in Dgcr8 Knockout Mouse Embryonic Stem Cells. International Journal of Molecular Sciences. 2019; 20(18):4345. https://doi.org/10.3390/ijms20184345
Chicago/Turabian StyleShi, Ming, Jing Hao, Xi-Wen Wang, Le-Qi Liao, Huiqing Cao, and Yangming Wang. 2019. "Functional Dissection of pri-miR-290~295 in Dgcr8 Knockout Mouse Embryonic Stem Cells" International Journal of Molecular Sciences 20, no. 18: 4345. https://doi.org/10.3390/ijms20184345
APA StyleShi, M., Hao, J., Wang, X. -W., Liao, L. -Q., Cao, H., & Wang, Y. (2019). Functional Dissection of pri-miR-290~295 in Dgcr8 Knockout Mouse Embryonic Stem Cells. International Journal of Molecular Sciences, 20(18), 4345. https://doi.org/10.3390/ijms20184345






