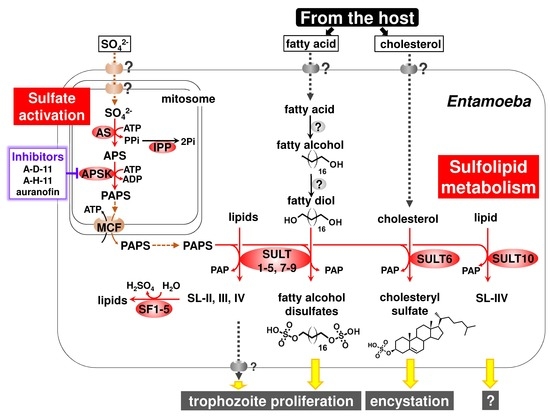
Unique Features of Entamoeba Sulfur Metabolism; Compartmentalization, Physiological Roles of Terminal Products, Evolution and Pharmaceutical Exploitation
Abstract
1. Introduction
1.1. Sulfur Metabolism
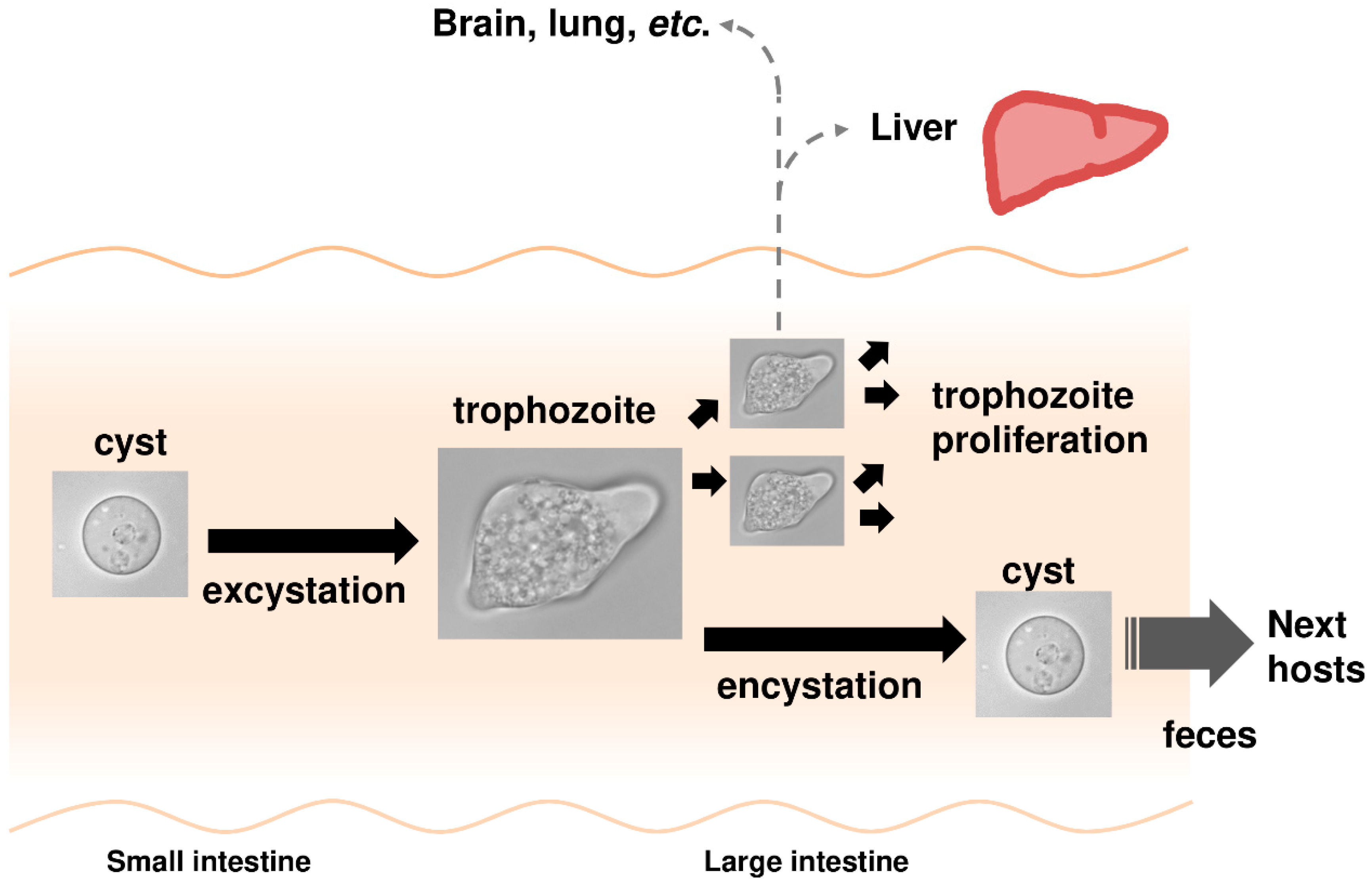
1.2. Entamoeba Histolytica Infection Causes Amoebiasis
2. Biosynthesis and Degradation of Sulfated Biomolecules That Are Crucial for Homeostasis
2.1. General Features of Sulfate Activation
2.2. Sulfotransferases (SULTs)
2.3. Sulfatases (SFs)
3. Unique Features of Sulfated Molecule Metabolism in Entamoeba
3.1. Atypical Localization of Sulfate Activation in Mitosomes
3.2. Sulfolipids Are the Major Products Synthesized by Entamoeba SULTs
3.3. Two Previously Unrecognized Entamoeba Enzymes Responsible for Fatty Alcohol Disulfate Synthesis
3.4. SF
3.5. Lateral Gene Transfer (LGT) Acquisition of Unique Sulfolipid Metabolism in Entamoeba
4. Pharmaceutical Exploitation of Entamoeba Sulfolipid Metabolism
5. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bradley, M.E.; Rest, J.S.; Li, W.H.; Schwartz, N.B. Sulfate activation enzymes: phylogeny and association with pyrophosphatase. J. Mol. Evol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Patron, N.J.; Durnford, D.G.; Kopriva, S. Sulfate assimilation in eukaryotes: fusions, relocations and lateral transfers. BMC Evol. Biol. 2008, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Lensmire, J.M.; Hammer, N.D. Nutrient sulfur acquisition strategies employed by bacterial pathogens. Curr. Opin. Microbiol. 2019, 47, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Gunal, S.; Hardman, R.; Kopriva, S.; Mueller, J.W. Sulfation pathways from red to green. J. Biol. Chem. 2019, 294, 12293–12312. [Google Scholar] [CrossRef] [PubMed]
- Mueller, J.W.; Gilligan, L.C.; Idkowiak, J.; Arlt, W.; Foster, P.A. The regulation of steroid action by sulfation and desulfation. Endocr. Rev. 2015, 36, 526–563. [Google Scholar] [CrossRef]
- Quach, J.; St-Pierre, J.; Chadee, K. The future for vaccine development against Entamoeba histolytica. Hum. Vaccines Immunother. 2014, 10, 1514–1521. [Google Scholar] [CrossRef] [PubMed]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the global burden of disease study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Haque, R.; Huston, C.D.; Hughes, M.; Houpt, E.; Petri, W.A., Jr. Amebiasis. N. Engl. J. Med. 2003, 348, 1565–1573. [Google Scholar] [CrossRef]
- Watanabe, K.; Petri, W.A., Jr. Molecular biology research to benefit patients with Entamoeba histolytica infection. Mol. Microbiol. 2015, 98, 208–217. [Google Scholar] [CrossRef]
- Mi-Ichi, F.; Yoshida, H.; Hamano, S. Entamoeba Encystation: New Targets to Prevent the Transmission of Amebiasis. PLoS Pathog. 2016, 12, e1005845. [Google Scholar] [CrossRef]
- Mi-ichi, F.; Miyamoto, T.; Takao, S.; Jeelani, G.; Hashimoto, T.; Hara, H.; Nozaki, T.; Yoshida, H. Entamoeba mitosomes play an important role in encystation by association with cholesteryl sulfate synthesis. Proc. Natl. Acad. Sci. USA 2015, 112, E2884–E2890. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.; Best, M.D.; Hanson, S.R.; Wong, C.H. Sulfotransferases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew. Chem. 2004, 43, 3526–3548. [Google Scholar] [CrossRef] [PubMed]
- Hirschmann, F.; Krause, F.; Papenbrock, J. The multi-protein family of sulfotransferases in plants: composition, occurrence, substrate specificity, and functions. Front. Plant Sci. 2014, 5, 556. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.W.; Backstrom, I.; Bally, M.B. Sulfonation, an underexploited area: from skeletal development to infectious diseases and cancer. Oncotarget 2016, 7, 55811–55827. [Google Scholar] [CrossRef] [PubMed]
- Elias, P.M.; Williams, M.L.; Choi, E.H.; Feingold, K.R. Role of cholesterol sulfate in epidermal structure and function: lessons from X-linked ichthyosis. Biochim. Biophys. Acta 2014, 1841, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Mougous, J.D.; Green, R.E.; Williams, S.J.; Brenner, S.E.; Bertozzi, C.R. Sulfotransferases and sulfatases in mycobacteria. Chem. Biol. 2002, 9, 767–776. [Google Scholar] [CrossRef]
- Geerdink, D.; Minnaard, A.J. Total synthesis of sulfolipid-1. Chem. Commun. 2014, 50, 2286–2288. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, Y.; Fujita, Y.; Naka, T.; Hirai, M.; Tomiyasu, I.; Yano, I. Mycobacterial sulfolipid shows a virulence by inhibiting cord factor induced granuloma formation and TNF-alpha release. Microb. Pathog. 2006, 40, 245–253. [Google Scholar] [CrossRef]
- Diez-Roux, G.; Ballabio, A. Sulfatases and human disease. Annu. Rev. Genomics Hum. Genet. 2005, 6, 355–379. [Google Scholar] [CrossRef] [PubMed]
- Toesch, M.; Schober, M.; Faber, K. Microbial alkyl- and aryl-sulfatases: Mechanism, occurrence, screening and stereoselectivities. Appl. Microbiol. Biotechnol. 2014, 98, 1485–1496. [Google Scholar] [CrossRef] [PubMed]
- Claus, S.P.; Guillou, H.; Ellero-Simatos, S. The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2016, 2, 16003. [Google Scholar] [CrossRef] [PubMed]
- Benjdia, A.; Berteau, O. Sulfatases and radical SAM enzymes: Emerging themes in glycosaminoglycan metabolism and the human microbiota. Biochem. Soc. Trans. 2016, 44, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Dallinga, M.G.; Dallinga-Thie, G.M. Role of sulfatase 2 in lipoprotein metabolism and angiogenesis. Curr. Opin. Lipidol. 2016, 27, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Rizner, T.L. The Important Roles of Steroid Sulfatase and Sulfotransferases in Gynecological Diseases. Front. Pharmacol. 2016, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Besset, S.; Vincourt, J.B.; Amalric, F.; Girard, J.P. Nuclear localization of PAPS synthetase 1: A sulfate activation pathway in the nucleus of eukaryotic cells. FASEB J. 2000, 14, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Saidha, T.; Na, S.Q.; Li, J.Y.; Schiff, J.A. A sulphate metabolizing centre in Euglena mitochondria. Biochem. J. 1988, 253, 533–539. [Google Scholar] [CrossRef]
- Brunold, C.; Schiff, J.A. Studies of sulfate utilization of algae: 15. Enzymes of assimilatory sulfate reduction in euglena and their cellular localization. Plant Physiol. 1976, 57, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Mi-ichi, F.; Abu Yousuf, M.; Nakada-Tsukui, K.; Nozaki, T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc. Natl. Acad. Sci. USA 2009, 106, 21731–21736. [Google Scholar] [CrossRef]
- Mi-ichi, F.; Makiuchi, T.; Furukawa, A.; Sato, D.; Nozaki, T. Sulfate activation in mitosomes plays an important role in the proliferation of Entamoeba histolytica. PLoS Negl. Trop. Dis. 2011, 5, e1263. [Google Scholar] [CrossRef]
- Santos, H.J.; Makiuchi, T.; Nozaki, T. Reinventing an Organelle: The Reduced Mitochondrion in Parasitic Protists. Trends Parasitol. 2018, 34, 1038–1055. [Google Scholar] [CrossRef]
- Stairs, C.W.; Leger, M.M.; Roger, A.J. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Philos. Trans. R. Soc. B 2015, 370, 20140326. [Google Scholar] [CrossRef] [PubMed]
- Shiflett, A.M.; Johnson, P.J. Mitochondrion-related organelles in eukaryotic protists. Annu. Rev. Microbiol. 2010, 64, 409–429. [Google Scholar] [CrossRef] [PubMed]
- van der Giezen, M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J. Eukaryot. Microbiol. 2009, 56, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Nyvltova, E.; Stairs, C.W.; Hrdy, I.; Ridl, J.; Mach, J.; Paces, J.; Roger, A.J.; Tachezy, J. Lateral gene transfer and gene duplication played a key role in the evolution of Mastigamoeba balamuthi hydrogenosomes. Mol. Biol. Evol. 2015, 32, 1039–1055. [Google Scholar] [CrossRef] [PubMed]
- Mi-ichi, F.; Nozawa, A.; Yoshida, H.; Tozawa, Y.; Nozaki, T. Evidence that the Entamoeba histolytica, itochondrial carrier family links mitosomal and cytosolic pathways through exchange of 3′-phosphoadenosine 5′-phosphosulfate and ATP. Eukaryot. Cell 2015, 14, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Mi-Ichi, F.; Miyamoto, T.; Yoshida, H. Uniqueness of Entamoeba sulfur metabolism: sulfolipid metabolism that plays pleiotropic roles in the parasitic life cycle. Mol. Microbiol. 2017, 106, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Bracha, R.; Nuchamowitz, Y.; Anbar, M.; Mirelman, D. Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog. 2006, 2, e48. [Google Scholar] [CrossRef] [PubMed]
- Eichinger, D. Encystation in parasitic protozoa. Curr. Opin. Microbiol. 2001, 4, 421–426. [Google Scholar] [CrossRef]
- Clark, C.G.; Alsmark, U.C.; Tazreiter, M.; Saito-Nakano, Y.; Ali, V.; Marion, S.; Weber, C.; Mukherjee, C.; Bruchhaus, I.; Tannich, E.; et al. Structure and content of the Entamoeba histolytica genome. Adv. Parasitol. 2007, 65, 51–190. [Google Scholar]
- Eirin-Lopez, J.M.; Rebordinos, L.; Rooney, A.P.; Rozas, J. The birth-and-death evolution of multigene families revisited. Genome Dyn. 2012, 7, 170–196. [Google Scholar]
- Pinot, F.; Beisson, F. Cytochrome P450 metabolizing fatty acids in plants: characterization and physiological roles. FEBS J. 2011, 278, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Bjelica, A.; Haggitt, M.L.; Woolfson, K.N.; Lee, D.P.; Makhzoum, A.B.; Bernards, M.A. Fatty acid omega-hydroxylases from Solanum tuberosum. Plant Cell Rep. 2016, 35, 2435–2448. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.W.; Weedall, G.D.; Lorenzi, H.; Howcroft, T.; Hon, C.C.; Deloger, M.; Guillen, N.; Paterson, S.; Clark, C.G.; Hall, N. Genetic diversity and gene family expansions in members of the genus Entamoeba. Genome Biol. Evol. 2019, 11, 688–705. [Google Scholar] [CrossRef] [PubMed]
- Loftus, B.; Anderson, I.; Davies, R.; Alsmark, U.C.; Samuelson, J.; Amedeo, P.; Roncaglia, P.; Berriman, M.; Hirt, R.P.; Mann, B.J.; et al. The genome of the protist parasite Entamoeba histolytica. Nature 2005, 433, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Hagelueken, G.; Adams, T.M.; Wiehlmann, L.; Widow, U.; Kolmar, H.; Tummler, B.; Heinz, D.W.; Schubert, W.D. The crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, defines a third class of sulfatases. Proc. Natl. Acad. Sci. USA 2006, 103, 7631–7636. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Krinsky, B.H.; Long, M. New genes as drivers of phenotypic evolution. Nat. Rev. Genet. 2013, 14, 645–660. [Google Scholar] [CrossRef] [PubMed]
- Mi-Ichi, F.; Ishikawa, T.; Tam, V.K.; Deloer, S.; Hamano, S.; Hamada, T.; Yoshida, H. Characterization of Entamoeba histolytica adenosine 5′-phosphosulfate (APS) kinase; validation as a target and provision of leads for the development of new drugs against amoebiasis. PLoS Negl. Trop. Dis. 2019, 13, e0007633. [Google Scholar] [CrossRef] [PubMed]
- Andrade, R.M.; Chaparro, J.D.; Capparelli, E.; Reed, S.L. Auranofin is highly efficacious against Toxoplasma gondii in vitro and in an in vivo experimental model of acute toxoplasmosis. PLoS Negl. Trop. Dis. 2014, 8, e2973. [Google Scholar] [CrossRef]
- Debnath, A.; Parsonage, D.; Andrade, R.M.; He, C.; Cobo, E.R.; Hirata, K.; Chen, S.; Garcia-Rivera, G.; Orozco, E.; Martinez, M.B.; et al. A high-throughput drug screen for Entamoeba histolytica identifies a new lead and target. Nat. Med. 2012, 18, 956–960. [Google Scholar] [CrossRef]
- Capparelli, E.V.; Bricker-Ford, R.; Rogers, M.J.; McKerrow, J.H.; Reed, S.L. Phase I clinical trial results of auranofin, a novel antiparasitic agent. Antimicrob. Agents Chemother. 2017, 61, e01947-16. [Google Scholar] [CrossRef] [PubMed]
- Parsonage, D.; Sheng, F.; Hirata, K.; Debnath, A.; McKerrow, J.H.; Reed, S.L.; Abagyan, R.; Poole, L.B.; Podust, L.M. X-ray structures of thioredoxin and thioredoxin reductase from Entamoeba histolytica and prevailing hypothesis of the mechanism of Auranofin action. J. Struct. Biol. 2016, 194, 180–190. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mi-ichi, F.; Yoshida, H. Unique Features of Entamoeba Sulfur Metabolism; Compartmentalization, Physiological Roles of Terminal Products, Evolution and Pharmaceutical Exploitation. Int. J. Mol. Sci. 2019, 20, 4679. https://doi.org/10.3390/ijms20194679
Mi-ichi F, Yoshida H. Unique Features of Entamoeba Sulfur Metabolism; Compartmentalization, Physiological Roles of Terminal Products, Evolution and Pharmaceutical Exploitation. International Journal of Molecular Sciences. 2019; 20(19):4679. https://doi.org/10.3390/ijms20194679
Chicago/Turabian StyleMi-ichi, Fumika, and Hiroki Yoshida. 2019. "Unique Features of Entamoeba Sulfur Metabolism; Compartmentalization, Physiological Roles of Terminal Products, Evolution and Pharmaceutical Exploitation" International Journal of Molecular Sciences 20, no. 19: 4679. https://doi.org/10.3390/ijms20194679
APA StyleMi-ichi, F., & Yoshida, H. (2019). Unique Features of Entamoeba Sulfur Metabolism; Compartmentalization, Physiological Roles of Terminal Products, Evolution and Pharmaceutical Exploitation. International Journal of Molecular Sciences, 20(19), 4679. https://doi.org/10.3390/ijms20194679