Dynamic Regulation of miRNA Expression by Functionally Enhanced Placental Mesenchymal Stem Cells Promotes Hepatic Regeneration in a Rat Model with Bile Duct Ligation
Abstract
1. Introduction
2. Results
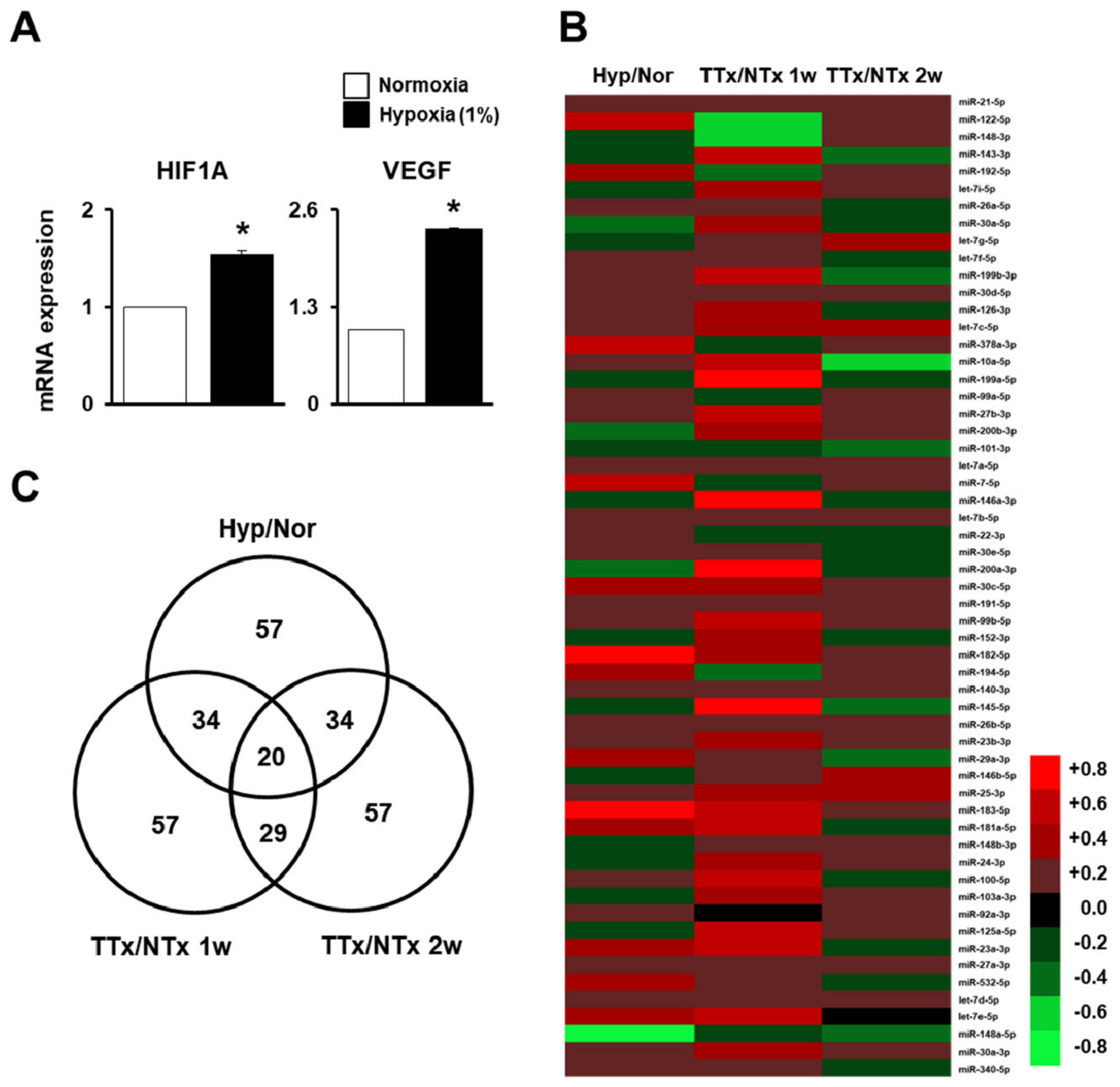
2.1. MiRNA Profiling of Naïve PD-MSCs under Hypoxic Conditions and in BDL-Injured Liver in Rats
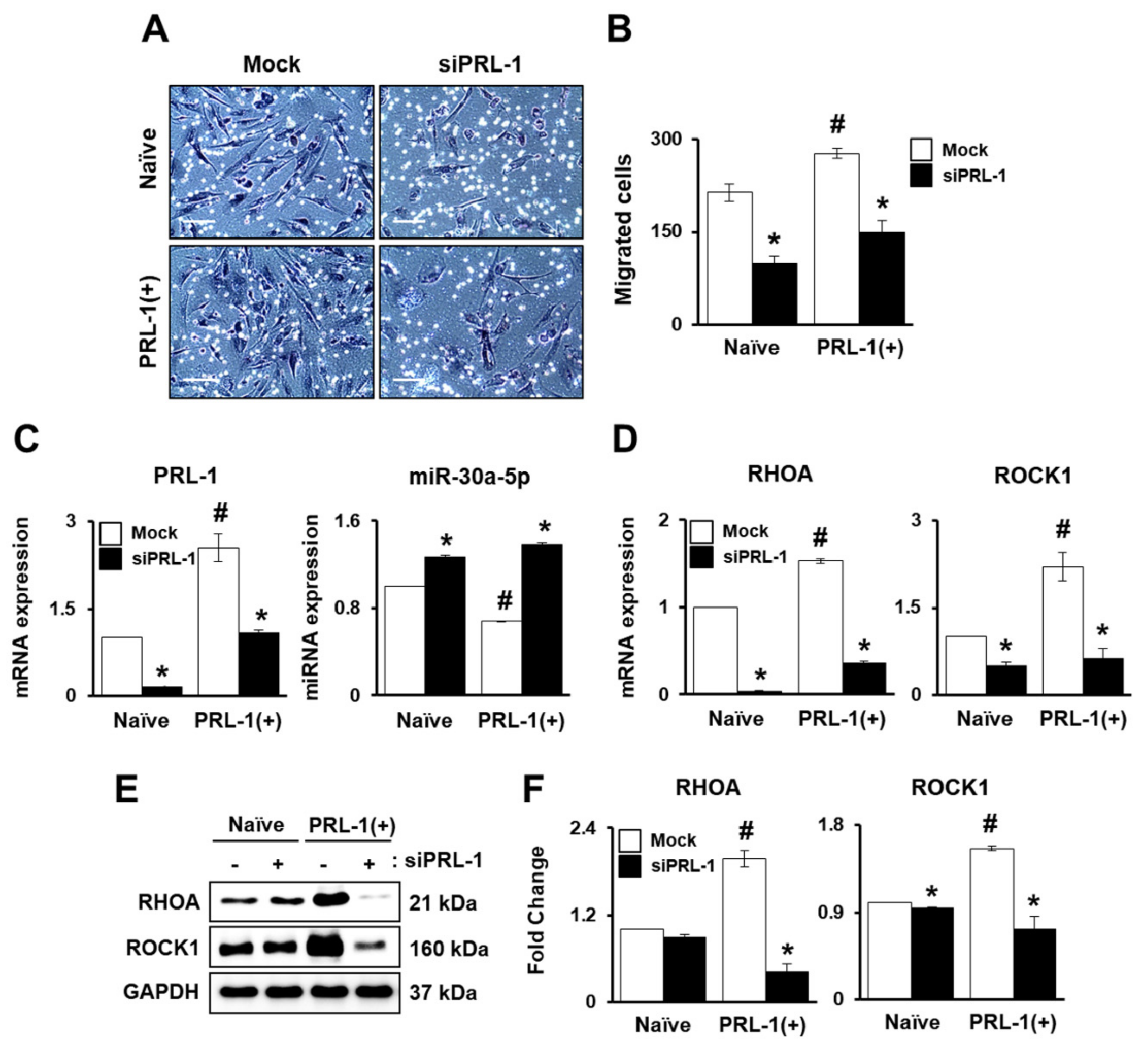
2.2. PRL-1-Dependent Migration Ability under Hypoxic Conditions Regulatedby miRNAs Targeting the Integrin Family
2.3. PRL-1-Targeted miRNA Expression Regulates Migration Ability through the RHO Family
2.4. miRNA Expression Regulates Integrin Family for PRL-1(+) PD-MSC Homing In Vivo in a Rat Model with BDL
2.5. Improved Vascular Remodeling by PRL-1(+) PD-MSCs through the Regulation of miRNA Expression by Platelet-Derived Growth Factor Receptor A(PDGFRA) in a BDL-injured Rat Model
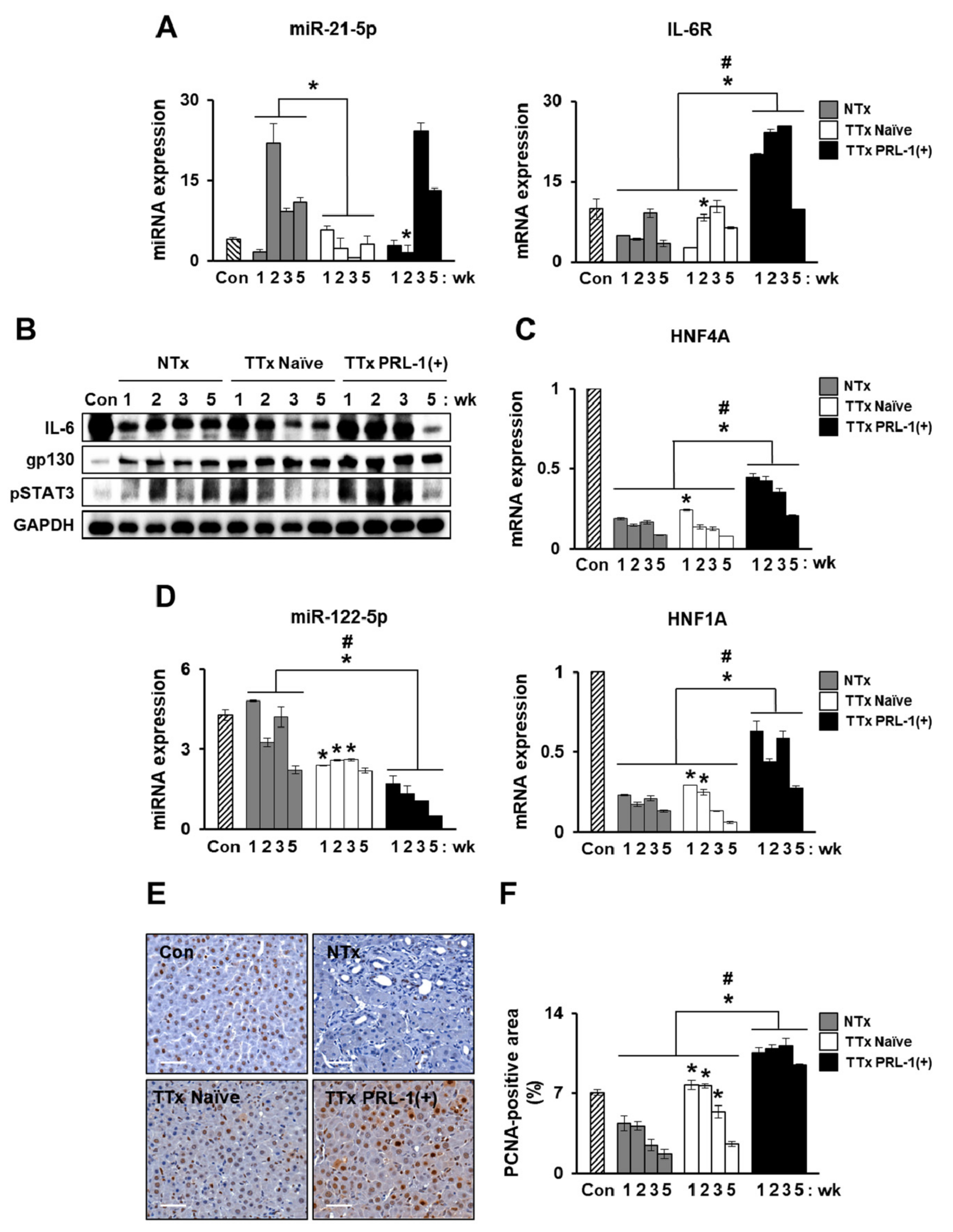
2.6. miRNAs Mediate Hepatic Regeneration by PRL-1(+) PD-MSCs in a Rat Model with BDL through IL-6/STAT3 Signaling
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Gene Transfection
4.2. Animal Models and MSC Transplantation
4.3. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.4. Immunohistochemistry
4.5. Immunofluorescence
4.6. Western Blotting
4.7. Transwell Migration Assay Using Transfection
4.8. Dual Luciferase Assay
4.9. Deep Sequencing and Analysis of Small RNAs
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Friedman, S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology 2008, 134, 1655–1669. [Google Scholar] [CrossRef] [PubMed]
- Praktiknjo, M.; Lehmann, J.; Nielsen, M.J.; Schierwagen, R.; Uschner, F.E.; Meyer, C.; Thomas, D.; Strassburg, C.P.; Bendtsen, F.; Moller, S.; et al. Acute decompensation boosts hepatic collagen type III deposition and deteriorates experimental and human cirrhosis. Hepatol. Commun. 2018, 2, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Sheppard, D.; Duffield, J.S.; Violette, S. Therapy for fibrotic diseases: Nearing the starting line. Sci. Transl. Med. 2013, 5, 167sr161. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yannam, G.R.; Nishikawa, T.; Yamamoto, T.; Basma, H.; Ito, R.; Nagaya, M.; Dutta-Moscato, J.; Stolz, D.B.; Duan, F.; et al. The microenvironment in hepatocyte regeneration and function in rats with advanced cirrhosis. Hepatology 2012, 55, 1529–1539. [Google Scholar] [CrossRef]
- Arany, Z.; Foo, S.Y.; Ma, Y.; Ruas, J.L.; Bommi-Reddy, A.; Girnun, G.; Cooper, M.; Laznik, D.; Chinsomboon, J.; Rangwala, S.M.; et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature 2008, 451, 1008–1012. [Google Scholar] [CrossRef]
- Yukawa, H.; Noguchi, H.; Oishi, K.; Takagi, S.; Hamaguchi, M.; Hamajima, N.; Hayashi, S. Cell transplantation of adipose tissue-derived stem cells in combination with heparin attenuated acute liver failure in mice. Cell Transplant. 2009, 18, 611–618. [Google Scholar] [CrossRef]
- Abdel aziz, M.T.; El Asmar, M.F.; Atta, H.M.; Mahfouz, S.; Fouad, H.H.; Roshdy, N.K.; Rashed, L.A.; Sabry, D.; Hassouna, A.A.; Taha, F.M. Efficacy of mesenchymal stem cells in suppression of hepatocarcinorigenesis in rats: Possible role of Wnt signaling. J.Exp.Clin. Cancer Res. 2011, 30, 49. [Google Scholar] [CrossRef]
- Jung, J.; Choi, J.H.; Lee, Y.; Park, J.W.; Oh, I.H.; Hwang, S.G.; Kim, K.S.; Kim, G.J. Human placenta-derived mesenchymal stem cells promote hepatic regeneration in CCl4 -injured rat liver model via increased autophagic mechanism. Stem Cells 2013, 31, 1584–1596. [Google Scholar] [CrossRef]
- Jung, J.; Moon, J.W.; Choi, J.H.; Lee, Y.W.; Park, S.H.; Kim, G.J. Epigenetic Alterations of IL-6/STAT3 Signaling by Placental Stem Cells Promote Hepatic Regeneration in a Rat Model with CCl4-induced Liver Injury. Int. J. Stem Cells 2015, 8, 79–89. [Google Scholar] [CrossRef]
- Haider, H.; Jiang, S.; Idris, N.M.; Ashraf, M. IGF-1-overexpressing mesenchymal stem cells accelerate bone marrow stem cell mobilization via paracrine activation of SDF-1alpha/CXCR4 signaling to promote myocardial repair. Circ. Res. 2008, 103, 1300–1308. [Google Scholar] [CrossRef]
- Teo, G.S.; Ankrum, J.A.; Martinelli, R.; Boetto, S.E.; Simms, K.; Sciuto, T.E.; Dvorak, A.M.; Karp, J.M.; Carman, C.V. Mesenchymal stem cells transmigrate between and directly through tumor necrosis factor-alpha-activated endothelial cells via both leukocyte-like and novel mechanisms. Stem Cells 2012, 30, 2472–2486. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Lim, S.M.; Yoo, Y.I.; Jung, J.; Park, J.W.; Kim, G.J. Microenvironmental Interaction Between Hypoxia and Endothelial Cells Controls the Migration Ability of Placenta-Derived Mesenchymal Stem Cells via alpha4 Integrin and Rho Signaling. J. Cell Biochem. 2016, 117, 1145–1157. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, L.K.; Dai, T.; Wang, A.; Li, S. Adult Stem Cells in Vascular Remodeling. Theranostics 2018, 8, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Mueller, L.; Broering, D.C.; Meyer, J.; Vashist, Y.; Goettsche, J.; Wilms, C.; Rogiers, X. The induction of the immediate-early-genes Egr-1, PAI-1 and PRL-1 during liver regeneration in surgical models is related to increased portal flow. J. Hepatol. 2002, 37, 606–612. [Google Scholar] [CrossRef]
- Peng, Y.; Du, K.; Ramirez, S.; Diamond, R.H.; Taub, R. Mitogenic up-regulation of the PRL-1 protein-tyrosine phosphatase gene by Egr-1. Egr-1 activation is an early event in liver regeneration. J. Biol. Chem. 1999, 274, 4513–4520. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Luo, Y.; Liu, S.; Zhang, L.; Shen, K.; Dong, Y.; Walls, C.D.; Quilliam, L.A.; Wells, C.D.; Cao, Y.; et al. PRL-1 protein promotes ERK1/2 and RhoA protein activation through a non-canonical interaction with the Src homology 3 domain of p115 Rho GTPase-activating protein. J. Biol. Chem. 2011, 286, 42316–42324. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Sui, L.; Wang, Q.; Chen, M.; Sun, H. MicroRNA-26a inhibits cell proliferation and invasion of cervical cancer cells by targeting protein tyrosine phosphatase type IVA 1. Mol. Med. Rep. 2014, 10, 1426–1432. [Google Scholar] [CrossRef]
- Flores-Perez, A.; Marchat, L.A.; Rodriguez-Cuevas, S.; Bautista, V.P.; Fuentes-Mera, L.; Romero-Zamora, D.; Maciel-Dominguez, A.; de la Cruz, O.H.; Fonseca-Sanchez, M.; Ruiz-Garcia, E.; et al. Suppression of cell migration is promoted by miR-944 through targeting of SIAH1 and PTP4A1 in breast cancer cells. BMC Cancer 2016, 16, 379. [Google Scholar] [CrossRef]
- Hu, J.Y.; Yi, W.; Wei, X.; Zhang, M.Y.; Xu, R.; Zeng, L.S.; Huang, Z.J.; Chen, J.S. miR-601 is a prognostic marker and suppresses cell growth and invasion by targeting PTP4A1 in breast cancer. Biomed.Pharm. 2016, 79, 247–253. [Google Scholar] [CrossRef]
- Li, Z.; Rana, T.M. Therapeutic targeting of microRNAs: Current status and future challenges. Nat. Rev. Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef]
- Li, N.; Long, B.; Han, W.; Yuan, S.; Wang, K. microRNAs: Important regulators of stem cells. Stem Cell Res. Ther. 2017, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.; Ge, J.; Xiu, L.; Zhao, Z.; Duan, X.; Tian, L.; Xie, J.; Yang, L.; Li, L. HuR mediates motility of human bone marrow-derived mesenchymal stem cells triggered by sphingosine 1-phosphate in liver fibrosis. J. Mol. Med. (Berl) 2017, 95, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Bi, Z.M.; Zhou, Q.F.; Geng, Y.; Zhang, H.M. Human umbilical cord mesenchymal stem cells ameliorate experimental cirrhosis through activation of keratinocyte growth factor by suppressing microRNA-199. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4905–4912. [Google Scholar] [PubMed]
- Nitzsche, F.; Muller, C.; Lukomska, B.; Jolkkonen, J.; Deten, A.; Boltze, J. Concise Review: MSC Adhesion Cascade-Insights into Homing and Transendothelial Migration. Stem Cells 2017, 35, 1446–1460. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Song, G.; Ju, Y.; Li, X.; Song, Y.; Watanabe, S. RhoA/ROCK, cytoskeletal dynamics, and focal adhesion kinase are required for mechanical stretch-induced tenogenic differentiation of human mesenchymal stem cells. J. Cell Physiol. 2012, 227, 2722–2729. [Google Scholar] [CrossRef] [PubMed]
- Eom, Y.W.; Shim, K.Y.; Baik, S.K. Mesenchymal stem cell therapy for liver fibrosis. Korean J. Intern. Med. 2015, 30, 580–589. [Google Scholar] [CrossRef]
- Alfaifi, M.; Eom, Y.W.; Newsome, P.N.; Baik, S.K. Mesenchymal stromal cell therapy for liver diseases. J. Hepatol. 2018, 68, 1272–1285. [Google Scholar] [CrossRef]
- Clark, E.A.; Kalomoiris, S.; Nolta, J.A.; Fierro, F.A. Concise review: MicroRNA function in multipotent mesenchymal stromal cells. Stem Cells 2014, 32, 1074–1082. [Google Scholar] [CrossRef]
- Saller, M.M.; Prall, W.C.; Docheva, D.; Schonitzer, V.; Popov, T.; Anz, D.; Clausen-Schaumann, H.; Mutschler, W.; Volkmer, E.; Schieker, M.; et al. Increased stemness and migration of human mesenchymal stem cells in hypoxia is associated with altered integrin expression. Biochem. Biophys. Res. Commun. 2012, 423, 379–385. [Google Scholar] [CrossRef]
- Choi, J.H.; Lee, Y.B.; Jung, J.; Hwang, S.G.; Oh, I.H.; Kim, G.J. Hypoxia Inducible Factor-1alpha Regulates the Migration of Bone Marrow Mesenchymal Stem Cells via Integrin alpha 4. Stem Cells Int. 2016, 2016, 7932185. [Google Scholar] [CrossRef]
- Luo, Y.; Liang, F.; Zhang, Z.Y. PRL1 promotes cell migration and invasion by increasing MMP2 and MMP9 expression through Src and ERK1/2 pathways. Biochemistry 2009, 48, 1838–1846. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lazo, J.S. Metastasis-associated phosphatase PRL-2 regulates tumor cell migration and invasion. Oncogene 2012, 31, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Gari, H.H.; DeGala, G.D.; Ray, R.; Lucia, M.S.; Lambert, J.R. PRL-3 engages the focal adhesion pathway in triple-negative breast cancer cells to alter actin structure and substrate adhesion properties critical for cell migration and invasion. Cancer Lett. 2016, 380, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.L. Integrin signaling through FAK in the regulation of mammary stem cells and breast cancer. IUBMB Life 2010, 62, 268–276. [Google Scholar] [CrossRef]
- Vitillo, L.; Kimber, S.J. Integrin and FAK Regulation of Human Pluripotent Stem Cells. Curr. Stem Cell Rep. 2017, 3, 358–365. [Google Scholar] [CrossRef]
- Drescher, H.K.; Schippers, A.; Clahsen, T.; Sahin, H.; Noels, H.; Hornef, M.; Wagner, N.; Trautwein, C.; Streetz, K.L.; Kroy, D.C. β7-Integrin and MAdCAM-1 play opposing roles during the development of non-alcoholic steatohepatitis. J. Hepatol. 2017, 66, 1251–1264. [Google Scholar] [CrossRef]
- Aldridge, V.; Garg, A.; Davies, N.; Bartlett, D.C.; Youster, J.; Beard, H.; Kavanagh, D.P.; Kalia, N.; Frampton, J.; Lalor, P.F.; et al. Human mesenchymal stem cells are recruited to injured liver in a beta1-integrin and CD44 dependent manner. Hepatology 2012, 56, 1063–1073. [Google Scholar] [CrossRef]
- Chute, J.P. Stem cell homing. Curr. Opin Hematol. 2006, 13, 399–406. [Google Scholar] [CrossRef]
- Amiri, F.; Molaei, S.; Bahadori, M.; Nasiri, F.; Deyhim, M.R.; Jalili, M.A.; Nourani, M.R.; Habibi Roudkenar, M. Autophagy-Modulated Human Bone Marrow-Derived Mesenchymal Stem Cells Accelerate Liver Restoration in Mouse Models of Acute Liver Failure. Iran. Biomed. J. 2016, 20, 135–144. [Google Scholar]
- Yang, L.; Dong, C.; Yang, J.; Yang, L.; Chang, N.; Qi, C.; Li, L. MicroRNA-26b-5p Inhibits Mouse Liver Fibrogenesis and Angiogenesis by Targeting PDGF Receptor-Beta. Mol. Ther. Nucleic Acids 2019, 16, 206–217. [Google Scholar] [CrossRef]
- Jiao, Y.; Ye, D.Z.; Li, Z.; Teta-Bissett, M.; Peng, Y.; Taub, R.; Greenbaum, L.E.; Kaestner, K.H. Protein tyrosine phosphatase of liver regeneration-1 is required for normal timing of cell cycle progression during liver regeneration. Am.J. Physiol. Gastrointest Liver Physiol. 2015, 308, G85–G91. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Xin, J.; Jiang, L.; Zhou, Q.; Wu, T.; Shi, D.; Lin, B.; Li, L.; Li, J. Characterisation of peripheral blood mononuclear cell microRNA in hepatitis B-related acute-on-chronic liver failure. Sci. Rep. 2015, 5, 13098. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Xu, Y.; Hao, J.; Wang, S.; Li, C.; Meng, S. MiR-122 in hepatic function and liver diseases. Protein Cell 2012, 3, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Lou, G.; Yang, Y.; Liu, F.; Ye, B.; Chen, Z.; Zheng, M.; Liu, Y. MiR-122 modification enhances the therapeutic efficacy of adipose tissue-derived mesenchymal stem cells against liver fibrosis. J. Cell Mol.Med. 2017, 21, 2963–2973. [Google Scholar] [CrossRef] [PubMed]
- Baranova, A.; Maltseva, D.; Tonevitsky, A. Adipose may actively delay progression of NAFLD by releasing tumor-suppressing, anti-fibrotic miR-122 into circulation. Obes. Rev. 2019, 20, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Shin, K.S.; Jeon, J.H.; Lee, D.R.; Shim, S.H.; Kim, J.K.; Cha, D.H.; Yoon, T.K.; Kim, G.J. Human chorionic-plate-derived mesenchymal stem cells and Wharton’s jelly-derived mesenchymal stem cells: A comparative analysis of their potential as placenta-derived stem cells. Cell Tissue Res. 2011, 346, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Jung, J.; Cho, K.J.; Lee, C.K.; Hwang, S.G.; Kim, G.J. Comparison of in vitro hepatogenic differentiation potential between various placenta-derived stem cells and other adult stem cells as an alternative source of functional hepatocytes. Differentiation 2012, 84, 223–231. [Google Scholar] [CrossRef]
- Kountouras, J.; Billing, B.H.; Scheuer, P.J. Prolonged bile duct obstruction: A new experimental model for cirrhosis in the rat. Br. J. Exp. Pathol. 1984, 65, 305–311. [Google Scholar]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.Y.; Jun, J.H.; Park, S.Y.; Yang, S.W.; Bae, S.H.; Kim, G.J. Dynamic Regulation of miRNA Expression by Functionally Enhanced Placental Mesenchymal Stem Cells Promotes Hepatic Regeneration in a Rat Model with Bile Duct Ligation. Int. J. Mol. Sci. 2019, 20, 5299. https://doi.org/10.3390/ijms20215299
Kim JY, Jun JH, Park SY, Yang SW, Bae SH, Kim GJ. Dynamic Regulation of miRNA Expression by Functionally Enhanced Placental Mesenchymal Stem Cells Promotes Hepatic Regeneration in a Rat Model with Bile Duct Ligation. International Journal of Molecular Sciences. 2019; 20(21):5299. https://doi.org/10.3390/ijms20215299
Chicago/Turabian StyleKim, Jae Yeon, Ji Hye Jun, Soo Young Park, Seong Wook Yang, Si Hyun Bae, and Gi Jin Kim. 2019. "Dynamic Regulation of miRNA Expression by Functionally Enhanced Placental Mesenchymal Stem Cells Promotes Hepatic Regeneration in a Rat Model with Bile Duct Ligation" International Journal of Molecular Sciences 20, no. 21: 5299. https://doi.org/10.3390/ijms20215299
APA StyleKim, J. Y., Jun, J. H., Park, S. Y., Yang, S. W., Bae, S. H., & Kim, G. J. (2019). Dynamic Regulation of miRNA Expression by Functionally Enhanced Placental Mesenchymal Stem Cells Promotes Hepatic Regeneration in a Rat Model with Bile Duct Ligation. International Journal of Molecular Sciences, 20(21), 5299. https://doi.org/10.3390/ijms20215299





