The Roles of MicroRNAs (miRNAs) in Avian Response to Viral Infection and Pathogenesis of Avian Immunosuppressive Diseases
Abstract
:1. Introduction
2. miRNA in Marek’s Disease (MD)
2.1. Marek’s Disease Virus (MDV)-Encoded miRNA
2.2. The Role of MDV1-Encoded miRNAs in MD
2.3. The Role of Cellular miRNAs in MD
3. miRNA in Avian Leukemia (AL)
3.1. Avian Leukosis Virus (ALV)-Encoded miRNA
3.2. The Role of Cellular miRNAs in AL
4. miRNA in infectious bursal disease (IBD)
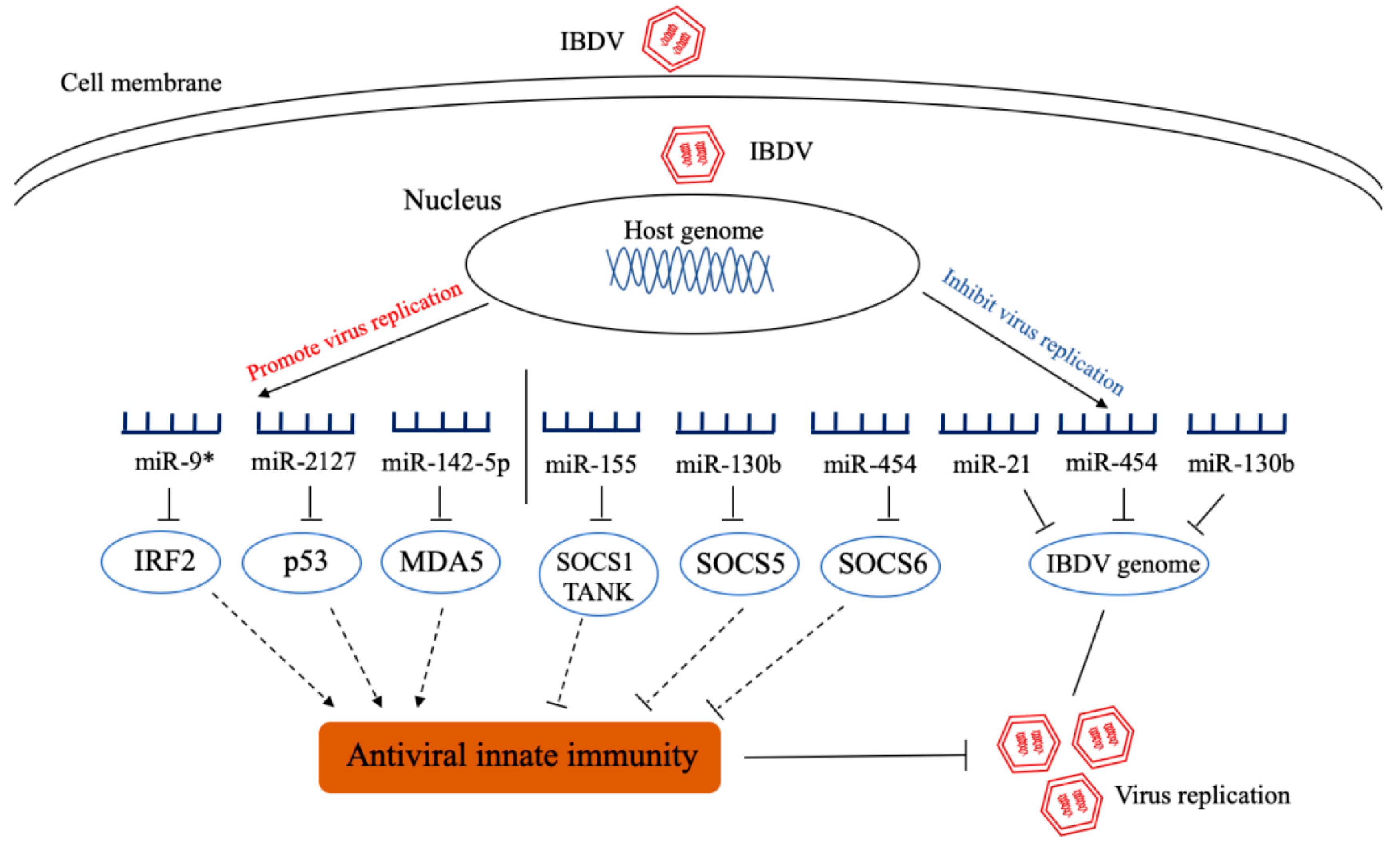
4.1. The Role of miRNA in the Pathogenesis of Infectious Bursal Disease Virus (IBDV)
4.2. The Role of miRNA in the Control of IBDV
5. miRNA in Reticuloendotheliosis (RE)
6. Conclusions
Funding
Conflicts of Interest
References
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Sewer, A.; Lagos-Quintana, M.; Sheridan, R.; Sander, C.; Grässer, F.A.; van Dyk, L.F.; Ho, C.K.; Shuman, S.; Chien, M.; et al. Identification of microRNAs of the herpesvirus family. Nat. Methods 2005, 2, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Clayton, S.A.; Jones, S.W.; Kurowska-Stolarska, M.; Clark, A.R. The role of microRNAs in glucocorticoid action. J. Biol. Chem. 2018, 293, 1865–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, É.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Filipowicz, W.; Jaskiewicz, L.; Kolb, F.A.; Pillai, R.S. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005, 15, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Siomi, H.; Siomi, M.C. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell 2010, 38, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Nair, V. Role of virus-encoded microRNAs in Avian viral diseases. Viruses 2014, 6, 1379–1394. [Google Scholar] [CrossRef] [PubMed]
- Schat, K.A.; Skinner, M.A. Avian immunosuppressive diseases and immunoevasion. In Avian immunology, 2nd ed.; Schat, K.A., Kaspers, B., Skinner, M.A., Eds.; Elsevier Ltd., Academic Press: Great Britain, 2014; pp. 275–297. [Google Scholar]
- Bondada, M.S.; Yao, Y.; Nair, V. Multifunctional miR-155 Pathway in Avian Oncogenic Virus-Induced Neoplastic Diseases. Noncoding RNA 2019, 5, 24. [Google Scholar] [CrossRef]
- Schat, K.A. Marek’s disease immunosuppression. In Marek’s Disease; Davison, F., Nair, V., Eds.; Elsevier Ltd., Academic Press: Great Britain, 2004; pp. 142–155, vii. [Google Scholar]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Burnside, J.; Bernberg, E.; Anderson, A.; Lu, C.; Meyers, B.C.; Green, P.J.; Jain, N.; Isaacs, G.; Morgan, R.W. Marek’s disease virus encodes MicroRNAs that map to meq and the latency-associated transcript. J. Virol. 2006, 80, 8778–8786. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Y.; Xu, H.; Smith, L.P.; Lawrie, C.H.; Watson, M.; Nair, V. MicroRNA profile of Marek’s disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J. Virol. 2008, 82, 4007–4015. [Google Scholar] [CrossRef]
- Boss, I.W.; Plaisance, K.B.; Renne, R. Role of virus-encoded microRNAs in herpesvirus biology. Trends Microbiol. 2009, 17, 544–553. [Google Scholar] [CrossRef] [Green Version]
- Hicks, J.A.; Liu, H.-C. Current state of Marek’s disease virus microRNA research. Avian dis. 2013, 57, 332–339. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Y.; Xu, H.; Smith, L.P.; Lawrie, C.H.; Sewer, A.; Zavolan, M.; Nair, V. Marek’s disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J. Virol. 2007, 81, 7164–7170. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Y.; Smith, L.P.; Watson, M.; Nair, V. Novel microRNAs (miRNAs) encoded by herpesvirus of Turkeys: evidence of miRNA evolution by duplication. J. Virol. 2009, 83, 6969–6973. [Google Scholar] [CrossRef] [PubMed]
- Waidner, L.A.; Morgan, R.W.; Anderson, A.S.; Bernberg, E.L.; Kamboj, S.; Garcia, M.; Riblet, S.M.; Ouyang, M.; Isaacs, G.K.; Markis, M.; et al. MicroRNAs of Gallid and Meleagrid herpesviruses show generally conserved genomic locations and are virus-specific. Virology 2009, 388, 128–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Xu, H.; Yao, Y.; Smith, L.P.; Kgosana, L.; Green, J.; Petherbridge, L.; Baigent, S.J.; Nair, V. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathog. 2011, 7, e1001305. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.H.; Teng, M.; Sun, A.J.; Yu, L.L.; Hu, B.; Qu, L.H.; Ding, K.; Cheng, X.C.; Liu, J.X.; Cui, Z.Z.; et al. Virus-encoded miR-155 ortholog is an important potential regulator but not essential for the development of lymphomas induced by very virulent Marek’s disease virus. Virology 2014, 448, 55–64. [Google Scholar] [CrossRef]
- Morgan, R.; Anderson, A.; Bernberg, E.; Kamboj, S.; Huang, E.; Lagasse, G.; Isaacs, G.; Parcells, M.; Meyers, B.C.; Green, P.J.; et al. Sequence conservation and differential expression of Marek’s disease virus microRNAs. J. Virol. 2008, 82, 12213–12220. [Google Scholar] [CrossRef]
- Zhao, Y.; Yao, Y.; Xu, H.; Lambeth, L.; Smith, L.P.; Kgosana, L.; Wang, X.; Nair, V. A functional MicroRNA-155 ortholog encoded by the oncogenic Marek’s disease virus. J. Virol. 2009, 83, 489–492. [Google Scholar] [CrossRef]
- Muylkens, B.; Coupeau, D.; Dambrine, G.; Trapp, S.; Rasschaert, D. Marek’s disease virus microRNA designated Mdv1-pre-miR-M4 targets both cellular and viral genes. Arch. Virol. 2010, 155, 1823–1837. [Google Scholar] [CrossRef]
- Chi, J.Q.; Teng, M.; Yu, Z.H.; Xu, H.; Su, J.W.; Zhao, P.; Xing, G.X.; Liang, H.D.; Deng, R.G.; Qu, L.H.; et al. Marek’s disease virus-encoded analog of microRNA-155 activates the oncogene c-Myc by targeting LTBP1 and suppressing the TGF-beta signaling pathway. Virology 2015, 476, 72–84. [Google Scholar] [CrossRef]
- Dang, L.; Teng, M.; Li, H.Z.; Ma, S.M.; Lu, Q.X.; Hao, H.F.; Zhao, D.; Zhou, E.M.; Zhang, G.P.; Luo, J. Marek’s disease virus type 1 encoded analog of miR-155 promotes proliferation of chicken embryo fibroblast and DF-1 cells by targeting hnRNPAB. Vet. Microbiol. 2017, 207, 210–218. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, N.; Luo, J.; Teng, M.; Moffat, K.; Shen, Z.; Watson, M.; Nair, V.; Yao, Y. Marek’s disease virus-encoded miR-155 ortholog critical for the induction of lymphomas is not essential for the proliferation of transformed cell lines. J. Virol. 2019, 93, e00713-19. [Google Scholar] [CrossRef]
- Figueroa, T.; Boumart, I.; Coupeau, D.; Rasschaert, D. Hyperediting by ADAR1 of a new herpesvirus lncRNA during the lytic phase of the oncogenic Marek’s disease virus. J. Gen. Virol. 2016, 97, 2973–2988. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Su, R.; Ruan, J.; Shao, H.; Qian, K.; Ye, J.; Qin, A. Toll-like receptor 3 pathway restricts Marek’s disease virus infection. Oncotarget 2017, 8, 70847–70853. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zou, H.; Qin, A.; Qian, K.; Shao, H.; Ye, J. Activation of Toll-like receptor 3 inhibits Marek’s disease virus infection in chicken embryo fibroblast cells. Arch. Virol. 2016, 161, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Teng, M.; Yu, Z.H.; Sun, A.J.; Min, Y.J.; Chi, J.Q.; Zhao, P.; Su, J.W.; Cui, Z.Z.; Zhang, G.P.; Luo, J. The significance of the individual Meq-clustered miRNAs of Marek’s disease virus in oncogenesis. J. Gen. Virol. 2015, 96, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Xue, C.; Li, J.; Bi, Y.; Cao, Y. Marek’s disease virus type 1 microRNA miR-M3 suppresses cisplatin-induced apoptosis by targeting Smad2 of the transforming growth factor beta signal pathway. J. Virol. 2011, 85, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Boumart, I.; Figueroa, T.; Dambrine, G.; Muylkens, B.; Pejakovic, S.; Rasschaert, D.; Dupuy, C. GaHV-2 ICP22 protein is expressed from a bicistronic transcript regulated by three GaHV-2 microRNAs. J. Gen. Virol. 2018, 99, 1286–1300. [Google Scholar] [CrossRef]
- Teng, M.; Yu, Z.H.; Zhao, P.; Zhuang, G.Q.; Wu, Z.X.; Dang, L.; Li, H.Z.; Ma, S.M.; Cui, Z.Z.; Zhang, G.P.; et al. Putative roles as oncogene or tumour suppressor of the Mid-clustered microRNAs in Gallid alphaherpesvirus 2 (GaHV2) induced Marek’s disease lymphomagenesis. J. Gen. Virol. 2017, 98, 1097–1112. [Google Scholar] [CrossRef]
- Strassheim, S.; Stik, G.; Rasschaert, D.; Laurent, S. mdv1-miR-M7-5p, located in the newly identified first intron of the latency-associated transcript of Marek’s disease virus, targets the immediate-early genes ICP4 and ICP27. J. Gen. Virol. 2012, 93, 1731–1742. [Google Scholar] [CrossRef]
- Burnside, J.; Ouyang, M.; Anderson, A.; Bernberg, E.; Lu, C.; Meyers, B.C.; Green, P.J.; Markis, M.; Isaacs, G.; Huang, E.; et al. Deep sequencing of chicken microRNAs. BMC Genom. 2008, 9, 185. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Y.; Smith, L.P.; Lawrie, C.H.; Saunders, N.J.; Watson, M.; Nair, V. Differential expression of microRNAs in Marek’s disease virus-transformed T-lymphoma cell lines. J. Gen. Virol. 2009, 90, 1551–1559. [Google Scholar] [CrossRef]
- Lambeth, L.S.; Yao, Y.; Smith, L.P.; Zhao, Y.; Nair, V. MicroRNAs 221 and 222 target p27Kip1 in Marek’s disease virus-transformed tumour cell line MSB-1. J. Gen. Virol. 2009, 90, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Stik, G.; Dambrine, G.; Pfeffer, S.; Rasschaert, D. The oncogenic microRNA OncomiR-21 overexpressed during Marek’s disease lymphomagenesis is transactivated by the viral oncoprotein Meq. J. Virol. 2013, 87, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yao, Y.; Smith, L.P.; Nair, V. MicroRNA-26a-mediated regulation of interleukin-2 expression in transformed avian lymphocyte lines. Cancer Cell Int. 2010, 10, 15. [Google Scholar] [CrossRef]
- Li, X.; Lian, L.; Zhang, D.; Qu, L.; Yang, N. gga-miR-26a targets NEK6 and suppresses Marek’s disease lymphoma cell proliferation. Poult Sci. 2014, 93, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Lian, L.; Zhang, D.; Wang, Q.; Yang, N.; Qu, L. The inhibitory effects of gga-miR-199-3p, gga-miR-140-3p, and gga-miR-221-5p in Marek’s disease tumorigenesis. Poultry Sci. 2015, 94, 2131–2135. [Google Scholar] [CrossRef] [PubMed]
- Lian, L.; Li, X.; Zhao, C.; Han, B.; Qu, L.; Song, J.; Liu, C.; Yang, N. Chicken gga-miR-181a targets MYBL1 and shows an inhibitory effect on proliferation of Marek’s disease virus-transformed lymphoid cell line. Poultry Sci. 2015, 94, 2616–2621. [Google Scholar] [CrossRef]
- Han, B.; Lian, L.; Li, X.; Zhao, C.; Qu, L.; Liu, C.; Song, J.; Yang, N. Chicken gga-miR-130a targets HOXA3 and MDFIC and inhibits Marek’s disease lymphoma cell proliferation and migration. Mol. Biol. Rep. 2016, 43, 667–676. [Google Scholar] [CrossRef]
- Zhao, C.; Li, X.; Han, B.; You, Z.; Qu, L.; Liu, C.; Song, J.; Lian, L.; Yang, N. Gga-miR-219b targeting BCL11B suppresses proliferation, migration and invasion of Marek’s disease tumor cell MSB1. Sci. Rep. 2017, 7, 4247. [Google Scholar] [CrossRef]
- Han, B.; Lian, L.; Li, X.; Zhao, C.; Qu, L.; Liu, C.; Song, J.; Yang, N. Chicken gga-miR-103-3p Targets CCNE1 and TFDP2 and Inhibits MDCC-MSB1 Cell Migration. G3 (Bethesda) 2016, 6, 1277–1285. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Li, X.; Han, B.; Qu, L.; Liu, C.; Song, J.; Lian, L.; Yang, N. Gga-miR-130b-3p inhibits MSB1 cell proliferation, migration, invasion, and its downregulation in MD tumor is attributed to hypermethylation. Oncotarget 2018, 9, 24187–24198. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Nair, V. Avian Leukosis Virus. In Molecular Detection of Animal Viral Pathogens; Liu, D., Ed.; CRC Press: Boca Raton, FL, USA, 2016; p. 145. [Google Scholar]
- Přikryl, D.; Plachý, J.; Kučerová, D.; Koslová, A.; Reinišová, M.; Šenigl, F.; Hejnar, J. The novel avian leukosis virus subgroup K shares its cellular chicken receptor with subgroup A. J. Virol. 2019, 93, e00580-19. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Smith, L.P.; Nair, V.; Watson, M. An avian retrovirus uses canonical expression and processing mechanisms to generate viral microRNA. J. Virol. 2014, 88, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Chesters, P.M.; Smith, L.P.; Nair, V. E (XSR) element contributes to the oncogenicity of Avian leukosis virus (subgroup J). J. Gen. Virol. 2006, 87, 2685–2692. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ji, J.; Xie, Q.; Shang, H.; Zhang, H.; Xin, X.; Chen, F.; Sun, B.; Xue, C.; Ma, J.; et al. Aberrant expression of liver microRNA in chickens infected with subgroup J avian leukosis virus. Virus Res. 2012, 169, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, Y.; Ji, X.; Qi, X.; Qin, L.; Gao, H.; Wang, Y.; Wang, X. Differential expression of microRNAs in avian leukosis virus subgroup J-induced tumors. Vet. Microbiol. 2013, 162, 232–238. [Google Scholar] [CrossRef]
- Yao, Y.; Charlesworth, J.; Nair, V.; Watson, M. MicroRNA expression profiles in avian haemopoietic cells. Front. Genet. 2013, 4, 153. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Zhou, D.; Meng, W.; Li, G.; Zhuang, P.; Pan, Z.; Wang, G.; Cheng, Z. Growth retardation induced by avian leukosis virus subgroup J associated with down-regulated Wnt/beta-catenin pathway. Microb. Pathog. 2017, 104, 48–55. [Google Scholar] [CrossRef]
- Zhou, D.; Xue, J.; He, S.; Du, X.; Zhou, J.; Li, C.; Huang, L.; Nair, V.; Yao, Y.; Cheng, Z. Reticuloendotheliosis virus and avian leukosis virus subgroup J synergistically increase the accumulation of exosomal miRNAs. Retrovirology 2018, 15, 45. [Google Scholar] [CrossRef]
- Dai, M.; Feng, M.; Xie, T.; Zhang, X. Long non-coding RNA and MicroRNA profiling provides comprehensive insight into non-coding RNA involved host immune responses in ALV-J-infected chicken primary macrophage. Dev. Comp. Immunol. 2019, 100, 103414. [Google Scholar] [CrossRef]
- Li, Z.; Chen, B.; Feng, M.; Ouyang, H.; Zheng, M.; Ye, Q.; Nie, Q.; Zhang, X. MicroRNA-23b Promotes Avian Leukosis Virus Subgroup J (ALV-J) Replication by Targeting IRF1. Sci. Rep. 2015, 5, 10294. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Ji, X.; Gao, Y.; Qi, X.; Wang, X.; Wang, Y.; Qin, L.; Gao, H.; Wang, X. Overexpression of microRNA gga-miR-1650 decreases the replication of avian leukosis virus subgroup J in infected cells. J. Gen. Virol. 2013, 94, 2287–2296. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Luo, Q.; Xu, H.; Zheng, M.; Abdalla, B.A.; Feng, M.; Cai, B.; Zhang, X.; Nie, Q.; Zhang, X. MiR-34b-5p Suppresses Melanoma Differentiation-Associated Gene 5 (MDA5) Signaling Pathway to Promote Avian Leukosis Virus Subgroup J (ALV-J)-Infected Cells Proliferaction and ALV-J Replication. Front. Cell Infect. Microbiol. 2017, 7, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Shang, H.; Shu, D.; Zhang, H.; Ji, J.; Sun, B.; Li, H.; Xie, Q. gga-miR-375 plays a key role in tumorigenesis post subgroup J avian leukosis virus infection. PLoS ONE 2014, 9, e90878. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Yu, M.; Zhang, Y.; Fan, M.; Chang, F.; Xing, L.; Liu, Y.; Wang, Y.; Qi, X.; Liu, C.; et al. Avian leukosis virus subgroup J promotes cell proliferation and cell cycle progression through miR-221 by targeting CDKN1B. Virology 2018, 519, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Withers, D.R.; Young, J.R.; Davison, T.F. Infectious bursal disease virus-induced immunosuppression in the chick is associated with the presence of undifferentiated follicles in the recovering bursa. Viral Immunol. 2005, 18, 127–137. [Google Scholar] [CrossRef]
- Fu, M.; Wang, B.; Chen, X.; He, Z.; Wang, Y.; Li, X.; Cao, H.; Zheng, S.J. MicroRNA gga-miR-130b Suppresses Infectious Bursal Disease Virus Replication via Targeting of the Viral Genome and Cellular Suppressors of Cytokine Signaling 5. J. Virol. 2018, 92, e01646-17. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, W.; Wang, Y.S.; Du, X.N.; Liu, H.J.; Zhang, H.B. gga-miR-9* inhibits IFN production in antiviral innate immunity by targeting interferon regulatory factor 2 to promote IBDV replication. Vet. Microbiol. 2015, 178, 41–49. [Google Scholar] [CrossRef]
- Ouyang, W.; Wang, Y.S.; Meng, K.; Pan, Q.X.; Wang, X.L.; Xia, X.X.; Zhu, Y.M.; Bi, Z.W.; Zhang, H.B.; Luo, K. gga-miR-2127 downregulates the translation of chicken p53 and attenuates chp53-mediated innate immune response against IBDV infection. Vet. Microbiol. 2017, 198, 34–42. [Google Scholar] [CrossRef]
- Ouyang, W.; Qian, J.; Pan, Q.X.; Wang, J.Y.; Xia, X.X.; Wang, X.L.; Zhu, Y.M.; Wang, Y.S. gga-miR-142-5p attenuates IRF7 signaling and promotes replication of IBDV by directly targeting the chMDA5’s 3′ untranslated region. Vet. Microbiol. 2018, 221, 74–80. [Google Scholar] [CrossRef]
- Fu, M.; Wang, B.; Chen, X.; He, Z.; Wang, Y.; Li, X.; Cao, H.; Zheng, S.J. gga-miR-454 suppresses infectious bursal disease virus (IBDV) replication via directly targeting IBDV genomic segment B and cellular Suppressors of Cytokine Signaling 6 (SOCS6). Virus Res. 2018, 252, 29–40. [Google Scholar] [CrossRef]
- Wang, B.; Fu, M.; Liu, Y.; Wang, Y.; Li, X.; Cao, H.; Zheng, S.J. gga-miR-155 Enhances Type I Interferon Expression and Suppresses Infectious Burse Disease Virus Replication via Targeting SOCS1 and TANK. Front. Cell Infect. Microbiol. 2018, 8, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.S.; Ouyang, W.; Pan, Q.X.; Wang, X.L.; Xia, X.X.; Bi, Z.W.; Wang, Y.Q.; Wang, X.M. Overexpression of microRNA gga-miR-21 in chicken fibroblasts suppresses replication of infectious bursal disease virus through inhibiting VP1 translation. Antiviral. Res. 2013, 100, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, H.; Shen, P.; Zhang, X.; Xia, X.; Xia, B. Effective inhibition of replication of infectious bursal disease virus by miRNAs delivered by vectors and targeting the VP2 gene. J. Virol. Methods. 2010, 165, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, H.; Shen, P.; Zhang, X.; Xia, X. Effective inhibition of infectious bursal disease virus replication by recombinant avian adeno-associated virus-delivered microRNAs. J. Gen. Virol. 2009, 90, 1417–1422. [Google Scholar] [CrossRef]
- Lin, T.W.; Lo, C.W.; Lai, S.Y.; Fan, R.J.; Lo, C.J.; Chou, Y.M.; Thiruvengadam, R.; Wang, A.H.; Wang, M.Y. Chicken heat shock protein 90 is a component of the putative cellular receptor complex of infectious bursal disease virus. J. Virol. 2007, 81, 8730–8741. [Google Scholar] [CrossRef]
- Payne, L.; Venugopal, K. Neoplastic diseases: Marek’s disease, avian leukosis and reticuloendotheliosis. Rev. Sci. Tech. 2000, 19, 544–560. [Google Scholar] [CrossRef]
- Gonda, T.J.; Sheiness, D.K.; Fanshier, L.; Bishop, J.M.; Moscovici, C.; Moscovici, M.G. The genome and the intracellular RNAs of avian myeloblastosis virus. Cell 1981, 23, 279–290. [Google Scholar] [CrossRef]
- Bolisetty, M.T.; Dy, G.; Tam, W.; Beemon, K.L. Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J. Virol. 2009, 83, 12009–12017. [Google Scholar] [CrossRef]
- Yu, Z.; Gao, X.; Liu, C.; Lv, X.; Zheng, S. Analysis of microRNA expression profile in specific pathogen-free chickens in response to reticuloendotheliosis virus infection. Appl. Microbiol. Biotechnol. 2017, 101, 2767–2777. [Google Scholar] [CrossRef]


| Cluster | miRNA | Target Gene | Characteristics/Functions | References |
|---|---|---|---|---|
| Meq-cluster | MDV1-miR-M9 | / | Promote tumorigenesis | [35] |
| MDV1-miR-M5 | ICP22 | Promote tumorigenesis | [35,37] | |
| MDV1-miR-M12 | / | Promote tumorigenesis | [35] | |
| MDV1-miR-M3 | Smad2 | Inhibit cisplatin-induced apoptosis; Promote tumorigenesis | [35,36] | |
| MDV1-miR-M2 | / | Promote tumorigenesis | [35] | |
| MDV1-miR-M4-5p | GPM6B, RREB1, c-Myb, MAP3K7IP2, PU.1, LTBP1, C/EBP, hnRNPAB | Mimics cellular mir-155; Promote cell proliferation and tumorigenesis | [24,25,28,29,30,31] | |
| SOCS1 | Inhibit interferon production and promote viral replication | [32] | ||
| MDV1-miR-M4-3p | UL28, UL32 | Prevent lytic replication and promote latency | [28] | |
| Mid-cluster | MDV1-miR-M11 | / | Inhibit tumorigenesis | [38] |
| MDV1-miR-M31 | / | Promote tumorigenesis | [38] | |
| MDV1-miR-M1 | ICP22 | Inhibit tumorigenesis | [37,38] | |
| LAT-cluster | MDV1-miR-M7 | ICP4, ICP27 | Establish and/or maintain latency | [39] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Zheng, S.J. The Roles of MicroRNAs (miRNAs) in Avian Response to Viral Infection and Pathogenesis of Avian Immunosuppressive Diseases. Int. J. Mol. Sci. 2019, 20, 5454. https://doi.org/10.3390/ijms20215454
Zhou L, Zheng SJ. The Roles of MicroRNAs (miRNAs) in Avian Response to Viral Infection and Pathogenesis of Avian Immunosuppressive Diseases. International Journal of Molecular Sciences. 2019; 20(21):5454. https://doi.org/10.3390/ijms20215454
Chicago/Turabian StyleZhou, Linyi, and Shijun J. Zheng. 2019. "The Roles of MicroRNAs (miRNAs) in Avian Response to Viral Infection and Pathogenesis of Avian Immunosuppressive Diseases" International Journal of Molecular Sciences 20, no. 21: 5454. https://doi.org/10.3390/ijms20215454
APA StyleZhou, L., & Zheng, S. J. (2019). The Roles of MicroRNAs (miRNAs) in Avian Response to Viral Infection and Pathogenesis of Avian Immunosuppressive Diseases. International Journal of Molecular Sciences, 20(21), 5454. https://doi.org/10.3390/ijms20215454





