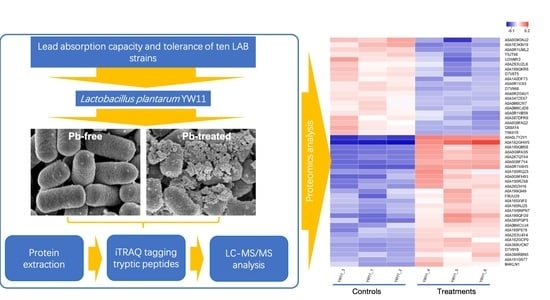
Evaluation and Proteomic Analysis of Lead Adsorption by Lactic Acid Bacteria
Abstract
1. Introduction
2. Results
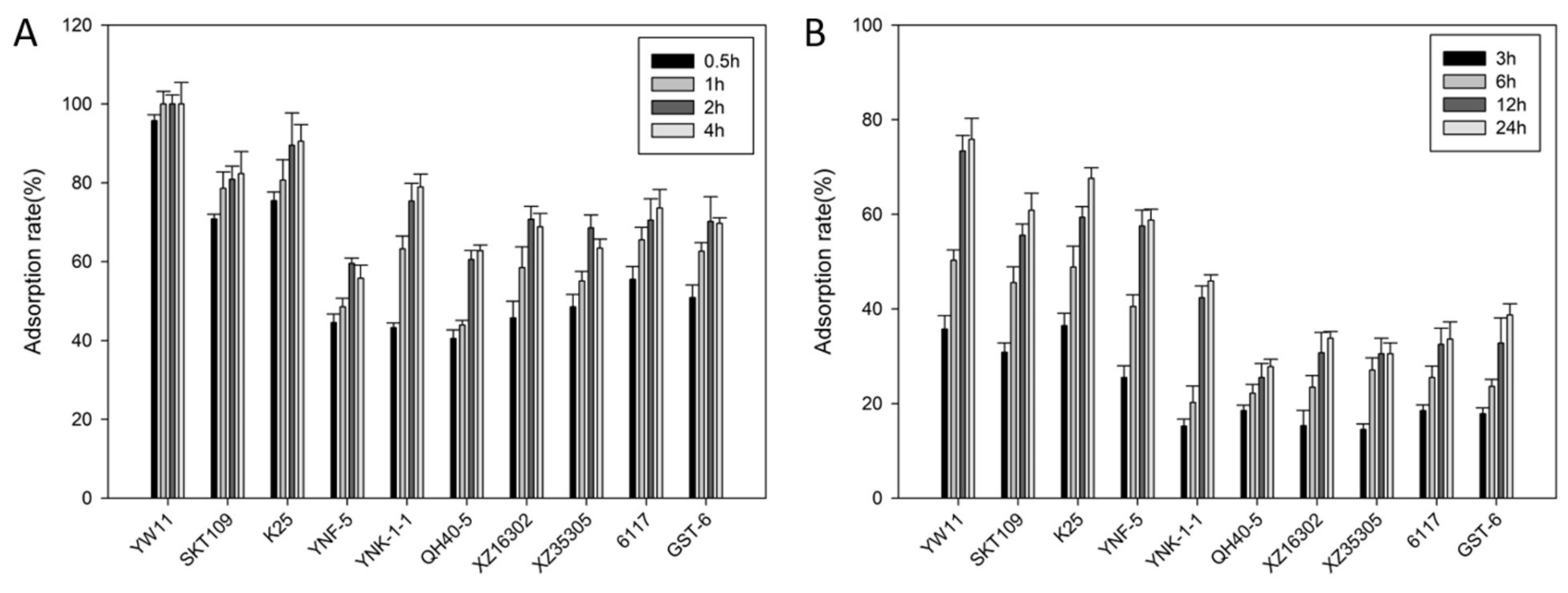
2.1. Lead Absorption Capacity and Tolerance of LAB
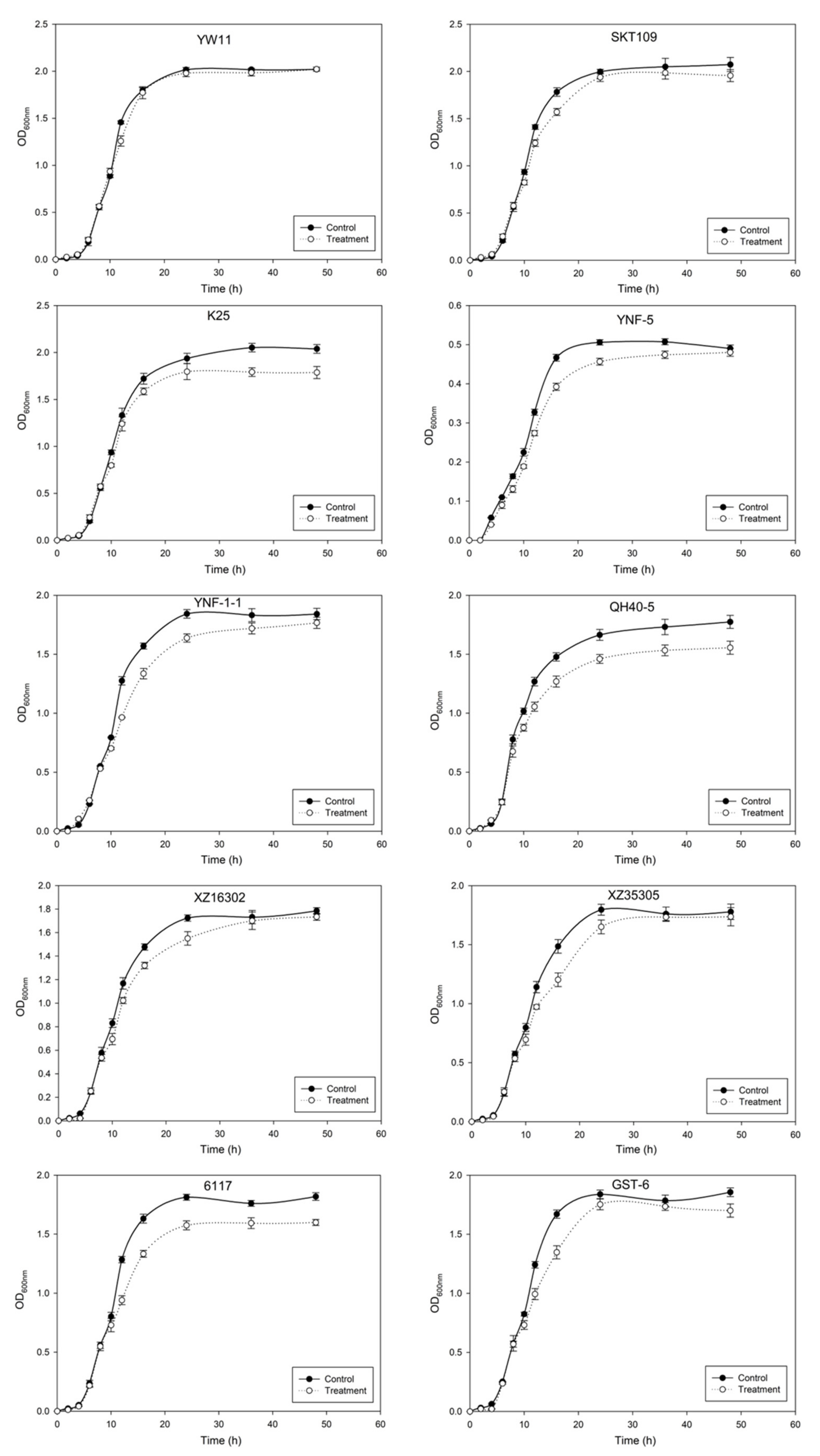
2.2. Effect of Lead on the Growth of LAB
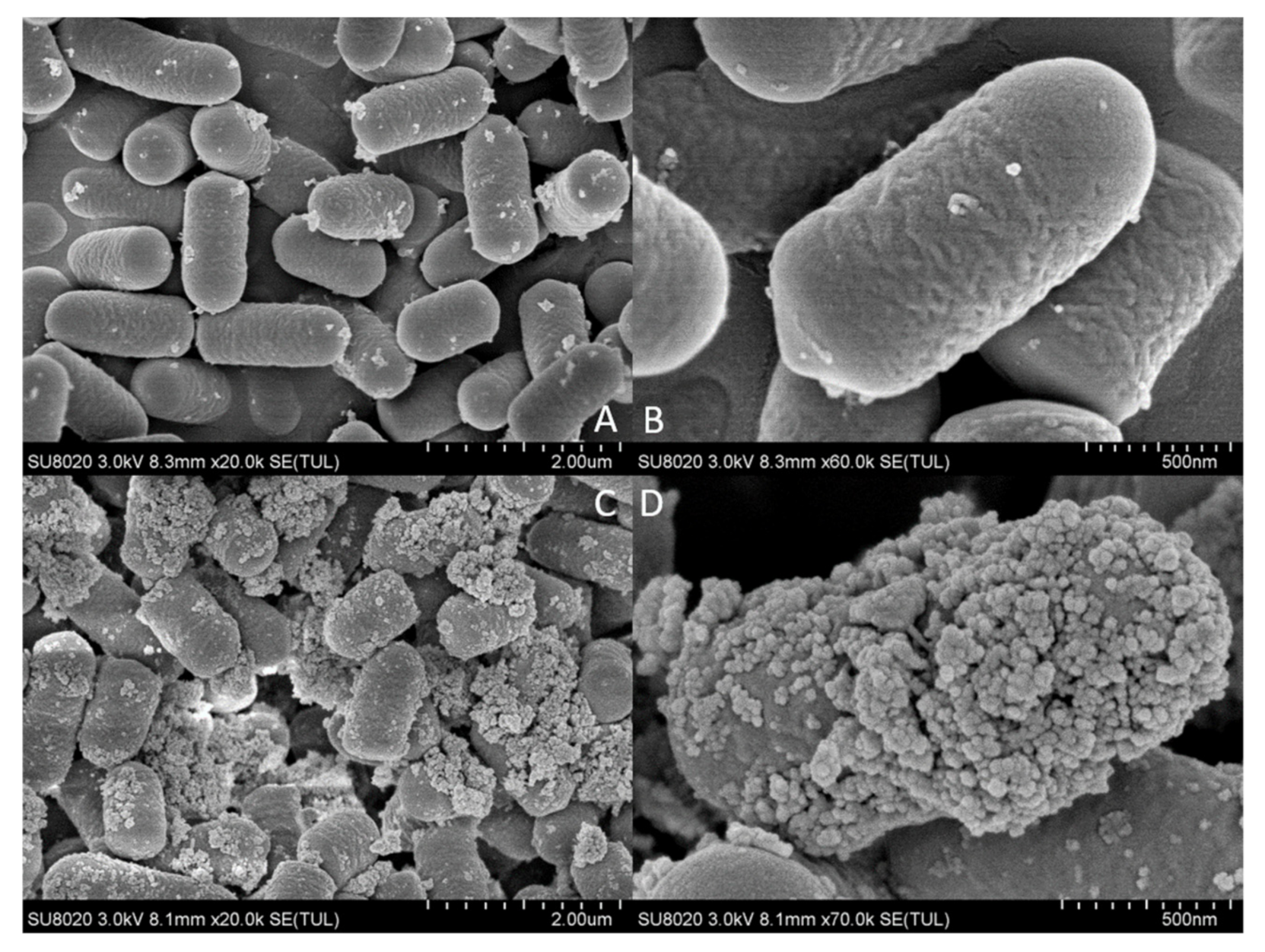
2.3. Scanning Electron Microscopy and Energy Spectrum Scanning
2.4. Quality Control Evaluation and Statistics of Proteomics
2.5. Statistics of Differentially Expressed Proteins
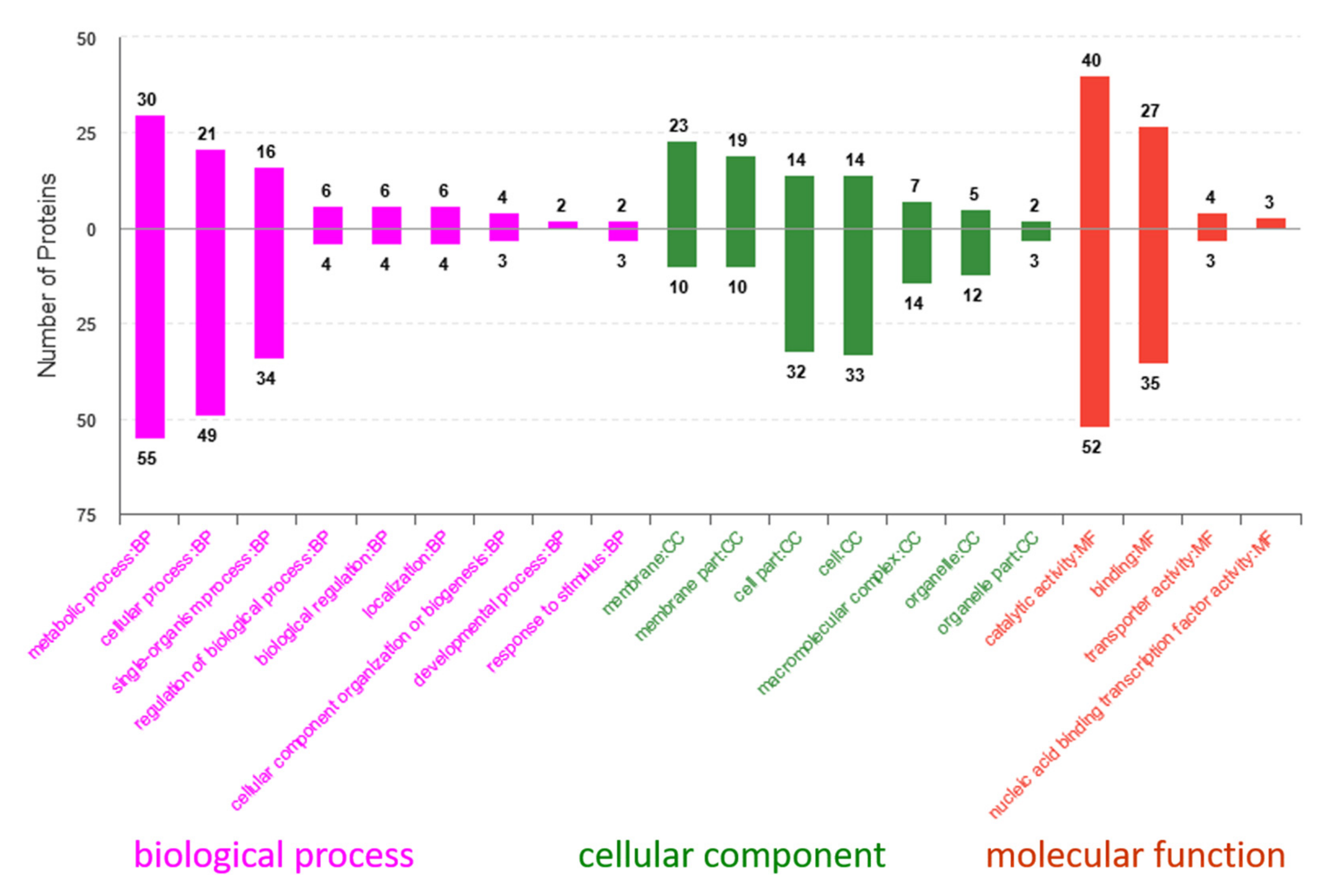
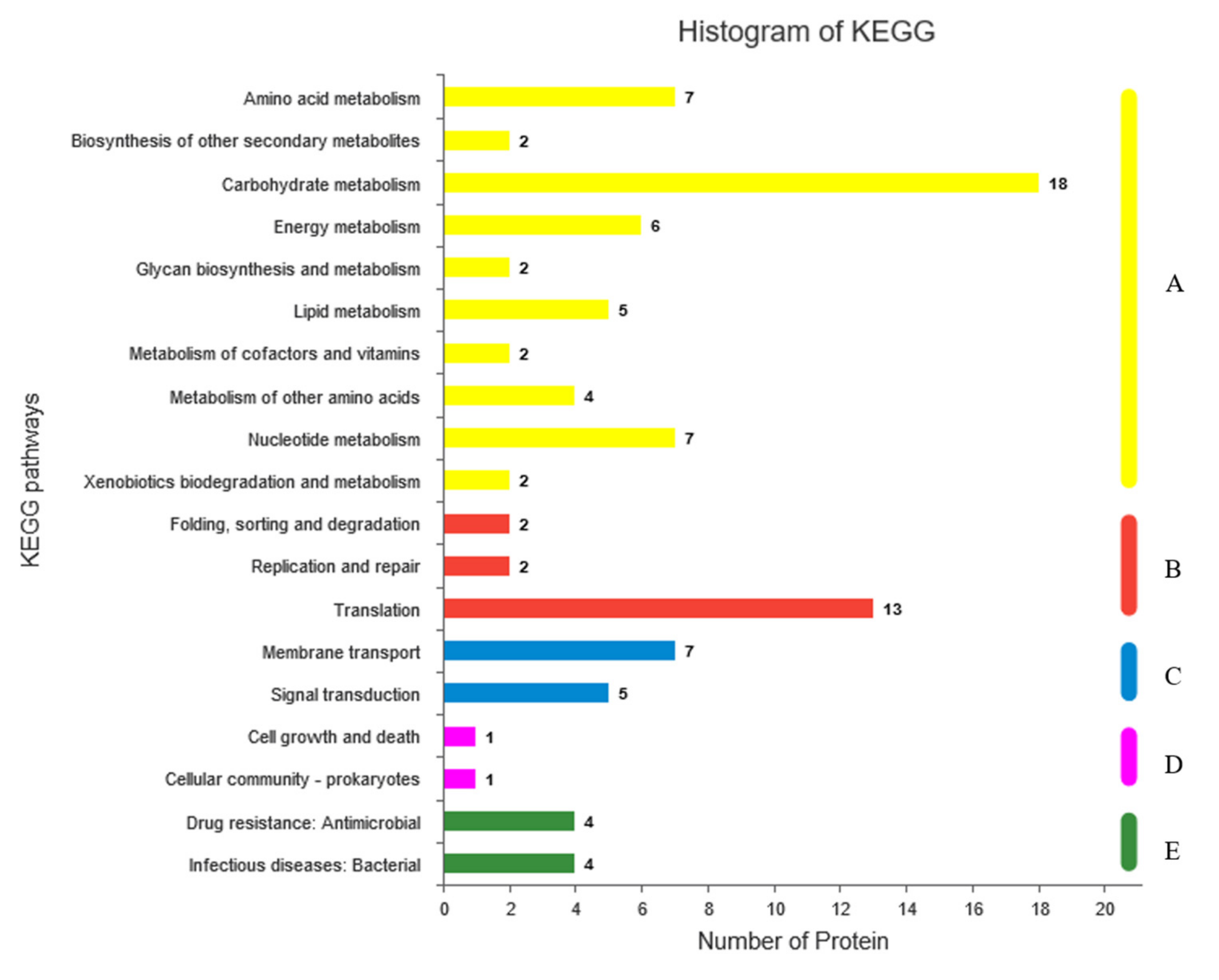
2.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analyses of Differentially Expressed Proteins
2.7. Differential Protein Expression Pattern Analysis
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Lead Absorption Experiment
4.3. Determination of Bacterial Tolerance to Lead
4.4. Effect of Lead on the Growth of LAB
4.5. Preparation of Samples for Scanning Electron Microscopy (SEM) and Proteomic Analysis
4.6. SEM Observation
4.7. Protein Extraction and Determination
4.8. Bioinformatic Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Viard, B.; Pihan, F.; Promeyrat, S.; Pihan, J.C. Integrated assessment of heavy metal (Pb, Zn, Cd) highway pollution: bioaccumulation in soil, Graminaceae and land snails. Chemosphere 2004, 55, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Feng, C.Y.; Zeng, G.M.; Gao, X.; Zhong, M.Z.; Li, X.D.; Li, X.; He, X.Y.; Fang, Y.L. Spatial distribution and source identification of heavy metals in surface soils in a typical coal mine city, Lianyuan, China. Environ. Pollut. 2017, 225, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Gupta, U.C.; Gupta, S.C. Trace element toxicity relationships to crop production and livestock and human health: implications for management. Commun. Soil Sci. Plan. 1998, 29, 1491–1522. [Google Scholar] [CrossRef]
- Mason, L.H.; Harp, J.P.; Han, D.Y. Pb neurotoxicity: neuropsychological effects of lead toxicity. Biomed Res. Int. 2014, 2014, 840547. [Google Scholar] [CrossRef]
- Bohumil, V. Biosorption and me. Water Res. 2007, 41, 4017–4029. [Google Scholar]
- Tan, S.Y.; Praveena, S.M.; Abidin, E.Z.; Cheema, M.S. Heavy metal quantification of classroom dust in school environment and its impacts on children health from Rawang (Malaysia). Environ. Sci. Pollut. Res. Int. 2018, 25, 34623–34635. [Google Scholar] [CrossRef]
- Friberg, L. Distribution and concentration of cadmium in human kidney. Envrion. Res. 1986, 39, 1–7. [Google Scholar]
- Schwartz, B.S.; Stewart, W.F.; Bolla, K.I.; Simon, D.; Bandeen-Roche, K.; Gordon, B.; Links, J.M.; Todd, A.C. Past adult lead exposure is associated with longitudinal decline in cognitive function. Neurology 2000, 55, 1144–1150. [Google Scholar] [CrossRef]
- Khalil, N.; Morrow, L.A.; Needleman, H.; Talbott, E.O.; Wilson, J.W.; Cauley, J.A. Association of cumulative lead and neurocognitive function in an occupational cohort. Neuropsychology 2009, 23, 10–19. [Google Scholar] [CrossRef]
- Garza, A.; Vega, R.; Soto, E. Cellular mechanisms of lead neurotoxicity. Med. Sci. Monit. 2006, 12, RA57–RA65. [Google Scholar]
- Monachese, M.; Burton, J.P.; Reid, G. Bioremediation and Tolerance of Humans to Heavy Metals through Microbial Processes: A Potential Role for Probiotics? Appl. Environ. Microbiol. 2012, 18, 6397–6404. [Google Scholar] [CrossRef] [PubMed]
- Porova, N.; Botvinnikova, V.; Krasulya, O.; Cherepanov, P.; Potoroko, I. Effect of ultrasonic treatment on heavy metal decontamination in milk. Ultrason. Sonochem. 2014, 21, 2107–2111. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Michihata, T.; Katsuyama, Y.; Take, H.; Nakamura, S.; Aburatani, M.; Tokuda, K.; Koyanagi, T.; Taniguchi, H.; Enomoto, T. Effective Removal of Cadmium from Fish Sauce Using Tannin. J. Agric. Food Chem. 2013, 61, 1184–1188. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Oh, S.J.; Shin, Y.J.; Han, S.; Nam, I.; So, J. Bioremediation of lead by ureolytic bacteria isolated from soil at abandoned metal mines in South Korea. Ecol. Eng. 2015, 74, 402–407. [Google Scholar] [CrossRef]
- Ibrahim, F.; Halttunen, T.; Tahvonen, R.; Salminen, S. Probiotic bacteria as potential detoxification tools: assessing their heavy metal binding isotherms. Can. J. Microbiol. 2006, 52, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.X.; Yin, R.J.; Yu, L.L.; Gang, W.; Tian, F.W.; Yu, R.P.; Zhao, J.X.; Liu, X.M.; Chen, Y.Q.; Zhang, H. Screening of lactic acid bacteria with potential protective effects against cadmium toxicity. Food Control 2015, 54, 23–30. [Google Scholar] [CrossRef]
- Halttunen, T.; Collado, M.; El-Nezami, H.; Meriluoto, J.; Salminen, S. Combining strains of lactic acid bacteria may reduce their toxin and heavy metal removal efficiency from aqueous solution. Lett. Appl. Microbiol. 2008, 46, 160–165. [Google Scholar] [CrossRef]
- Topcu, A.E.; Bulat, T. Removal of cadmium and lead from aqueous solution by Enterococcus faecium strains. J. Food Sci. 2010, 75, T13–T17. [Google Scholar] [CrossRef]
- Bhakta, J.N.; Ohnishi, K.; Munekage, Y.; Iwasaki, K.; Wei, M.Q. Characterization of lactic acid bacteria-based probiotics as potential heavy metal sorbents. J. Appl. Microbiol. 2012, 112, 1193–1206. [Google Scholar] [CrossRef]
- Zeidan, A.A.; Poulsen, V.K.; Janzen, T.; Buldo, P.; Derkx, P.M.F.; Regaard, G.; Neves, A.R. Polysaccharide production by lactic acid bacteria: from genes to industrial applications. FEMS Microbiol. Rev. 2017, 41, S168–S200. [Google Scholar] [CrossRef]
- De Vuyst, L.; Leroy, F. Bacteriocins from Lactic Acid Bacteria: Production, Purification, and Food Applications. J. Mol. Microbiol. Biotechnol. 2007, 13, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Hati, S.; Mandal, S.; Prajapati, J.B. Novel Starters for Value Added Fermented Dairy Products. Curr. Res. Nutr. Food Sci. 2013, 1, 83–91. [Google Scholar] [CrossRef]
- Belicová, A.; Mikulá Ová, M.; Du Insky, R. Probiotic Potential and Safety Properties of Lactobacillus plantarum from Slovak Bryndza Cheese. Biomed Res. Int. 2013, 2013, 760298. [Google Scholar] [CrossRef] [PubMed]
- Giri, S.S.; Yun, S.; Jun, J.W.; Kim, H.J.; Kim, S.G.; Kang, J.W.; Kim, S.W.; Han, S.J.; Sukumaran, V.; Park, S.C. Corrigendum: Therapeutic Effect of Intestinal Autochthonous Lactobacillus reuteri P16 Against Waterborne Lead Toxicity in Cyprinus carpio. Front. Immunol. 2018, 9, 2208. [Google Scholar] [CrossRef]
- Tian, F.W.; Zhai, Q.X.; Zhao, J.X.; Liu, X.M.; Wang, G.; Zhang, H.; Zhang, H.P.; Chen, W. Lactobacillus plantarum CCFM8661 Alleviates Lead Toxicity in Mice. Biol. Trace Elem. Res. 2012, 150, 264–271. [Google Scholar] [CrossRef]
- Halttunen, T.; Salminen, S.; Tahvonen, R. Rapid removal of lead and cadmium from water by specific lactic acid bacteria. Int. J. Food Microbiol. 2007, 114, 30–35. [Google Scholar] [CrossRef]
- Bhakta, J.N.; Ohnishi, K.; Jana, B.B. Isolation and identification of cadmium- and lead-resistant lactic acid bacteria for application as metal removing probiotic. Int. J. Environ. Sci. Tecnol. 2012, 9, 433–440. [Google Scholar] [CrossRef]
- Feng, M.Q.; Chen, X.H.; Li, C.C.; Rahman, N.; Dong, M.S. Isolation and Identification of an Exopolysaccharide-Producing Lactic Acid Bacterium Strain from Chinese Paocai and Biosorption of Pb(II) by Its Exopolysaccharide. J. Food Sci. 2012, 77, T111–T117. [Google Scholar] [CrossRef]
- Montazer-Rahmati, M.M.; Rabbani, P.; Abdolali, A.; Keshtkar, A.R. Kinetics and equilibrium studies on biosorption of cadmium, lead, and nickel ions from aqueous solutions by intact and chemically modified brown algae. J. Hazard. Mater. 2011, 185, 401–407. [Google Scholar] [CrossRef]
- Kirillova, A.V.; Danilushkina, A.A.; Irisov, D.S.; Bruslik, N.L.; Fakhrullin, R.F.; Zakharov, Y.A.; Bukhmin, V.S.; Yarullina, D.R. Assessment of Resistance and Bioremediation Ability of Lactobacillus Strains to Lead and Cadmium. Int. J. Microbiol. 2017, 2017, 9869145. [Google Scholar] [CrossRef]
- Sánchez, B.; Schmitter, J.M.; Urdaci, M.C. Identification of novel proteins secreted by Lactobacillus plantarum that bind to mucin and fibronectin. J. Mol. Microbiol. Biotechnol. 2009, 17, 158–162. [Google Scholar] [PubMed]
- Hynönen, U.; Palva, A. Lactobacillus surface layer proteins: structure, function and applications. Appl. Microbiol. Biotechnol. 2013, 97, 5225–5243. [Google Scholar] [CrossRef] [PubMed]
- Gerbino, E.; Mobili, P.; Tymczyszyn, E.; Fausto, R.; Gómez-Zavaglia, A. FTIR spectroscopy structural analysis of the interaction between Lactobacillus kefir S-layers and metal ions. J. Mol. Struct. 2011, 987, 186–192. [Google Scholar] [CrossRef]
- Gerbino, E.; Carasi, P.; Araujo-Andrade, C.; Tymczyszyn, E.E.; Gómez-Zavaglia, A. Role of S-layer proteins in the biosorption capacity of lead by Lactobacillus kefir. World J. Microbiol. Biotechnol. 2015, 31, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Garrote, G.L.; Serradell, M.A.; Abraham, A.G.; Anon, M.C.; Fossati, C.A.; de Antoni, G.L. Development of an immunochemical method to detect Lactobacillus kefir. Food Agr. Immunol. 2005, 16, 221–233. [Google Scholar] [CrossRef][Green Version]
- Sundararajan, K.; Goley, E.D. Cytoskeletal Proteins in Caulobacter crescentus: Spatial Orchestrators of Cell Cycle Progression, Development, and Cell Shape. Subcell. Biochem. 2017, 84, 103–137. [Google Scholar] [PubMed]
- O’May, G.A.; Jacobsen, S.M.; Longwell, M.; Stoodley, P.; Mobley, H.L.T.; Shirtliff, M.E. The high-affinity phosphate transporter Pst in Proteus mirabilis HI4320 and its importance in biofilm formation. Microbiology 2009, 155, 1523–1535. [Google Scholar] [CrossRef]
- Blus-Kadosh, I.; Zilka, A.; Yerushalmi, G.; Banin, E. The Effect of pstS and phoB on Quorum Sensing and Swarming Motility in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e74444. [Google Scholar] [CrossRef]
- Luz, D.E.; Nepomuceno, R.S.L.; Spira, B.; Ferreira, R.C.C. The Pst system of Streptococcus mutans is important for phosphate transport and adhesion to abiotic surfaces. Mol. Oral Microbiol. 2012, 27, 172–181. [Google Scholar] [CrossRef]
- Bourdineaud, J.P.; Nehmé, B.; Tesse, S.; Lonvaud-Funel, A. A bacterial gene homologous to ABC transporters protect Oenococcus oeni from ethanol and other stress factors in wine. Int. J. Food Microbiol. 2004, 92, 1–14. [Google Scholar] [CrossRef]
- Manson, J.M.; Stefanie, K.; Smith, J.M.B.; Cook, G.M. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob. Agents Chemother. 2004, 48, 3743–3748. [Google Scholar] [CrossRef] [PubMed]
- Obis, D.; Guillot, A.; Gripon, J.C.; Renault, P.; Bolotin, A.; Mistou, M.Y. Genetic and biochemical characterization of a high-affinity betaine uptake system (BusA) in Lactococcus lactis reveals a new functional organization within bacterial ABC transporters. J. Bacteriol. 1999, 181, 6238–6246. [Google Scholar] [PubMed]
- Wang, J.; Zhao, X.; Tian, Z.; Yang, Y.; Yang, Z. Characterization of an exopolysaccharide produced by Lactobacillus plantarum YW11 isolated from Tibet Kefir. Carbohydr. Polym. 2015, 125, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Booth, S.C.; Workentine, M.L.; Wen, J.; Shaykhutdinov, R.; Vogel, H.J.; Ceri, H.; Turner, R.J.; Weljie, A.M. Differences in metabolism between the biofilm and planktonic response to metal stress. J. Proteome Res. 2011, 10, 3190–3199. [Google Scholar] [CrossRef] [PubMed]
- Fomina, M.; Gadd, G.M. Biosorption: current perspectives on concept, definition and application. Bioresour. Technol. 2014, 160, 3–14. [Google Scholar] [CrossRef]
- Lesmana, S.O.; Febriana, N.; Soetaredjo, F.E.; Sunarso, J.; Ismadji, S. Studies on potential applications of biomass for the separation of heavy metals from water and wastewater. Biochem. Eng. J. 2009, 44, 19–41. [Google Scholar] [CrossRef]
- Gupta, V.K.; Nayak, A.; Agarwal, S. Bioadsorbents for remediation of heavy metals: Current status and their future prospects. Environ. Eng. Res. 2015, 20, 1–18. [Google Scholar] [CrossRef]
- Mosa, K.A.; Saadoun, I.; Kumar, K.; Helmy, M.; Dhankher, O.P. Potential Biotechnological Strategies for the Cleanup of Heavy Metals and Metalloids. Front. Plant Sci. 2016, 7, 303. [Google Scholar] [CrossRef]
- De Man, J.C.; Rogosa, M.; Sharpe, M.E. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Argun, M.E.; Dursun, S.; Ozdemir, C.; Karatas, M. Heavy metal adsorption by modified oak sawdust: thermodynamics and kinetics. J. Hazard. Mater. 2007, 141, 77–85. [Google Scholar] [CrossRef]
- Abou-Shanab, R.A.I.; van Berkum, P.; Angle, J.S. Heavy metal resistance and genotypic analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere 2007, 68, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Pajarillo, E.A.; Kim, M.J.; Chae, J.P.; Kang, D.K. Proteomic and transcriptional analysis of Lactobacillus johnsonii PF01 during bile salt exposure by iTRAQ shotgun proteomics and quantitative RT-PCR. J. Proteome Res. 2013, 12, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed Protein Quantitation in Saccharomyces cerevisiae Using Amine-reactive Isobaric Tagging Reagents. Mol. Cell. Proteomics 2004, 3, 1154–1169. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.H.; Huang, C.; Yuan, J.; Li, W.; Yin, R.L.; Li, H.S.; Dong, Q.; Li, X.T.; Bai, W.L. iTRAQ-based proteomics analysis reveals the deregulated proteins related to liver toxicity induced by melamine with or without cyanuric acid in mice. Ecotoxicol. Environ. Saf. 2019, 174, 618–629. [Google Scholar] [CrossRef] [PubMed]

| Stains | MIC (mg/L) |
|---|---|
| L. plantarum YW11 | >1000 |
| L. plantarum SKT109 | >100 |
| L. plantarum K25 | >100 |
| L. plantarum YNF-5 | <100 |
| Lactococcus lactis YNK-1-1 | >100 |
| Lactococcus lactis XZ16302 | >100 |
| Lactococcus lactis XZ35305 | <100 |
| Lactococcus lactis QH40-5 | <100 |
| L. casei 6117 | >100 |
| S. thermophilus GST-6 | >100 |
| Category | Accession | Description | Subcellular Localization | FC |
|---|---|---|---|---|
| Amino acid metabolism | A0A0R1VB59 | dtd: d-aminoacyl-tRNA deacylase | cytoplasm | −1.57 |
| Transporter | D7V9Y8 | pstS: phosphate-binding protein | membrane | 1.7 |
| A0A151G577 | AYO51_05730: hemin ATP-binding cassette (ABC) transporter | — | 1.54 | |
| Mismatch repair | A0A2S3U4Y4 | xseB: exodeoxyribonuclease 7 small subunit | cytoplasm | 1.52 |
| Transcriptional regulation | A0A199QFG9 | nrdR: transcriptional repressor | — | 2.03 |
| Membrane protein and cell surface protein | A0A199QM58 | A0U96_08550: peptidoglycan-binding protein | — | 2.17 |
| A0A2K7QYX4 | A0U96_06145: peptidoglycan-binding protein | — | 2.17 | |
| A0A199QI49 | A0U96_08550: adhesin | membrane | 1.56 | |
| A0A369UCN7 | DVK84_07570: cell wall anchor domain-containing protein | — | 1.76 | |
| A0A162GCP0 | Nizo2802_2963: membrane occupation and recognition nexus (MORN) motif family protein | membrane | 1.64 | |
| Global stress response | A0A0L7Y2V1 | A8704_12230: flavin mononucleotide (FMN)-binding protein | — | 3.08 |
| A0A385PQP5 | CFI98_11100: DNA replicationg protein D (DnaD) domain protein | — | 1.84 | |
| A0A1W6NPN7 | BIZ32_04340: FMN-binding protein | membrane | 1.57 | |
| D7V968 | adh: chaperonin 10 (GroES)-like protein | — | −1.78 | |
| A0A0M0CIV7 | hemH: ferrochelatase | cytoplasm | −1.51 | |
| A0A0M0CJD8 | AYO51_13390: macro domain ADP–ribose-binding module | — | −1.59 | |
| A0A0R1UML2 | FD10_GL000592: deoxycytidine (dCMP) deaminase | — | −2.22 | |
| A0A1E3KN19 | LPJSA22_03294: putative transposon Tn552 DNA-invertase bin3 | — | −2.56 | |
| A0A0R1V3I3 | FD10_GL001348: anaerobic ribonucleoside-triphosphate reductase large subunit | — | −1.69 | |
| Extracellular protein | A0A0G9FAG5 | AVR82_06000: extracellular protein | — | 2.28 |
| A0A165XVF0 | Nizo1839_1013: cell-shape-determining protein | — | 1.55 | |
| A0A2I0ZH16 | CUR48_01040: Lysin motif (LysM) domain-containing protein | — | 1.96 | |
| A0A165P076 | Nizo2802_0557: extracellular protein | — | 1.58 | |
| Carbohydrate metabolism | A0A1S0RZ68 | AVR82_00090: glycosyl hydrolase family | membrane | 1.89 |
| T5JT98 | N692_15475: formate acetyltransferase | cytoplasm | −1.82 | |
| Translation | A0A2S3U2L6 | rplO: 50S ribosomal protein L15 | — | −1.52 |
| A0A1A0DF73 | rpmG: 50S ribosomal protein L33 | intracellular | −1.56 | |
| A0A199QKR6 | rpsI: 50S ribosomal protein S9 | — | −1.72 | |
| T5K018 | rplU: 50S ribosomal protein L21 | intracellular | −1.75 | |
| D7V8T5 | rpmI: 50S ribosomal protein L35 | intracellular | −1.82 | |
| Q88XY4 | rplW: 50S ribosomal protein L23 | intracellular | −1.89 | |
| U2WMY2 | rpsU: 30S ribosomal protein S21 | intracellular | −2.83 | |
| A0A0G9FAQ2 | ybaK: cys-tRNA(Pro)/Cys-tRNA(Cys) deacylase | — | −1.67 | |
| A0A0R2GAU1 | rsmG: ribosomal RNA small subunit methyltransferase G | cytoplasm | −1.49 | |
| Uncharacterized protein | A0A162GHW5 | Lp19_2585 | membrane | 3.71 |
| A0A0G9F7Y4 | DVK84_02520 | — | 2.29 | |
| F9UU29 | lp_0444 | — | 1.57 | |
| A0A1S0RQZ5 | AVR82_00885 | membrane | 1.74 | |
| A0A165NJ25 | Nizo2802_1443 | — | 1.56 | |
| A0A0M4CUJ4 | AVR82_13000 | — | 1.58 | |
| M4KLN1 | zj316_3034 | — | 1.56 | |
| A0A386RBN5 | CO218_15800 | — | 1.52 | |
| A0A387DFR9 | CFI62_00270 | — | −1.64 | |
| A0A0G9GNJ2 | WP50_25770 | membrane | −2.78 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Zheng, Y.; Ma, Y.; Sarwar, A.; Zhao, X.; Luo, T.; Yang, Z. Evaluation and Proteomic Analysis of Lead Adsorption by Lactic Acid Bacteria. Int. J. Mol. Sci. 2019, 20, 5540. https://doi.org/10.3390/ijms20225540
Liu S, Zheng Y, Ma Y, Sarwar A, Zhao X, Luo T, Yang Z. Evaluation and Proteomic Analysis of Lead Adsorption by Lactic Acid Bacteria. International Journal of Molecular Sciences. 2019; 20(22):5540. https://doi.org/10.3390/ijms20225540
Chicago/Turabian StyleLiu, Shaoli, Yi Zheng, Yimiao Ma, Abid Sarwar, Xiao Zhao, Tianqi Luo, and Zhennai Yang. 2019. "Evaluation and Proteomic Analysis of Lead Adsorption by Lactic Acid Bacteria" International Journal of Molecular Sciences 20, no. 22: 5540. https://doi.org/10.3390/ijms20225540
APA StyleLiu, S., Zheng, Y., Ma, Y., Sarwar, A., Zhao, X., Luo, T., & Yang, Z. (2019). Evaluation and Proteomic Analysis of Lead Adsorption by Lactic Acid Bacteria. International Journal of Molecular Sciences, 20(22), 5540. https://doi.org/10.3390/ijms20225540