HLA-F Allele-Specific Peptide Restriction Represents an Exceptional Proteomic Footprint
Abstract
1. Introduction
2. Results
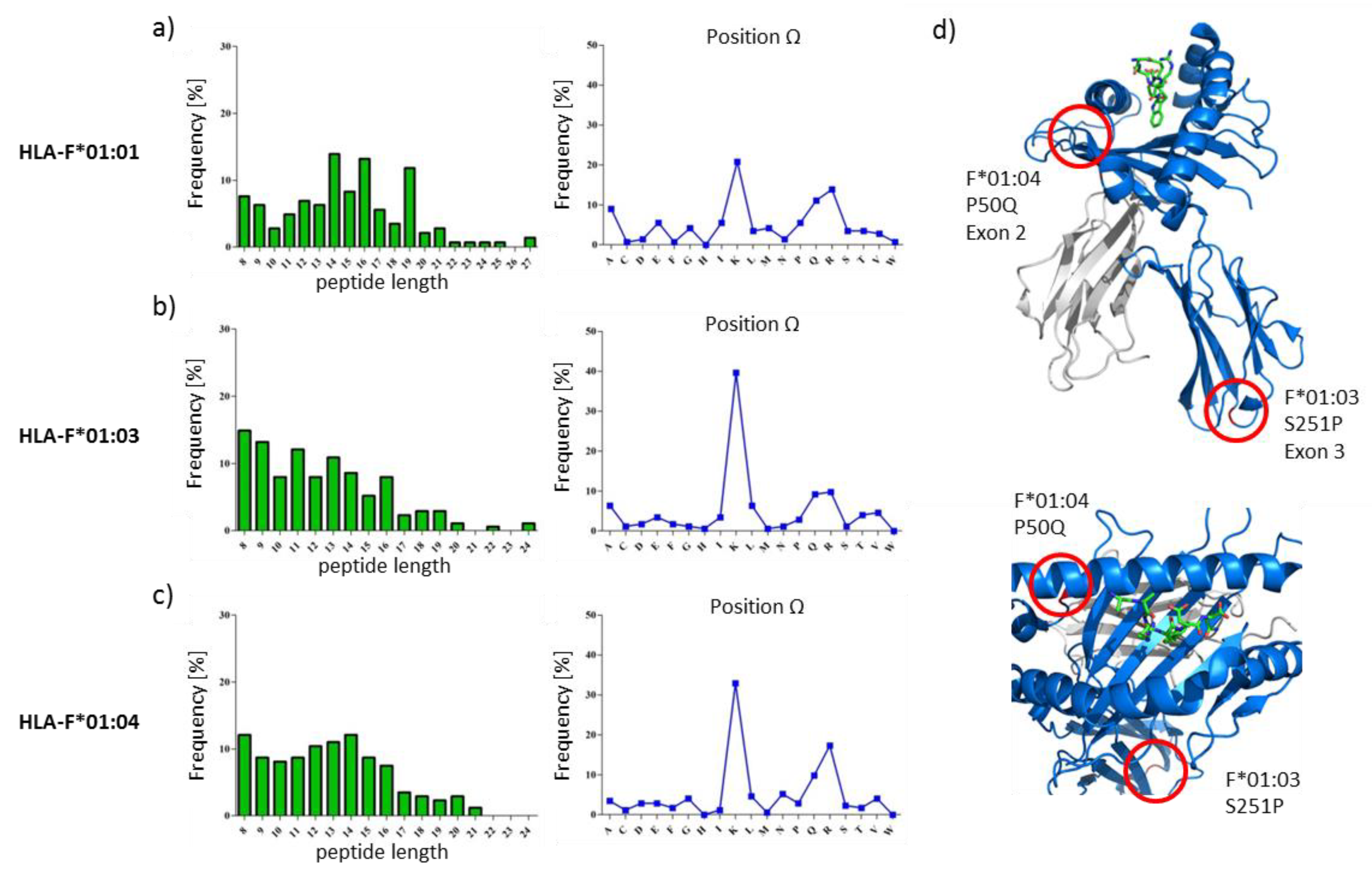
2.1. HLA-F Restricted Peptides of All Allelic Variants Exhibite Non-Canonical Length
2.2. The Peptide Binding Motif of HLA-F*01:0x Exhibits a High Frequency of Polar and Positively Charged AAs at pΩ
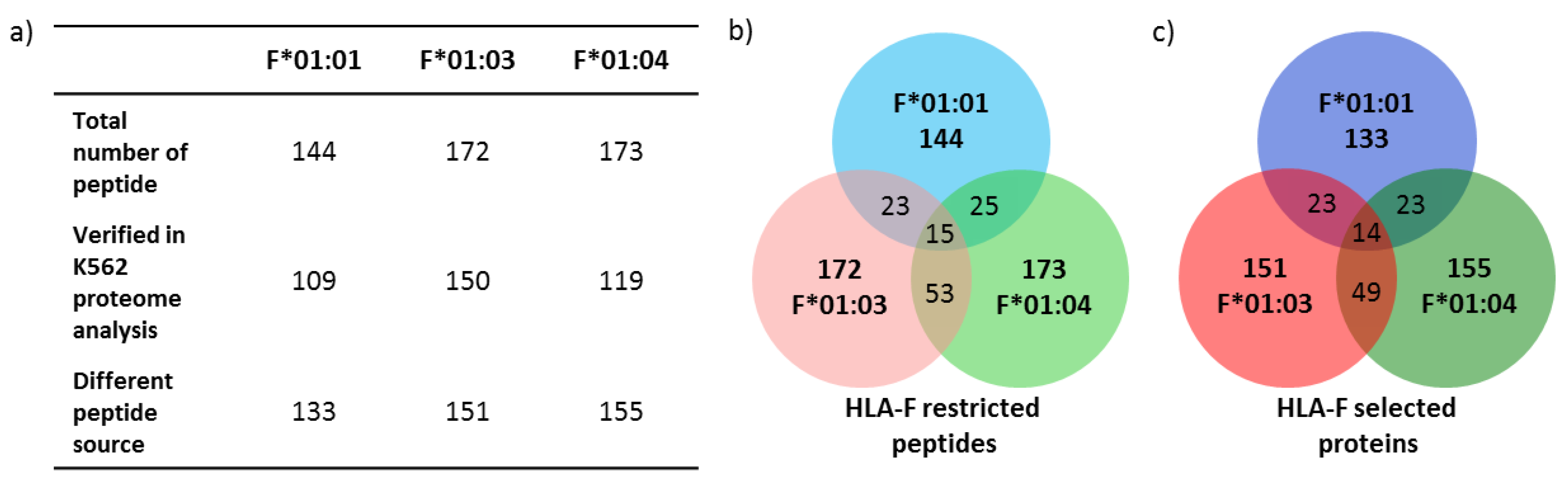
2.3. Only 9.2% of Peptides Are Shared between the Three Allelic HLA-F Variants
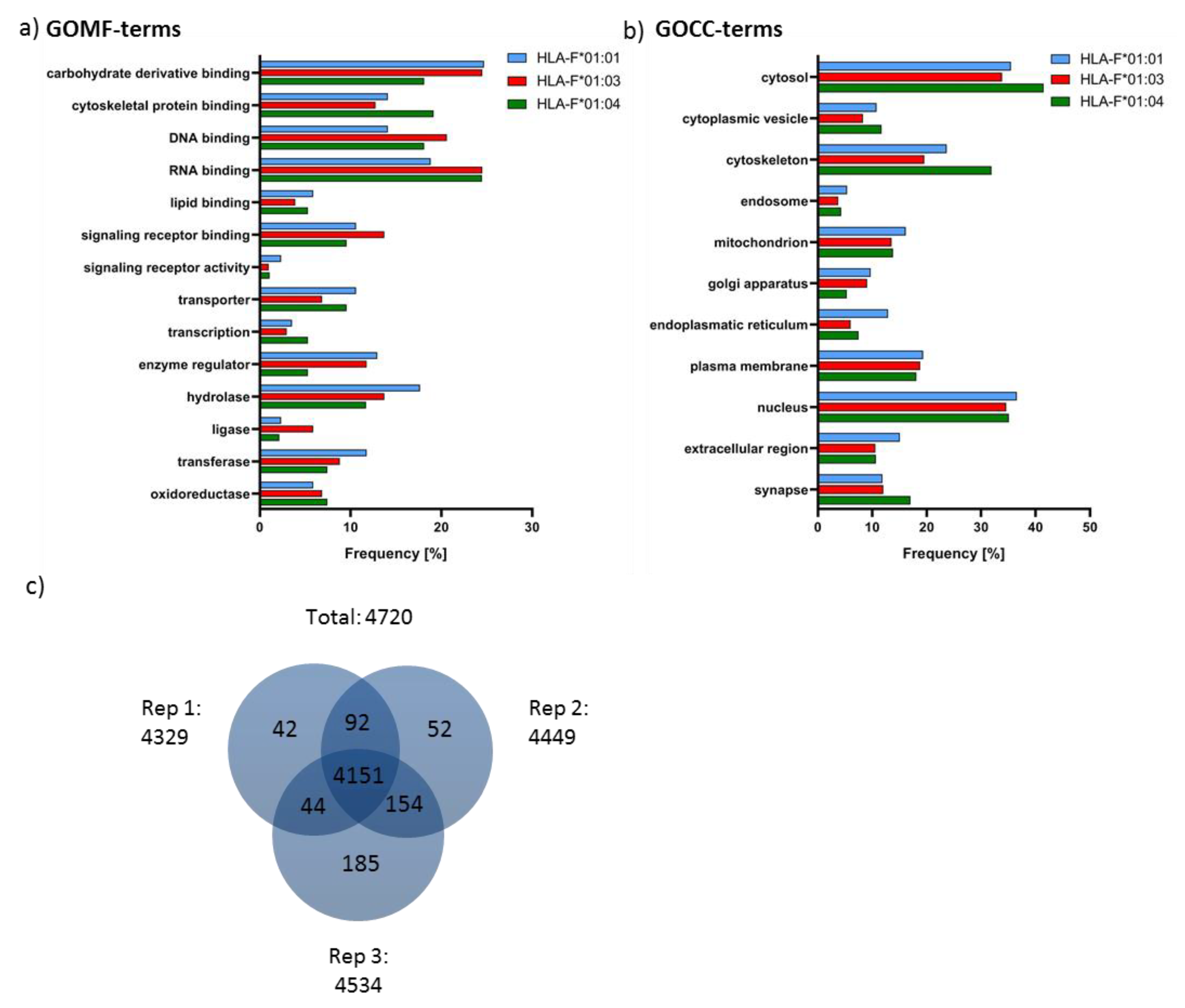
2.4. Features of HLA-F Selected Proteins
3. Discussion
4. Material and Methods
4.1. Maintenance of Cell Lines
4.2. Cloning of HLA-F Encoding Contructs
4.3. Stable Transduction of K562 Cells with Lentivirus Encoding for HLA-F*01:0x Molecules
4.4. Large-scale Production of sHLA-F*01:0x Molecules
4.5. LC-MS Analysis of sHLA-F*01:0x Restricted Peptides and the Proteome
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Garcia-Beltran, W.F.; Holzemer, A.; Martrus, G.; Chung, A.W.; Pacheco, Y.; Simoneau, C.R.; Rucevic, M.; Lamothe-Molina, P.A.; Pertel, T.; Kim, T.E.; et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat. Immunol. 2016, 17, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- .Burian, A.; Wang, K.L.; Finton, K.A.; Lee, N.; Ishitani, A.; Strong, R.K.; Geraghty, D.E. HLA-F and MHC-I Open Conformers Bind Natural Killer Cell Ig-Like Receptor KIR3DS1. Plos ONE 2016, 11, e0163297. [Google Scholar] [CrossRef] [PubMed]
- Dulberger, C.L.; McMurtrey, C.P.; Holzemer, A.; Neu, K.E.; Liu, V.; Steinbach, A.M.; Garcia-Beltran, W.F.; Sulak, M.; Jabri, B.; Lynch, V.J.; et al. Human Leukocyte Antigen F Presents Peptides and Regulates Immunity through Interactions with NK Cell Receptors. Immunity 2017, 46, 1018–1029. [Google Scholar] [CrossRef] [PubMed]
- .Ho, G.T.; Heinen, F.J.; Huyton, T.; Blasczyk, R.; Bade-Doding, C. HLA-F*01:01 presents peptides with N-terminal flexibility and a preferred length of 16 residues. Immunogenetics 2019, 71, 353–360. [Google Scholar] [CrossRef]
- Abels, W.C.; Celik, A.A.; Simper, G.S.; Blasczyk, R.; Bade-Döding, C. Peptide Presentation Is the Key to Immunotherapeutical Success. In Polypeptide—New Insight into Drug Discovery and Development; Tambunan, U.S.F., Ed.; IntechOpen: London, UK, 2018; pp. 7–25. [Google Scholar] [CrossRef]
- .Chan, K.F.; Gully, B.S.; Gras, S.; Beringer, D.X.; Kjer-Nielsen, L.; Cebon, J.; McCluskey, J.; Chen, W.; Rossjohn, J. Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide. Nat. Commun. 2018, 9, 1026. [Google Scholar] [CrossRef]
- Beringer, D.X.; Kleijwegt, F.S.; Wiede, F.; van der Slik, A.R.; Loh, K.L.; Petersen, J.; Dudek, N.L.; Duinkerken, G.; Laban, S.; Joosten, A.; et al. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat. Immunol. 2015, 16, 1153–1161. [Google Scholar] [CrossRef]
- .Badrinath, S.; Kunze-Schumacher, H.; Blasczyk, R.; Huyton, T.; Bade-Doeding, C. A Micropolymorphism Altering the Residue Triad 97/114/156 Determines the Relative Levels of Tapasin Independence and Distinct Peptide Profiles for HLA-A(*)24 Allotypes. J. Immunol. Res. 2014, 2014, 298145. [Google Scholar] [CrossRef]
- Badrinath, S.; Saunders, P.; Huyton, T.; Aufderbeck, S.; Hiller, O.; Blasczyk, R.; Bade-Doeding, C. Position 156 influences the peptide repertoire and tapasin dependency of human leukocyte antigen B*44 allotypes. Haematologica 2012, 97, 98–106. [Google Scholar] [CrossRef]
- .Rammensee, H.; Bachmann, J.; Emmerich, N.P.; Bachor, O.A.; Stevanovic, S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics 1999, 50, 213–219. [Google Scholar] [CrossRef]
- Huyton, T.; Ladas, N.; Schumacher, H.; Blasczyk, R.; Bade-Doeding, C. Pocketcheck: Updating the HLA class I peptide specificity roadmap. Tissue Antigens 2012, 80, 239–248. [Google Scholar] [CrossRef]
- .Meyer, D.; VR, C.A.; Bitarello, B.D.; DY, C.B.; Nunes, K. A genomic perspective on HLA evolution. Immunogenetics 2018, 70, 5–27. [Google Scholar] [CrossRef] [PubMed]
- Ho, G.T.; Heinen, F.J.; Stieglitz, F.; Blasczyk, R.; Bade-Döding, C. Dynamic Interaction between Immune Escape Mechanism and HLA-Ib Regulation. In Immunogenetics; Rezaei, N., Ed.; IntechOpen: London, UK, 2018; pp. 1–25. [Google Scholar] [CrossRef]
- Celik, A.A.; Simper, G.S.; Huyton, T.; Blasczyk, R.; Bade-Döding, C. HLA-G mediated immune regulation is impaired by a single amino acid exchange in the alpha 2 domain. Hum. Immunol. 2018, 79, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, T.; Celik, A.A.; Huyton, T.; Kunze-Schumacher, H.; Blasczyk, R.; Bade-Doding, C. HLA-E: Presentation of a Broader Peptide Repertoire Impacts the Cellular Immune Response-Implications on HSCT Outcome. Stem Cells Int. 2015, 2015, 346714. [Google Scholar] [CrossRef] [PubMed]
- .Celik, A.A.; Kraemer, T.; Huyton, T.; Blasczyk, R.; Bade-Doding, C. The diversity of the HLA-E-restricted peptide repertoire explains the immunological impact of the Arg107Gly mismatch. Immunogenetics 2016, 68, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Stieglitz, F.; Celik, A.A.; von Kaisenberg, C.; Camps, M.A.; Blasczyk, R.; Bade-Doding, C. The microstructure in the placenta is influenced by the functional diversity of HLA-G allelic variants. Immunogenetics 2019, 71, 455–463. [Google Scholar] [CrossRef]
- .Wagner, B.; da Silva Nardi, F.; Schramm, S.; Kraemer, T.; Celik, A.A.; Durig, J.; Horn, P.A.; Duhrsen, U.; Nuckel, H.; Rebmann, V. HLA-E allelic genotype correlates with HLA-E plasma levels and predicts early progression in chronic lymphocytic leukemia. Cancer 2017, 123, 814–823. [Google Scholar] [CrossRef]
- Lunemann, S.; Schobel, A.; Kah, J.; Fittje, P.; Holzemer, A.; Langeneckert, A.E.; Hess, L.U.; Poch, T.; Martrus, G.; Garcia-Beltran, W.F.; et al. Interactions Between KIR3DS1 and HLA-F Activate Natural Killer Cells to Control HCV Replication in Cell Culture. Gastroenterology 2018, 155, 1366–1371. [Google Scholar] [CrossRef]
- Kiani, Z.; Bruneau, J.; Geraghty, D.E.; Bernard, N.F. HLA-F on autologous HIV infected cells activates primary NK cells expressing the activating killer immunoglobulin-like receptor KIR3DS1. J. Virol. 2019. [Google Scholar] [CrossRef]
- Kiani, Z.; Dupuy, F.P.; Bruneau, J.; Lebouche, B.; Zhang, C.X.; Jackson, E.; Lisovsky, I.; da Fonseca, S.; Geraghty, D.E.; Bernard, N.F. HLA-F on HLA-Null 721.221 Cells Activates Primary NK Cells Expressing the Activating Killer Ig-like Receptor KIR3DS1. J. Immunol. 2018, 201, 113. [Google Scholar] [CrossRef]
- .Buttura, R.V.; Ramalho, J.; Lima, T.H.A.; Donadi, E.A.; Veiga-Castelli, L.C.; Mendes-Junior, C.T.; Castelli, E.C. HLA-F displays highly divergent and frequent haplotype lineages associated with different mRNA expression levels. Hum. Immunol. 2019, 80, 112–119. [Google Scholar] [CrossRef]
- Laaribi, A.B.; Hannachi, N.; Ben Yahia, H.; Marzouk, M.; Mehri, A.; Belhadj, M.; Yacoub, S.; Letaief, A.; Ouzari, H.I.; Boudabous, A.; et al. Human leukocyte antigen (HLA-F) polymorphism is associated with chronic HBV infection. 3 Biotech 2018, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- .Lepin, E.J.; Bastin, J.M.; Allan, D.S.; Roncador, G.; Braud, V.M.; Mason, D.Y.; van der Merwe, P.A.; McMichael, A.J.; Bell, J.I.; Powis, S.H.; et al. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur. J. Immunol. 2000, 30, 3552–3561. [Google Scholar] [CrossRef]
- Goodridge, J.P.; Burian, A.; Lee, N.; Geraghty, D.E. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J. Immunol. 2013, 191, 3553–3562. [Google Scholar] [CrossRef] [PubMed]
- .Robinson, J.; Halliwell, J.A.; Hayhurst, J.D.; Flicek, P.; Parham, P.; Marsh, S.G. The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 2015, 43, D423–D431. [Google Scholar] [CrossRef] [PubMed]
- Paganini, J.; Ramdane, A.; Gouret, P.; Chiaroni, J.; Di Cristofaro, J. Validation of new HLA-F alleles assigned by next-generation sequencing. HLA 2019, 93, 131–132. [Google Scholar] [CrossRef]
- .Geraghty, D.E.; Wei, X.H.; Orr, H.T.; Koller, B.H. Human leukocyte antigen F (HLA-F). An expressed HLA gene composed of a class I coding sequence linked to a novel transcribed repetitive element. J. Exp. Med. 1990, 171, 1–18. [Google Scholar] [CrossRef]
- Burrows, C.K.; Kosova, G.; Herman, C.; Patterson, K.; Hartmann, K.E.; Velez Edwards, D.R.; Stephenson, M.D.; Lynch, V.J.; Ober, C. Expression Quantitative Trait Locus Mapping Studies in Mid-secretory Phase Endometrial Cells Identifies HLA-F and TAP2 as Fecundability-Associated Genes. Plos Genet. 2016, 12, e1005858. [Google Scholar] [CrossRef]
- Song, S.; Miranda, C.J.; Braun, L.; Meyer, K.; Frakes, A.E.; Ferraiuolo, L.; Likhite, S.; Bevan, A.K.; Foust, K.D.; McConnell, M.J.; et al. Major histocompatibility complex class I molecules protect motor neurons from astrocyte-induced toxicity in amyotrophic lateral sclerosis. Nat. Med. 2016, 22, 397–403. [Google Scholar] [CrossRef]
- Lin, A.; Zhang, X.; Ruan, Y.Y.; Wang, Q.; Zhou, W.J.; Yan, W.H. HLA-F expression is a prognostic factor in patients with non-small-cell lung cancer. Lung Cancer 2011, 74, 504–509. [Google Scholar] [CrossRef]
- .Feng, E.; Liang, T.; Wang, X.; Du, J.; Tang, K.; Wang, X.; Wang, F.; You, G. Correlation of alteration of HLA-F expression and clinical characterization in 593 brain glioma samples. J. Neuroinflammation 2019, 16, 33. [Google Scholar] [CrossRef]
- Kraemer, T.; Blasczyk, R.; Bade-Doeding, C. HLA-E: A novel player for histocompatibility. J. Immunol. Res. 2014, 2014, 352160. [Google Scholar] [CrossRef] [PubMed]
- .Pump, W.C.; Kraemer, T.; Huyton, T.; Ho, G.T.; Blasczyk, R.; Bade-Doeding, C. Between Innate and Adaptive Immune Responses: NKG2A, NKG2C, and CD8(+) T Cell Recognition of HLA-E Restricted Self-Peptides Acquired in the Absence of HLA-Ia. Int. J. Mol. Sci. 2019, 20, 1454. [Google Scholar] [CrossRef] [PubMed]
- Apps, R.; Murphy, S.P.; Fernando, R.; Gardner, L.; Ahad, T.; Moffett, A. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology 2009, 127, 26–39. [Google Scholar] [CrossRef] [PubMed]
- .Ferreira, L.M.R.; Meissner, T.B.; Tilburgs, T.; Strominger, J.L. HLA-G: At the Interface of Maternal-Fetal Tolerance. Trends Immunol. 2017, 38, 272–286. [Google Scholar] [CrossRef]
- Amiot, L.; Vu, N.; Samson, M. Immunomodulatory properties of HLA-G in infectious diseases. J. Immunol. Res. 2014, 2014, 298569. [Google Scholar] [CrossRef]
- .Celik, A.A.; Simper, G.S.; Hiemisch, W.; Blasczyk, R.; Bade-Döding, C. HLA-G peptide preferences change in transformed cells: Impact on the binding motif. Immunogenetics 2018, 70, 485–494. [Google Scholar] [CrossRef]
- Morandi, F.; Rizzo, R.; Fainardi, E.; Rouas-Freiss, N.; Pistoia, V. Recent Advances in Our Understanding of HLA-G Biology: Lessons from a Wide Spectrum of Human Diseases. J. Immunol. Res. 2016. [Google Scholar] [CrossRef]
- .Elsner, H.A.; DeLuca, D.; Strub, J.; Blasczyk, R. HistoCheck: Rating of HLA class I and II mismatches by an internet-based software tool. Bone Marrow Transplant. 2004, 33, 165–169. [Google Scholar] [CrossRef]
- Wahl, A.; Schafer, F.; Bardet, W.; Buchli, R.; Air, G.M.; Hildebrand, W.H. HLA class I molecules consistently present internal influenza epitopes. Proc. Natl. Acad. Sci. USA 2009, 106, 540–545. [Google Scholar] [CrossRef]
- Clemens, E.B.; van de Sandt, C.; Wong, S.S.; Wakim, L.M.; Valkenburg, S.A. Harnessing the Power of T Cells: The Promising Hope for a Universal Influenza Vaccine. Vaccines 2018, 6, 18. [Google Scholar] [CrossRef]
- .Crux, N.B.; Elahi, S. Human Leukocyte Antigen (HLA) and Immune Regulation: How Do Classical and Non-Classical HLA Alleles Modulate Immune Response to Human Immunodeficiency Virus and Hepatitis C Virus Infections? Front. Immunol. 2017, 8, 832. [Google Scholar] [CrossRef] [PubMed]
- .Bade-Doeding, C.; Cano, P.; Huyton, T.; Badrinath, S.; Eiz-Vesper, B.; Hiller, O.; Blasczyk, R. Mismatches outside exons 2 and 3 do not alter the peptide motif of the allele group B*44:02P. Hum. Immunol. 2011, 72, 1039–1044. [Google Scholar] [CrossRef] [PubMed]
- Kunze-Schumacher, H.; Blasczyk, R.; Bade-Doeding, C. Soluble HLA technology as a strategy to evaluate the impact of HLA mismatches. J. Immunol. Res. 2014, 2014, 246171. [Google Scholar] [CrossRef] [PubMed]
| Type of HLA-F | Allelic Variant | NK Cell Receptor | Method of Identification | Reference |
|---|---|---|---|---|
| pHLA-F tetramer | F*01:01 | ILT-2 | SPR | [24] |
| pHLA-F tetramer | F*01:01 | ILT-4 | SPR | [24] |
| HLA-F OC | F*01:01 | KIR3DL2 | rKIR-Fc binding to HLA-I coated beads; SPR; rKIRζ jurkat reporter cell assay | [1,3,25] |
| HLA-F OC | F*01:01 | KIR2DS4 | Pull-down precipitation; SPR | [25] |
| HLA-F OC | F*01:01 | KIR3DL1 | SPR; rKIR-Fc binding to HLA-I coated beads | [1,2] |
| HLA-F OC | F*01:01 | KIR3DS1 | pull-down precipitation; rKIR-Fc binding to HLA-I coated beads; SPR; rKIRζ jurkat reporter cell assay | [1,2,3] |
| pHLA-F | F*01:01 | ILT-2 | Biolayer interferometry assay; X-ray crystallography | [3] |
| Sequence | Length | Source |
|---|---|---|
| KVGDDIAK | 8 | 60S ribosomal protein L12 |
| MAHMASKE | 8 | Glyceraldehyde-3-phosphate dehydrogenase |
| APNHAVVTR | 9 | Serotransferrin |
| AVTKYTSAK | 9 | Histone H2B type 1-K |
| AGFAGDDAPR | 10 | Actin, cytoplasmic 1 |
| AGEKVEKPDTK | 11 | 60S ribosomal protein L6 |
| EITALAPSTMK | 11 | Actin, cytoplasmic 1 |
| IVTDRETGSSK | 11 | Nucleolin |
| MYLGYEYVTAIR | 12 | Serotransferrin |
| TVLIMELINNVAK | 13 | ATP synthase subunit beta, mitochondrial |
| VNVDEVGGEALGR | 13 | Hemoglobin subunit beta |
| VTGYNDPETGNII | 13 | Desmoplakin |
| SYELPDGQVITIGNER | 16 | Actin, cytoplasmic 1 |
| TGAIVDVPVGEELLGR | 16 | ATP synthase subunit alpha, mitochondrial |
| TITLEVEPSDTIENVK | 16 | Ubiquitin-40S ribosomal protein S27a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hò, G.-G.T.; Heinen, F.J.; Blasczyk, R.; Pich, A.; Bade-Doeding, C. HLA-F Allele-Specific Peptide Restriction Represents an Exceptional Proteomic Footprint. Int. J. Mol. Sci. 2019, 20, 5572. https://doi.org/10.3390/ijms20225572
Hò G-GT, Heinen FJ, Blasczyk R, Pich A, Bade-Doeding C. HLA-F Allele-Specific Peptide Restriction Represents an Exceptional Proteomic Footprint. International Journal of Molecular Sciences. 2019; 20(22):5572. https://doi.org/10.3390/ijms20225572
Chicago/Turabian StyleHò, Gia-Gia T., Funmilola J. Heinen, Rainer Blasczyk, Andreas Pich, and Christina Bade-Doeding. 2019. "HLA-F Allele-Specific Peptide Restriction Represents an Exceptional Proteomic Footprint" International Journal of Molecular Sciences 20, no. 22: 5572. https://doi.org/10.3390/ijms20225572
APA StyleHò, G.-G. T., Heinen, F. J., Blasczyk, R., Pich, A., & Bade-Doeding, C. (2019). HLA-F Allele-Specific Peptide Restriction Represents an Exceptional Proteomic Footprint. International Journal of Molecular Sciences, 20(22), 5572. https://doi.org/10.3390/ijms20225572






