Identification of B Cell Epitopes of Blo t 13 Allergen and Cross-Reactivity with Human Adipocytes and Heart Fatty Acid Binding Proteins
Abstract
:1. Introduction
2. Results
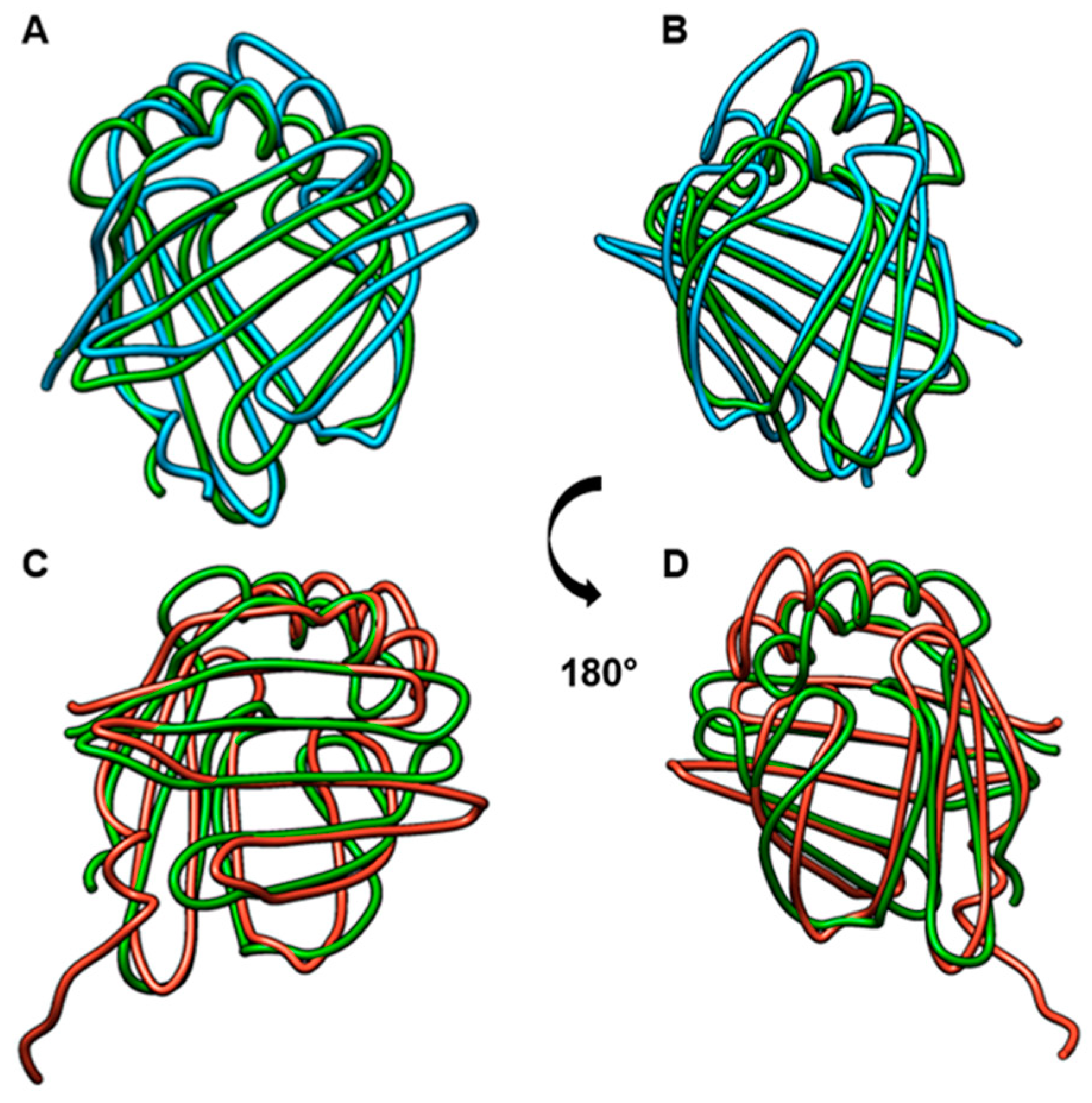
2.1. Blo t 13, FABP3, and FABP4 Share Two Conserved Regions
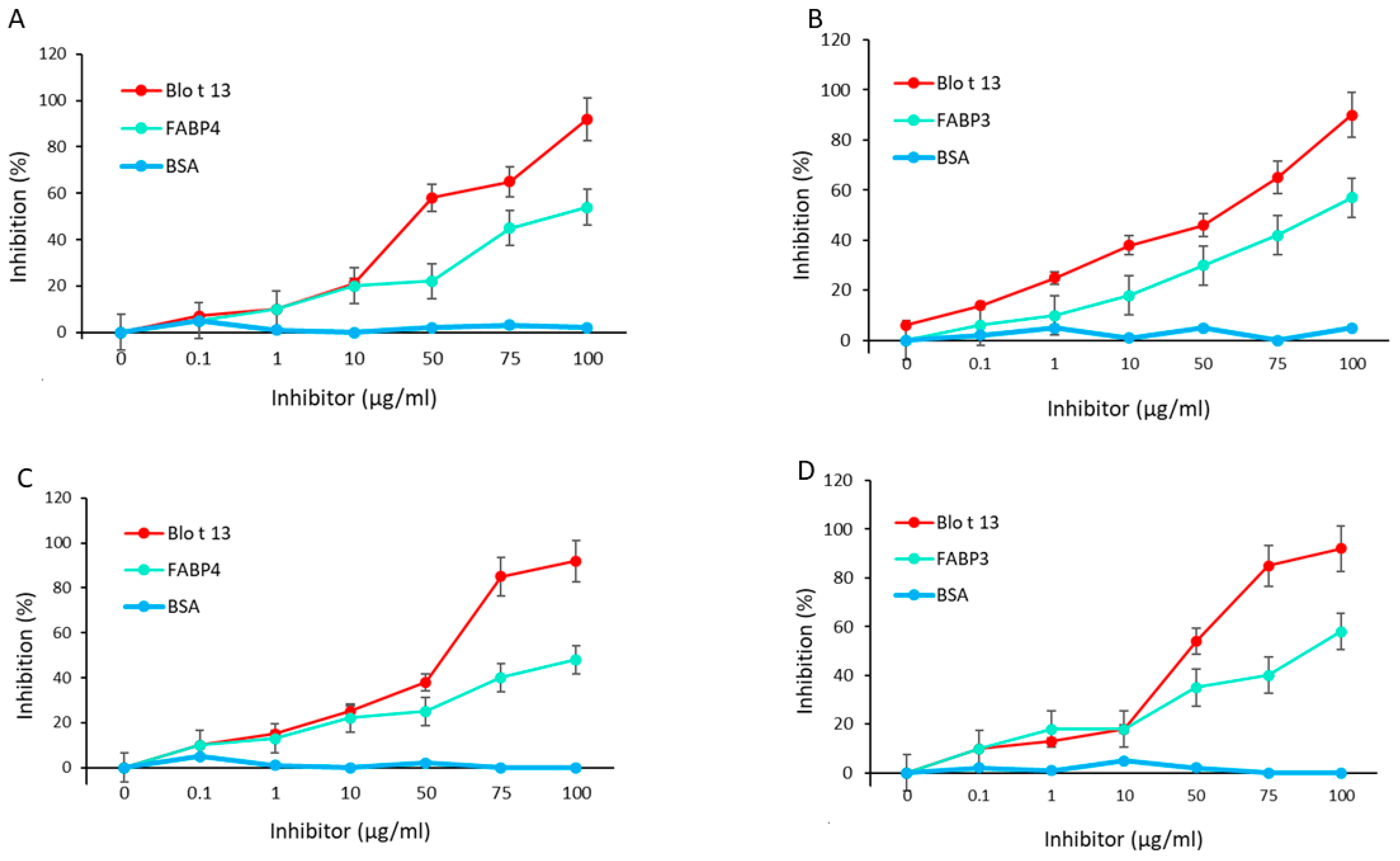
2.2. IgE and IgG Reactivity to FABP3 and FABP4 in Allergic Patients Sensitized to Blo t 13
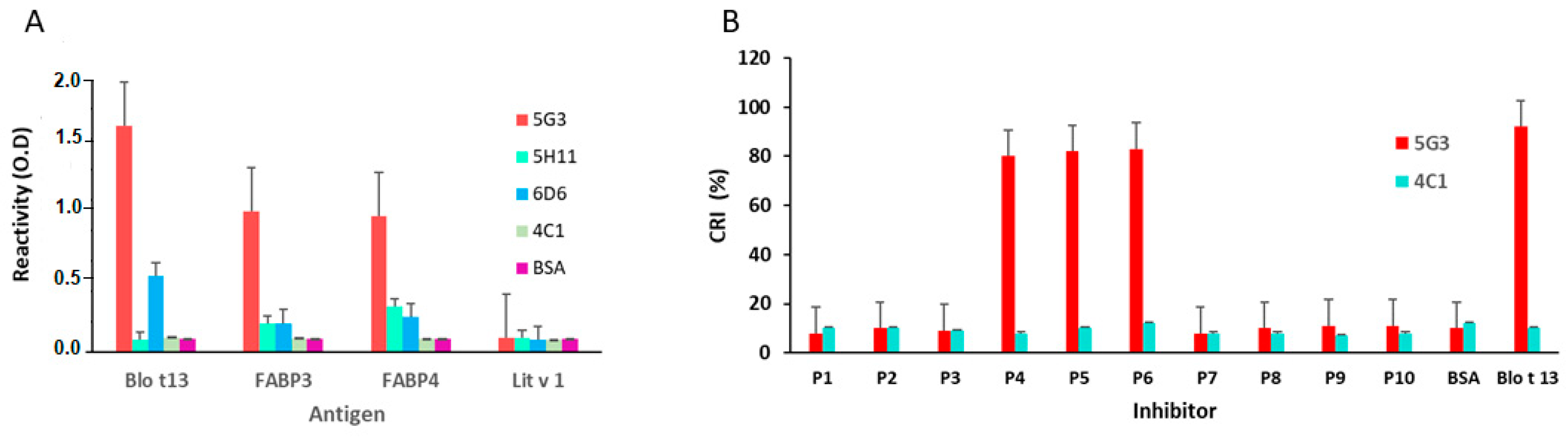
2.3. Blo t 13 Cross-reacts with Human FABPs
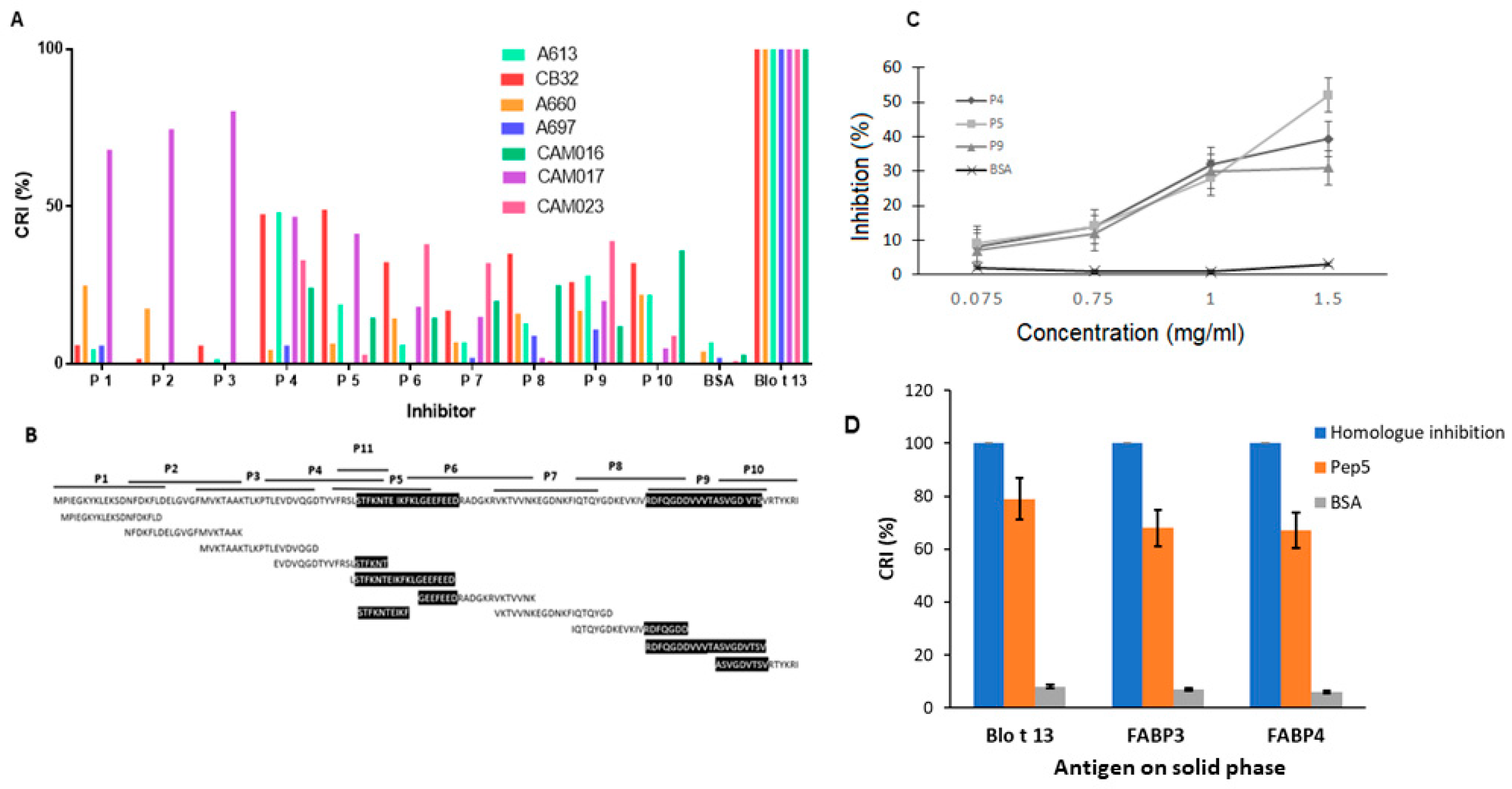
2.4. Epitope Mapping Reveals Two IgE Binding Regions in Blo t 13
3. Discussion
4. Materials and Methods
4.1. Recombinant Proteins
4.2. Serum Samples
4.3. Monoclonal Antibody Reactivity to Blo t 13 and Human FABPs
4.4. Escherichia coli Lysate Preparation for Serum Absorption
4.5. Determination of Serum Antibodies Against FABP and ELISA Inhibition
4.6. B Cell Epitope Mapping of Blo t 13
4.7. Cross-inhibition Assays by ELISA—Cross-Reactive Epitopes
4.8. Sequence Alignment and Structural Modeling
4.9. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AIT | Allergen immunotherapy |
| BSA | Bovine Serum Albumin |
| CRI | Cross-reactivity index |
| FABP | Fatty Acid Binding Protein |
| h | Hours |
| HDM | House Dust Mites |
| LB | Luria Bertani |
| M | Molar |
| mAb | Monoclonals antibodies |
| min | Minutes |
| mM | Milimolar |
| MnSOD | Manganese superoxide dismutase |
| nm | nanometers |
| OD | Optical density |
| PBS-T | Phosphate Buffer Saline Tween |
| RMSD | Root mean square deviation |
| rpm | Revolutions per minute |
| RT | Room temperature |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SPT | Skin prick test |
References
- Mackay, I.R. Autoantigens. In Encyclopedia of Immunology, 2nd ed.; Delves, P.J., Ed.; Elsevier: Oxford, UK, 1998; pp. 266–269. [Google Scholar]
- Crameri, R. Immunoglobulin E-binding autoantigens: Biochemical characterization and clinical relevance. Clin. Exp. Allergy. 2012, 42, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Zeller, S.; Rhyner, C.; Meyer, N.; Schmid-Grendelmeier, P.; Akdis, C.A.; Crameri, R. Exploring the repertoire of IgE-binding self-antigens associated with atopic eczema. J. Allergy Clin. Immunol. 2009, 124, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Glaser, A.G.; Limacher, A.; Fluckiger, S.; Scheynius, A.; Scapozza, L.; Crameri, R. Analysis of the cross-reactivity and of the 1.5 A crystal structure of the Malassezia sympodialis Mala s 6 allergen, a member of the cyclophilin pan-allergen family. Biochem. J. 2006, 396, 41–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limacher, A.; Glaser, A.G.; Meier, C.; Schmid-Grendelmeier, P.; Zeller, S.; Scapozza, L.; Crameri, R. Cross-reactivity and 1.4-A crystal structure of Malassezia sympodialis thioredoxin (Mala s 13), a member of a new pan-allergen family. J. Immunol. 2007, 178, 389–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crameri, R.; Faith, A.; Hemmann, S.; Jaussi, R.; Ismail, C.; Menz, G.; Blaser, K. Humoral and cell-mediated autoimmunity in allergy to Aspergillus fumigatus. J. Exp. Med. 1996, 184, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Schmid-Grendelmeier, P.; Fluckiger, S.; Disch, R.; Trautmann, A.; Wuthrich, B.; Blaser, K.; Scheynius, A.; Crameri, R. IgE-mediated and T cell-mediated autoimmunity against manganese superoxide dismutase in atopic dermatitis. J. Allergy Clin. Immunol. 2005, 115, 1068–1075. [Google Scholar] [CrossRef]
- Hradetzky, S.; Roesner, L.M.; Heratizadeh, A.; Crameri, R.; Garbani, M.; Scheynius, A.; Werfel, T. Differential cytokine induction by the human skin–associated autoallergen thioredoxin in sensitized patients with atopic dermatitis and healthy control subjects. J. Allergy Clin. Immunol. 2015, 135, 1378–1380.e5. [Google Scholar] [CrossRef]
- Hradetzky, S.; Werfel, T.; Rösner, L.M. Autoallergy in atopic dermatitis. Allergo J. Int. 2015, 24, 16–22. [Google Scholar] [CrossRef] [Green Version]
- Milian, E.; Diaz, A.M. Allergy to house dust mites and asthma. P. R. Health Sci. J. 2004, 23, 47–57. [Google Scholar]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, T.L.; Rasool, O.; Huecas, S.; Whitley, P.; Crameri, R.; Appenzeller, U.; Gafvelin, G.; van Hage-Hamsten, M. Cloning of three new allergens from the dust mite Lepidoglyphus destructor using phage surface display technology. Eur. J. Biochem. 2001, 268, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Satitsuksanoa, P.; Kennedy, M.; Gilis, D.; Le Mignon, M.; Suratannon, N.; Soh, W.T.; Wongpiyabovorn, J.; Chatchatee, P.; Vangveravong, M.; Rerkpattanapipat, T.; et al. The minor house dust mite allergen Der p 13 is a fatty acid-binding protein and an activator of a TLR2-mediated innate immune response. Allergy 2016, 71, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Puerta, L.; Kennedy, M.W.; Jim nez, S.; Caraballo, L. Structural and ligand binding analysis of recombinant blo t 13 allergen from Blomia tropicalis mite, a fatty acid binding protein. Int. Arch. Allergy Immunol. 1999, 119, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Shum, B.O.V.; Mackay, C.R.; Gorgun, C.Z.; Frost, M.J.; Kumar, R.K.; Hotamisligil, G.S.; Rolph, M.S. The adipocyte fatty acid–binding protein aP2 is required in allergic airway inflammation. J. Clin. Investig. 2006, 116, 2183–2192. [Google Scholar] [CrossRef] [Green Version]
- Ghelfi, E.; Yu, C.W.; Elmasri, H.; Terwelp, M.; Lee, C.G.; Bhandari, V.; Comhair, S.A.; Erzurum, S.C.; Hotamisligil, G.S.; Elias, J.A.; et al. Fatty acid binding protein 4 regulates VEGF-induced airway angiogenesis and inflammation in a transgenic mouse model: Implications for asthma. Am. J. Pathol. 2013, 182, 1425–1433. [Google Scholar] [CrossRef] [Green Version]
- Rolph, M.S.; Young, T.R.; Shum, B.O.V.; Gorgun, C.Z.; Schmitz-Peiffer, C.; Ramshaw, I.A.; Hotamisligil, G.S.; Mackay, C.R. Regulation of Dendritic Cell Function and T Cell Priming by the Fatty Acid-Binding Protein aP2. J. Immunol. 2006, 177, 7794. [Google Scholar] [CrossRef] [Green Version]
- Pfiffner, P.; Truffer, R.; Matsson, P.; Rasi, C.; Mari, A.; Stadler, B.M. Allergen cross reactions: A problem greater than ever thought? Allergy 2010, 65, 1536–1544. [Google Scholar] [CrossRef]
- Qian, Y.; Culton, D.A.; Jeong, J.S.; Trupiano, N.; Valenzuela, J.G.; Diaz, L.A. Non-infectious environmental antigens as a trigger for the initiation of an autoimmune skin disease. Autoimmun. Rev. 2016, 15, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Cipriani, F.; Ricci, G.; Leoni, M.C.; Capra, L.; Baviera, G.; Longo, G.; Maiello, N.; Galli, E. Autoimmunity in atopic dermatitis: Biomarker or simply epiphenomenon? J. Derm. 2014, 41, 569–576. [Google Scholar] [CrossRef] [Green Version]
- Peterson, L.K.; Fujinami, R.S. 3 - MOLECULAR MIMICRY. In Autoantibodies, 2nd ed.; Shoenfeld, Y., Gershwin, M.E., Meroni, P.L., Eds.; Elsevier: Burlington, NJ, USA, 2007; pp. 13–19. [Google Scholar]
- Chan, S.L.; Ong, S.T.; Ong, S.Y.; Chew, F.T.; Mok, Y.K. Nuclear Magnetic Resonance Structure-Based Epitope Mapping and Modulation of Dust Mite Group 13 Allergen as a Hypoallergen. J. Immunol. 2006, 176, 4852–4860. [Google Scholar] [CrossRef]
- Appenzeller, U.; Meyer, C.; Menz, G.; Blaser, K.; Crameri, R. IgE–Mediated Reactions to Autoantigens in Allergic Diseases. Int. Arch. Allergy Immunol. 1999, 118, 193–196. [Google Scholar] [CrossRef]
- Valenta, R.; Duchene, M.; Pettenburger, K.; Sillaber, C.; Valent, P.; Bettelheim, P.; Breitenbach, M.; Rumpold, H.; Kraft, D.; Scheiner, O. Identification of profilin as a novel pollen allergen; IgE autoreactivity in sensitized individuals. Science 1991, 253, 557–560. [Google Scholar] [CrossRef]
- Munera, M.; Gomez, L.; Puerta, L. Shrimp as an allergen source. Biomedica 2013, 33, 306–318. [Google Scholar]
- Chauhan, V.; Kaur, G.; Naveen, K.G.; Sehgal, R. Fatty Acid Binding Protein-1 (FABP-1) of Echinococcus granulosus- a promising vaccine candidate- An in silico analysis. Indo Am. J. Pharm. Res. 2016, 6, 6445–6451. [Google Scholar]
- Waine, G.J.; Scott, J.C.; Mazzer, D.; McManus, D.P. Mapping of linear B-cell epitopes on the 14-kDa fatty-acid binding protein of Chinese Schistosoma japonicum. Int. J. Parasitol. 1998, 28, 303–308. [Google Scholar] [CrossRef]
- Zakzuk, J.; Jimenez, S.; Cheong, N.; Puerta, L.; Lee, B.W.; Chua, K.Y.; Caraballo, L. Immunological characterization of a Blo t 12 isoallergen: Identification of immunoglobulin E epitopes. Clin. Exp. Allergy 2009, 39, 608–616. [Google Scholar] [CrossRef]
- Rottem, M.; Shoenfeld, Y. Asthma as a paradigm for autoimmune disease. Int. Arch. Allergy Immunol. 2003, 132, 210–214. [Google Scholar] [CrossRef]
- Tedeschi, A.; Asero, R. Asthma and autoimmunity: A complex but intriguing relation. Expert Rev. Clin. Immunol. 2008, 4, 767–776. [Google Scholar] [CrossRef]
- Agache, I.; Lau, S.; Akdis, C.A.; Smolinska, S.; Bonini, M.; Cavkaytar, O.; Flood, B.; Gajdanowicz, P.; Izuhara, K.; Kalayci, O.; et al. EAACI Guidelines on Allergen Immunotherapy: House dust mite-driven allergic asthma. Allergy 2019, 74, 855–873. [Google Scholar] [CrossRef] [Green Version]
- Linneberg, A.; Jacobsen, R.K.; Jespersen, L.; Abildstrøm, S.Z. Association of subcutaneous allergen-specific immunotherapy with incidence of autoimmune disease, ischemic heart disease, and mortality. J. Allergy Clin. Immunol. 2012, 129, 413–419. [Google Scholar] [CrossRef]
- Bozek, A.; Kozlowska, R.; Jarzab, J. The safety of specific immunotherapy for patients allergic to house-dust mites and pollen in relation to the development of neoplasia and autoimmune disease: A long-term, observational case-control study. Int. Arch. Allergy Immunol. 2014, 163, 307–312. [Google Scholar] [CrossRef]
- Labrada, M.; Uyema, K.; Sewer, M.; Labrada, A.; Gonzalez, M.; Caraballo, L.; Puerta, L. Monoclonal antibodies against Blo t 13, a recombinant allergen from Blomia tropicalis. Int. Arch. Allergy Immunol. 2002, 129, 212–218. [Google Scholar] [CrossRef]
- UniProt Consortium. The universal protein resource (UniProt). Nucleic Acids Res. 2008, 36, D190–D195. [Google Scholar]
- Brandt, B.W.; Heringa, J. Protein analysis tools and services at IBIVU. J. Integr. Bioinform. 2011, 8, 168. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [Green Version]
- Parasuraman, S. Protein data bank. J. Pharmacol. Pharmacother. 2012, 3, 351–352. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Gromiha, M.; Fawareh, H.; Sarai, A. ASAView: Database and tool for solvent accessibility representation in proteins. BMC Bioinform. 2004, 5, 51. [Google Scholar] [CrossRef] [Green Version]



| Patient | Sex | Prick Test > 3 mm | IgE Levels (O.D.) Median | ||
|---|---|---|---|---|---|
| Blo t 13 | FABP3 | FABP4 | |||
| 1 | M | Bt, Dp | 0.28 | 0.20 | 0.24 |
| 2 | M | Bt, Dp | 0.19 | 0.16 | 0.20 |
| 3 | F | Bt, Dp | 0.30 | 0.24 | 0.23 |
| 4 | F | Bt, Dp | 0.16 | 0.13 | 0.11 |
| 5 | F | Bt, Dp | 0.30 | 0.13 | 0.12 |
| 6 | F | Bt, Dp | 0.31 | 0.15 | 0.14 |
| 7 | M | Bt, Dp | 0.24 | 0.62 | 0.48 |
| 8 | F | Bt, Dp | 0.26 | 0.09 | 0.17 |
| 9 | M | Bt, Dp | 0.41 | 0.11 | 0.17 |
| 10 | M | Bt, Dp | 0.26 | 0.09 | 0.09 |
| 11 | F | Bt, Dp | 0.30 | 0.15 | 0.16 |
| 12 | M | Bt | 0.24 | 0.12 | 0.14 |
| 13 | F | Bt, Dp | 0.33 | 0.17 | 0.16 |
| 14 | M | Bt, Dp | 0.35 | 0.11 | 0.10 |
| 15 | M | Bt, Dp | 0.60 | 0.18 | 0.16 |
| 16 | F | Bt | 1.15 | 0.11 | 0.11 |
| 17 | M | Bt, Dp | 0.34 | 0.14 | 0.09 |
| 18 | M | Bt, Dp | 0.36 | 0.11 | 0.11 |
| 19 | F | Bt, Dp | 0.24 | 0.20 | 0.32 |
| 20 | M | Bt | 0.76 | 0.10 | 0.11 |
| 21 | M | Bt, Dp | 0.25 | 0.11 | 0.11 |
| 22 | F | Bt, Dp | 0.20 | 0.15 | 0.14 |
| 23 | M | Bt, Dp | 0.22 | 0.11 | 0.22 |
| 24 | M | Bt, Dp | 0.25 | 0.15 | 0.17 |
| 25 | M | Bt, Dp | 0.25 | 0.11 | 0.14 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Múnera, M.; Martínez, D.; Labrada, A.; Caraballo, L.; Puerta, L. Identification of B Cell Epitopes of Blo t 13 Allergen and Cross-Reactivity with Human Adipocytes and Heart Fatty Acid Binding Proteins. Int. J. Mol. Sci. 2019, 20, 6107. https://doi.org/10.3390/ijms20246107
Múnera M, Martínez D, Labrada A, Caraballo L, Puerta L. Identification of B Cell Epitopes of Blo t 13 Allergen and Cross-Reactivity with Human Adipocytes and Heart Fatty Acid Binding Proteins. International Journal of Molecular Sciences. 2019; 20(24):6107. https://doi.org/10.3390/ijms20246107
Chicago/Turabian StyleMúnera, Marlon, Dalgys Martínez, Alexis Labrada, Luis Caraballo, and Leonardo Puerta. 2019. "Identification of B Cell Epitopes of Blo t 13 Allergen and Cross-Reactivity with Human Adipocytes and Heart Fatty Acid Binding Proteins" International Journal of Molecular Sciences 20, no. 24: 6107. https://doi.org/10.3390/ijms20246107






