CRK5 Protein Kinase Contributes to the Progression of Embryogenesis of Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
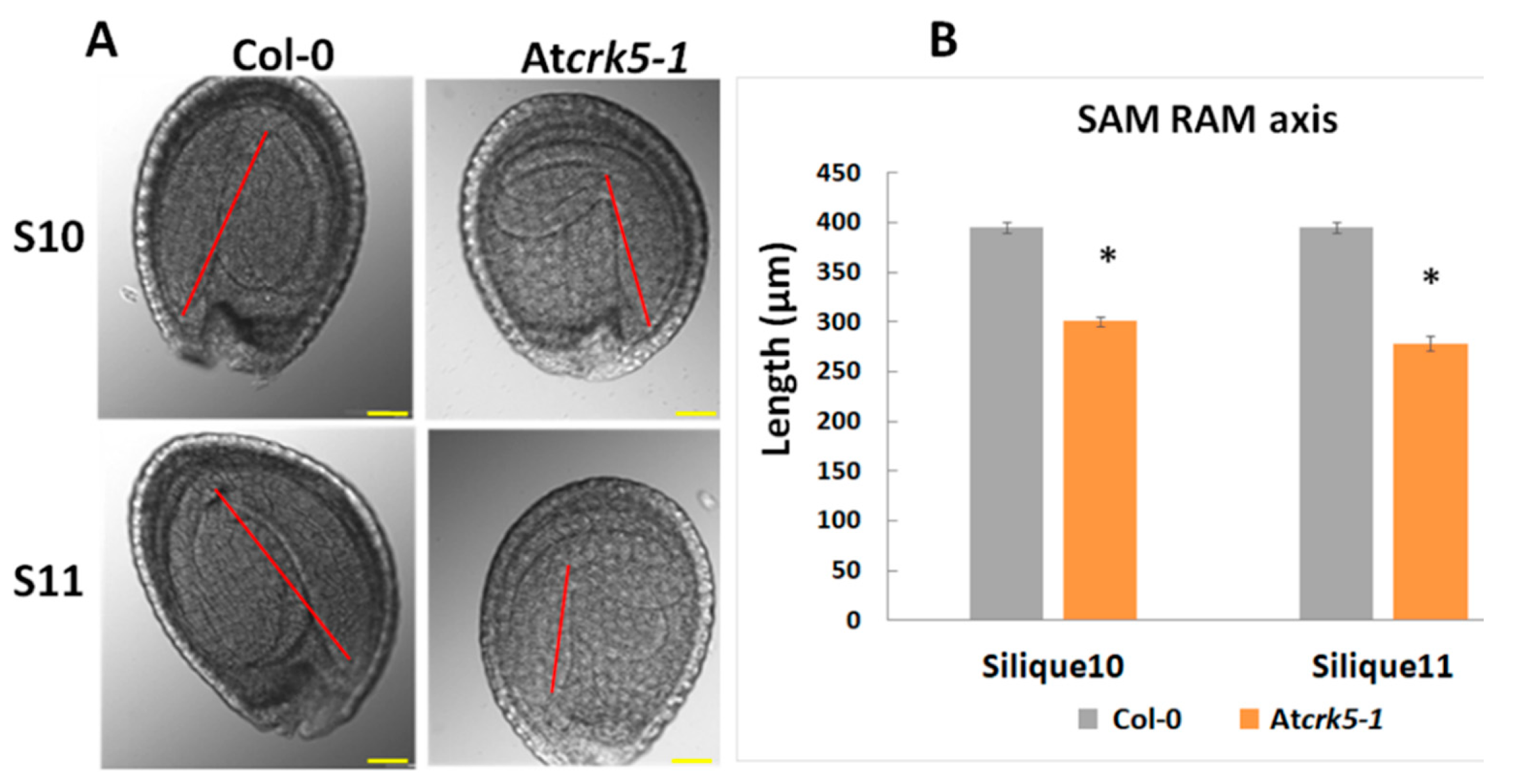
2.1. The Atcrk5-1 Mutant Exhibits a Considerable Delay in the Progression of the Phases of Embryogenesis
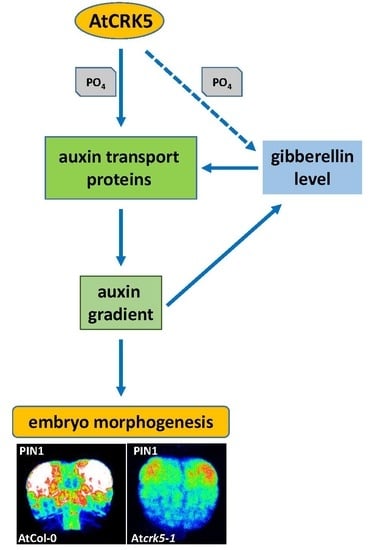
2.2. The Delayed Development of the Atcrk5-1 Embryos Is Linked to Their Decreased Gibberellin Synthesis and Level
2.3. The Auxin Level Is Decreased in the Atcrk5-1 Mutant Embryos in Comparison with the Wild Type Ones.
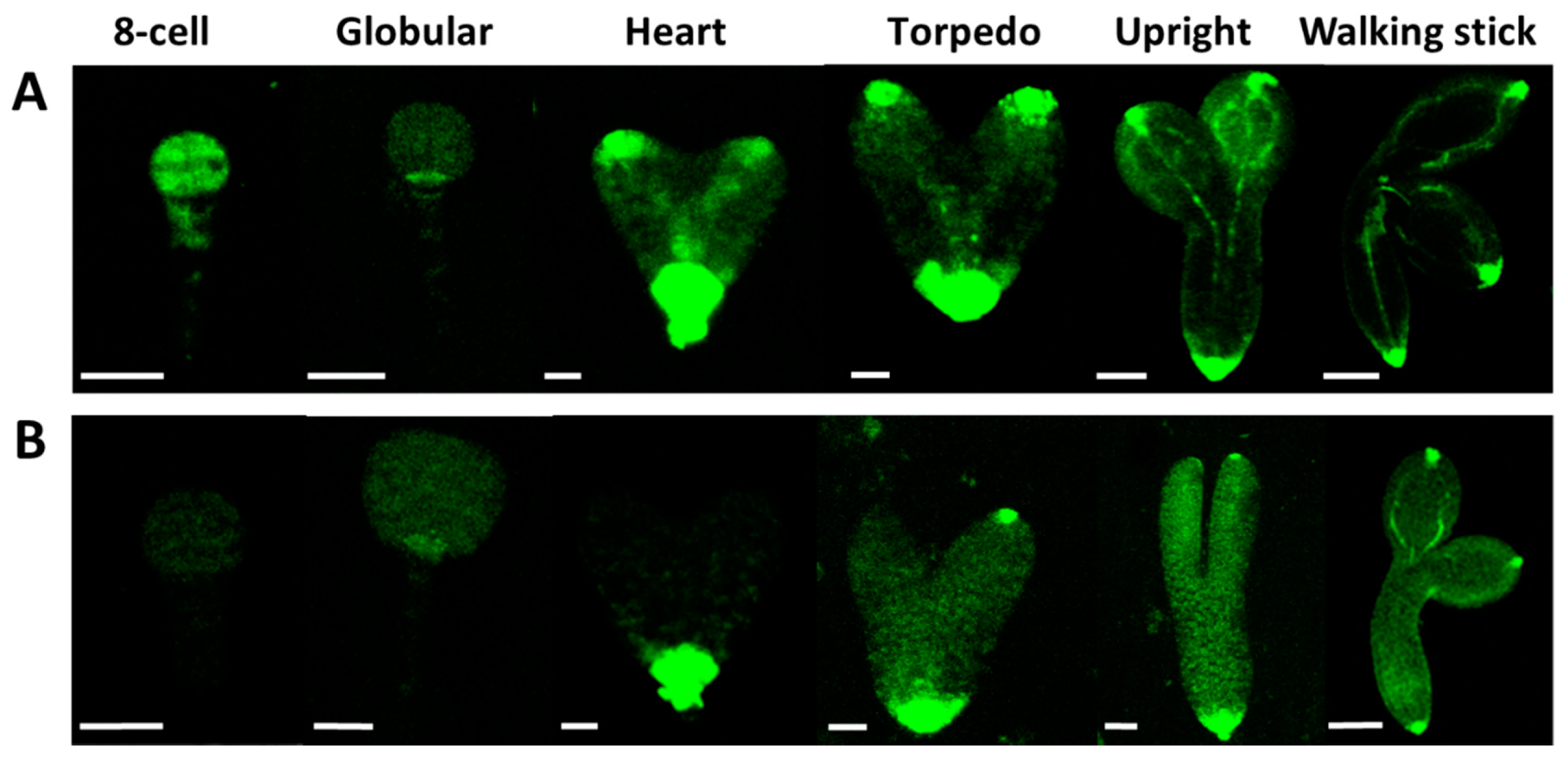
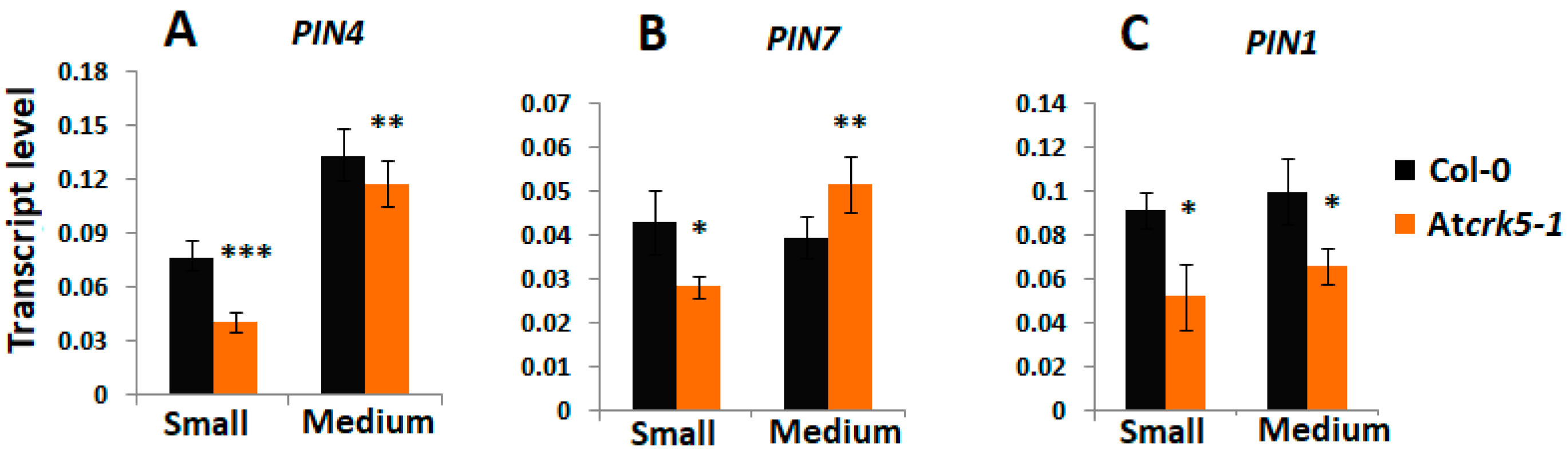
2.4. Expression and Abundance of Auxin Efflux Proteins During Embryogenesis of Wild Type and Atcrk5-1 Mutant Plants
2.5. Expression and Abundance of Auxin Influx (AUX1) Proteins During Embryogenesis of Wild Type and Atcrk5-1 Mutant Plants
2.6. AtCRK5 Can Phosphorylate the Auxin Efflux Proteins PIN1, PIN4 and PIN7 in vitro
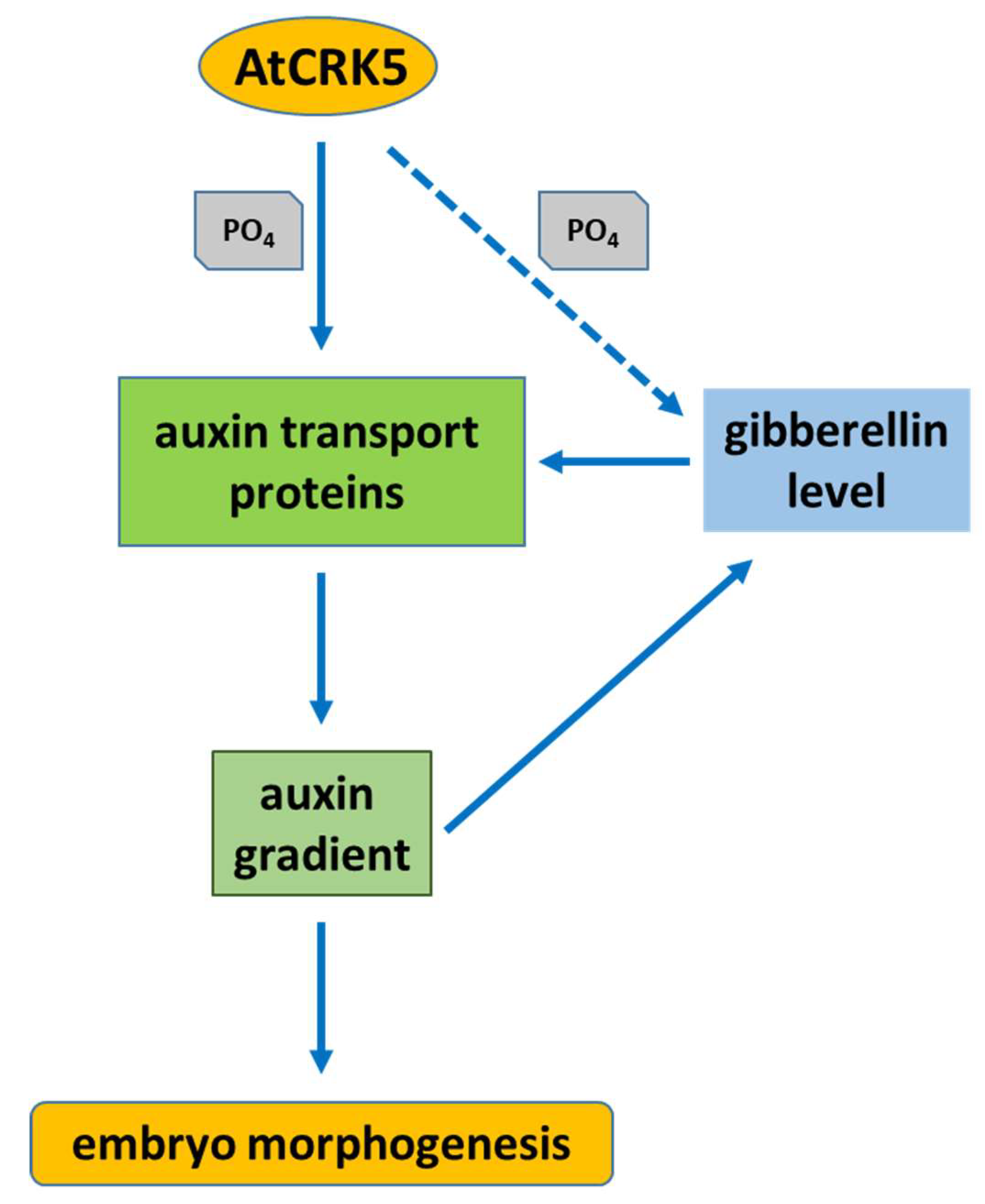
3. Discussion
3.1. The AtCRK5 Protein Kinase Controls the Gibberellin Level Influencing Seed Size and Embryogenesis
3.2. AtCRK5 is a General Regulator of Auxin Distribution Potentially via the Phosphorylation of Several PINs
3.3. The AtCRK5 Protein Kinase is Involved in Hormonal Crosstalk Influencing Embryogenesis
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Seed/Embryo Size/Axis Determination, Embryo Isolation and GA Rescue Experiments
4.3. Total GA Measurement by Competitive GAs Elisa Assay
4.4. Embryo Morphology Monitored by Cell-R Microscopy
4.5. Monitoring of Abundance of GFP/YFP Signals in Embryos by LSM Microscopy
4.6. RNA Isolation and Real Time Quantitative PCR (qRT-PCR) for Embryo Gene Expression
4.7. PIN4 and PIN7 Hydrophilic Loop Region Cloning
4.8. Purification of Tagged Proteins
4.9. In vitro Kinase Assays
4.10. Mass Spectrometry
4.11. Bioinformatic Analysis and Tools
4.12. Accession Numbers
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mayer, U.; Buttner, G.; Jurgens, G. Apical-basal pattern formation in the Arabidopsis embryo: Studies on the role of the gnom gene. Development 1993, 117, 149–162. [Google Scholar]
- Ten Hove, C.A.; Lu, K.-J.; Weijers, D. Building a plant: Cell fate specification in the early Arabidopsis embryo. Development 2015, 142, 420–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Zhou, L.; Huang, M.; He, X.; Yang, Y.; Liu, X.; Li, Y.; Hou, X. Gibberellins play an essential role in late embryogenesis of Arabidopsis. Nat. Plants 2018, 4, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Locascio, A.; Roig-Villanova, I.; Bernardi, J.; Varotto, S. Current perspectives on the hormonal control of seed development in Arabidopsis and maize: A focus on auxin. Front. Plant Sci. 2014, 5, 412. [Google Scholar] [CrossRef] [Green Version]
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef]
- Vanneste, S.; Friml, J. Auxin: A trigger for change in plant development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef]
- Vanstraelen, M.; Benkova, E. Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 2012, 28, 463–487. [Google Scholar] [CrossRef]
- Zádnikova, P.; Smet, D.; Zhu, Q.; Van der Straeten, D.; Benková, E. Strategies of seedlings to overcome their sessile nature: Auxin in mobility control. Front. Plant Sci. 2015, 6, 218. [Google Scholar] [CrossRef] [Green Version]
- Hamann, T.; Benkova, E.; Baurle, I.; Kientz, M.; Jurgens, G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002, 16, 1610–1615. [Google Scholar] [CrossRef] [Green Version]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef]
- Jenik, P.D.; Barton, M.K. Surge and destroy: The role of auxin in plant embryogenesis. Development 2005, 132, 3577–3585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wabnik, K.; Robert, H.S.; Smith, R.S.; Friml, J. Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr. Biol. 2013, 23, 2513–2518. [Google Scholar] [CrossRef]
- Bennett, M.J.; Marchant, A.; Green, H.G.; May, S.T.; Ward, S.P.; Millner, P.A.; Walker, A.R.; Schulz, B.F.; Feldmann, K.A. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 1996, 273, 948–950. [Google Scholar] [CrossRef] [PubMed]
- Petrasek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.J.; Abas, M.; Seifertova, D.; Wiśniewska, J.; Tadele, Z.; Kubeš, M.; Čovanová, M.; et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krecek, P.; Skupa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249–260. [Google Scholar] [CrossRef] [Green Version]
- Barbez, E.; Kubeš, M.; Rolčík, J.; Béziat, C.; Pěnčík, A.; Wang, B.; Rosquete, M.R.; Zhu, J.; Dobrev, P.I.; Lee, Y.; et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 2012, 485, 119–122. [Google Scholar] [CrossRef] [Green Version]
- Robert, H.S.; Grunewald, W.; Sauer, M.; Cannoot, B.; Soriano, M.; Swarup, R.; Weijers, D.; Bennett, M.; Boutilier, K.; Friml, J. Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 2015, 142, 702–711. [Google Scholar] [CrossRef] [Green Version]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Dong, Q.; Kita, D.; Huang, J.B.; Liu, G.; Wu, X.; Zhu, X.; Cheung, A.Y.; Wu, H.M.; Tao, L.Z. RopGEF1 Plays a Critical Role in Polar Auxin Transport in Early Development. Plant Physiol. 2017, 175, 157–171. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.J.; Luo, J. The PIN-FORMED auxin effux carriers in plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef] [Green Version]
- Wisniewska, J.; Xu, J.; Seifertova, D.; Brewer, P.B.; Ruzicka, K.; Blilou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Möller, B.; Weijers, D. Auxin Control of Embryo Patterning. Cold Spring Harb. Perspect. Biol. 2009, 1, a001545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, H.S.; Chulmin Park, C.; Gutierrez, C.L.; Wójcikowska, B.; Pěnčík, A.; Novák, O.; Chen, J.; Grunewald, W.; Dresselhaus, T.; Friml, J.; et al. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 2018, 4, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Ugartechea-Chirino, Y.; Swarup, Y.R.; Swarup, K.; Peret, B.; Whitworth, M.; Bennett, M.; Bougourd, S. The AUX1 LAX family of auxin influx carriers is required for the establishment of embryonic root cell organization in Arabidopsis thaliana. Ann. Bot. 2010, 105, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Swain, S.M.; Reid, J.B.; Kamiya, Y. Gibberellins are required for embryo growth and seed development in pea. Plant J. 1997, 12, 1329–1338. [Google Scholar] [CrossRef]
- Hays, D.B.; Yeung, E.C.; Pharis, R.P. The role of gibberellins in embryo axis development. J. Exp. Bot. 2002, 53, 1747–1751. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.P.; Jermakow, A.M.; Swain, S.M. Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 2002, 14, 3133–3147. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arab. Book 2008, 6, e0103. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef]
- Schwecheimer, C. Gibberellin signaling in plants—The extended version. Front. Plant Sci. 2012, 2, 107. [Google Scholar] [CrossRef] [Green Version]
- Abbas, M.; Alabadi, D.; Blazquez, M.A. Differential growth at the apical hook: All roads lead to auxin. Front. Plant Sci. 2013, 4, 441–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salanenka, Y.; Verstraeten, I.; Löfke, C.; Tabata, K.; Naramoto, S.; Glanc, M.; Friml, J. Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane. Proc. Natl. Acad. Sci. USA 2018, 115, 3716–3721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willige, B.C.; Isono, E.; Richter, R.; Zourelidou, M.; Schwechheimer, C. Gibberellin Regulates PIN-FORMED Abundance and Is Required for Auxin Transport–Dependent Growth and Development in Arabidopsis thaliana. Plant Cell 2011, 23, 2184–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.P.; Kamiya, Y. The Arabidopsis GAl Locus Encodes the Cyclase ent-Kaurene Synthetase A of Gibberellin Biosynthesis. Plant Cell 1994, 6, 1509–1518. [Google Scholar] [PubMed] [Green Version]
- Willige, C.V.; Ghosh, S.; Nill, C.; Zourelidou, M.; Dohmann, E.M.N.; Maier, A.; Schwechheimer, C. The DELLA Domain of GA INSENSITIVE Mediates the Interaction with the GA INSENSITIVE DWARF1A Gibberellin Receptor of Arabidopsis. Plant Cell 2007, 19, 1209–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreenivasulu, N.; Wobus, U. Seed-development programs: A systems biology-based comparison between dicots and monocots. Annu. Rev. Plant Biol. 2013, 64, 189–217. [Google Scholar] [CrossRef]
- Ye, N.; Zhang, J. Antagonism between abscisic acid and gibberellins is partially mediated by ascorbic acid during seed germination in rice. Plant Signal. Behav. 2012, 7, 563–565. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Hou, X. Antagonistic Regulation of ABA and GA in Metabolism and Signaling Pathways. Front. Plant Sci. 2018, 9, 251. [Google Scholar] [CrossRef] [Green Version]
- Shu, K.; Zhou, W.; Chen, F.; Luo, X.; Yang, W. Abscisic Acid and Gibberellins Antagonistically Mediate Plant Development and Abiotic Stress Responses. Front. Plant Sci. 2018, 9, 416. [Google Scholar] [CrossRef] [Green Version]
- Koornneef, M.; Jorna, M.L.; Brinkhorst-Van Der Swan, D.L.; Karssen, C.M. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 1982, 61, 385–393. [Google Scholar] [CrossRef]
- Roscoe, T.T.; Guilleminot, J.; Bessoule, J.-J.; Berger, F.; Devic, M. Complementation of Seed Maturation Phenotype by Ectopic Expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant Cell Physiol. 2015, 56, 1215–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, M.A.L.; Yee, K.M.; Danao, J.; Zimmermann, J.L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 1994, 6, 1731–1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luerssen, H.; Kirik, V.; Herrmann, P.; Misera, S. FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 1998, 15, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFYCOTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA 2003, 100, 2152–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braybrook, S.A.; Harada, J.J. LECs go crazy in embryo development. Trends Plant Sci. 2008, 13, 624–630. [Google Scholar] [CrossRef]
- Yamamoto, A.; Yoshii, M.; Murase, S.; Fujita, M.; Kurata, N.; Hobo, T.; Kagaya, Y.; Takeda, S.; Hattori, T. Cell-by-Cell Developmental Transition from Embryo to Post-Germination Phase Revealed by Heterochronic Gene Expression and ER-Body Formation in Arabidopsis leafy cotyledon Mutants. Plant Cell Physiol. 2014, 55, 2112–2125. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.Q.; Luan, S.; Li, J.; He, Z.H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef] [Green Version]
- Frigerio, M.; Alabadı, D.; APerez-Gomez, J.; Garcıa-Carcel, L.; Phillips, A.F.; Hedden, P.; Blazquez, M.A. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 2006, 142, 553–563. [Google Scholar] [CrossRef] [Green Version]
- Rieu, I.; Ruiz-Rivero, O.; Fernandez-Garcia, N.; Griffiths, J.; Powers, S.J.; Gong, F.; Linhartova, T.; Eriksson, S.; Nilsson, O.; Thomas, S.G.; et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008, 53, 488–504. [Google Scholar] [CrossRef] [PubMed]
- Dorcey, E.; Urbez, C.; Blazquez, M.A.; Carbonell, J.; Perez-Amador, M.A. Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J. 2009, 58, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998, 10, 1555–1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Cheng, H.; King, K.E.; Wang, W.; He, Y.; Hussain, A.; Lo, L.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002, 16, 646–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daviere, J.M.; Achard, P. A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 2016, 9, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Rigó, G.; Ayaydin, F.; Tietz, O.; Zsigmond, L.; Kovács, H.; Páy, A.; Salchert, K.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; et al. Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 2013, 25, 1592–1608. [Google Scholar]
- Baba, A.I.; Rigó, G.; Ayaydin, F.; Rehman, A.U.; Andrási, N.; Zsigmond, L.; Valkai, I.; Urbancsok, J.; Vass, I.; Pasternak, T.; et al. Functional Analysis of the Arabidopsis thaliana CDPK-Related Kinase Family: AtCRK1 Regulates Responses to Continuous Light. Int. J. Mol. Sci. 2018, 19, 1282. [Google Scholar] [CrossRef] [Green Version]
- Baba, A.I.; Andrási, N.; Valkai, I.; Gorcsa, T.; Koczka, L.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; Fehér, A.; Rigó, G.; et al. AtCRK5 Protein Kinase Exhibits a Regulatory Role in Hypocotyl Hook Development during Skotomorphogenesis. Int. J. Mol. Sci. 2019, 20, 3432. [Google Scholar] [CrossRef] [Green Version]
- Robert, S.H. Molecular communication for coordinated seed and fruit dvelopment: What can we learn from auxin and sugars? Int. J. Mol. Sci. 2019, 20, 936. [Google Scholar] [CrossRef] [Green Version]
- Ottenschläger, I.; Wolff, P.; Wolverton, C.; Bhalerao, R.P.; Sandberg, G.; Ishikawa, H.; Evans, M.; Palme, K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 2003, 100, 2987–2991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenik, P.D.; Gillmor, C.S.; Lukowitz, W. Embryonic patterning in Arabidopsis thaliana. Annu. Rev. Cell Dev. Biol. 2007, 23, 207–236. [Google Scholar] [CrossRef] [PubMed]
- Offringa, R.; Huang, F. Phosphorylation-dependent trafficking of plasma membrane proteins in animal and plant cells. J. Integr. Plant Biol. 2013, 55, 789–808. [Google Scholar] [CrossRef] [PubMed]
- Luschnig, C.; Vert, G. The dynamics of plant plasma membrane proteins: PINs and beyond. Development 2014, 141, 2924–2938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbosa, I.C.R.; Hammes, U.Z.; Schwechheimer, C. Activation and polarity control of PIN-FORMED auxin transporters by phosphorylation. Trends Plant Sci. 2018, 23, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Hrabak, E.M.; Chan, C.W.M.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK Superfamily of Protein Kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [Green Version]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, Y.-T. Calmodulin-binding protein kinases in plants. Trends Plant Sci. 2003, 8, 123–127. [Google Scholar] [CrossRef]
- Zhang, L.; Du, L.; Poovaiah, B.W. Calcium signaling and biotic defense responses in plants. Plant Signal Behav. 2014, 9, e973818. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Xu, L.; Singh, A.; Wang, H.; Du, L.; Poovaiah, B.W. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front Plant Sci. 2015, 6, 600. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.P.; Xu, Y.P.; Munyampundu, J.P.; Liu, T.Y.; Cai, X.Z. Calcium dependent protein kinase (CDPK) and CDPK related kinase (CRK) gene families in tomato: Genome wide identification and functional analyses in disease resistance. Mol. Genet. Genom. 2016, 291, 661–676. [Google Scholar] [CrossRef] [PubMed]
- Simeunovic, A.; Mair, A.; Wurzinger, B.; Teige, M. Know where your clients are: Subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 2016, 67, 3855–3872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delormel, T.Y.; Boudsocq, M. Properties and functions of calcium dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, A.I.; Rigó, G.; Andrási, N.; Tietz, O.; Palme, K.; Szabados, L.; Cséplő, Á. Striving Towards Abiotic Stresses: Role of the Plant CDPK Superfamily Members. In International Climate Protection; Palócz-Andresen, M., Szalay, D., Gosztom, A., Sípos, L., Taligás, T., Eds.; Springer: Cham, Switzerland, 2019; pp. 99–105. [Google Scholar]
- Leyser, O. Dynamic integration of auxin transport and signalling. Curr. Biol. 2006, 16, R424–R433. [Google Scholar] [CrossRef] [Green Version]
- Paponov, I.A.; Paponov, M.; Teale, W.; Menges, M.; Chakrabortee, S.; Murray, J.A.H.; Palme, K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 2008, 1, 321–337. [Google Scholar] [CrossRef]
- Ganguly, A.; Sasayama, D.; Cho, H.T. Regulation of the polarity of protein trafficking by phosphorylation. Mol. Cells. 2012, 33, 423–430. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, I.C.R.; Schwechheimer, C. Dynamic control of auxin transport-dependent growth by AGCVIII protein kinases. Curr. Opin. Plant Biol. 2014, 22, 108–115. [Google Scholar] [CrossRef]
- Mazzella, M.A.; Casal, J.J.; Muschietti, J.P.; Fox, A.R. Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 2014, 5, 52–65. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Harberd, N.P. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 2003, 421, 740–743. [Google Scholar] [CrossRef]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Zadnikova, P.; Petrasek, J.; Marhavy, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerová, K.; Morita, M.T.; Tasaka, M.; Hejátko, J.; et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607–617. [Google Scholar] [CrossRef] [Green Version]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]
- Swarup, R.; Friml, J.; Marchant, A.; Ljung, K.; Sandberg, G.; Palme, K.; Bennett, M. Localization of the auxin permease AUX 1suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 2001, 15, 2648–2653. [Google Scholar] [CrossRef] [Green Version]
- Bechtold, N.; Ellis, J.; Pelletier, G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 1993, 316, 1194–1199. [Google Scholar]
- Stangeland, B.; Salehian, Z. An Improved Clearing Method for GUS Assay in Arabidopsis Endosperm and Seeds. Plant Mol. Biol. Rep. 2002, 20, 107–114. [Google Scholar] [CrossRef]
- Jaakola, L.; Pirttilä, A.; Halonen, M.; Hohtola, A. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol 2001, 19, 201–203. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [Green Version]










© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baba, A.I.; Valkai, I.; Labhane, N.M.; Koczka, L.; Andrási, N.; Klement, É.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; Fehér, A.; et al. CRK5 Protein Kinase Contributes to the Progression of Embryogenesis of Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6120. https://doi.org/10.3390/ijms20246120
Baba AI, Valkai I, Labhane NM, Koczka L, Andrási N, Klement É, Darula Z, Medzihradszky KF, Szabados L, Fehér A, et al. CRK5 Protein Kinase Contributes to the Progression of Embryogenesis of Arabidopsis thaliana. International Journal of Molecular Sciences. 2019; 20(24):6120. https://doi.org/10.3390/ijms20246120
Chicago/Turabian StyleBaba, Abu Imran, Ildikó Valkai, Nitin M. Labhane, Lilla Koczka, Norbert Andrási, Éva Klement, Zsuzsanna Darula, Katalin F. Medzihradszky, László Szabados, Attila Fehér, and et al. 2019. "CRK5 Protein Kinase Contributes to the Progression of Embryogenesis of Arabidopsis thaliana" International Journal of Molecular Sciences 20, no. 24: 6120. https://doi.org/10.3390/ijms20246120
APA StyleBaba, A. I., Valkai, I., Labhane, N. M., Koczka, L., Andrási, N., Klement, É., Darula, Z., Medzihradszky, K. F., Szabados, L., Fehér, A., Rigó, G., & Cséplő, Á. (2019). CRK5 Protein Kinase Contributes to the Progression of Embryogenesis of Arabidopsis thaliana. International Journal of Molecular Sciences, 20(24), 6120. https://doi.org/10.3390/ijms20246120