A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana
Abstract
:1. Introduction
2. Molecular Regulation of Phase Change
2.1. miR156 As a Key Regulator
2.2. Regulation of miR156 by Endogenous Cues
2.2.1. Fine-Tuning Regulation of miR156 by GCT/CCT
2.2.2. Sugars Repress Expression of miR156 Genes
2.2.3. Epigenetic Regulation of miR156 Genes
3. The miR156-SPL Act as a Regulatory Hub
3.1. miR156-SPL Regulates Developmental Process
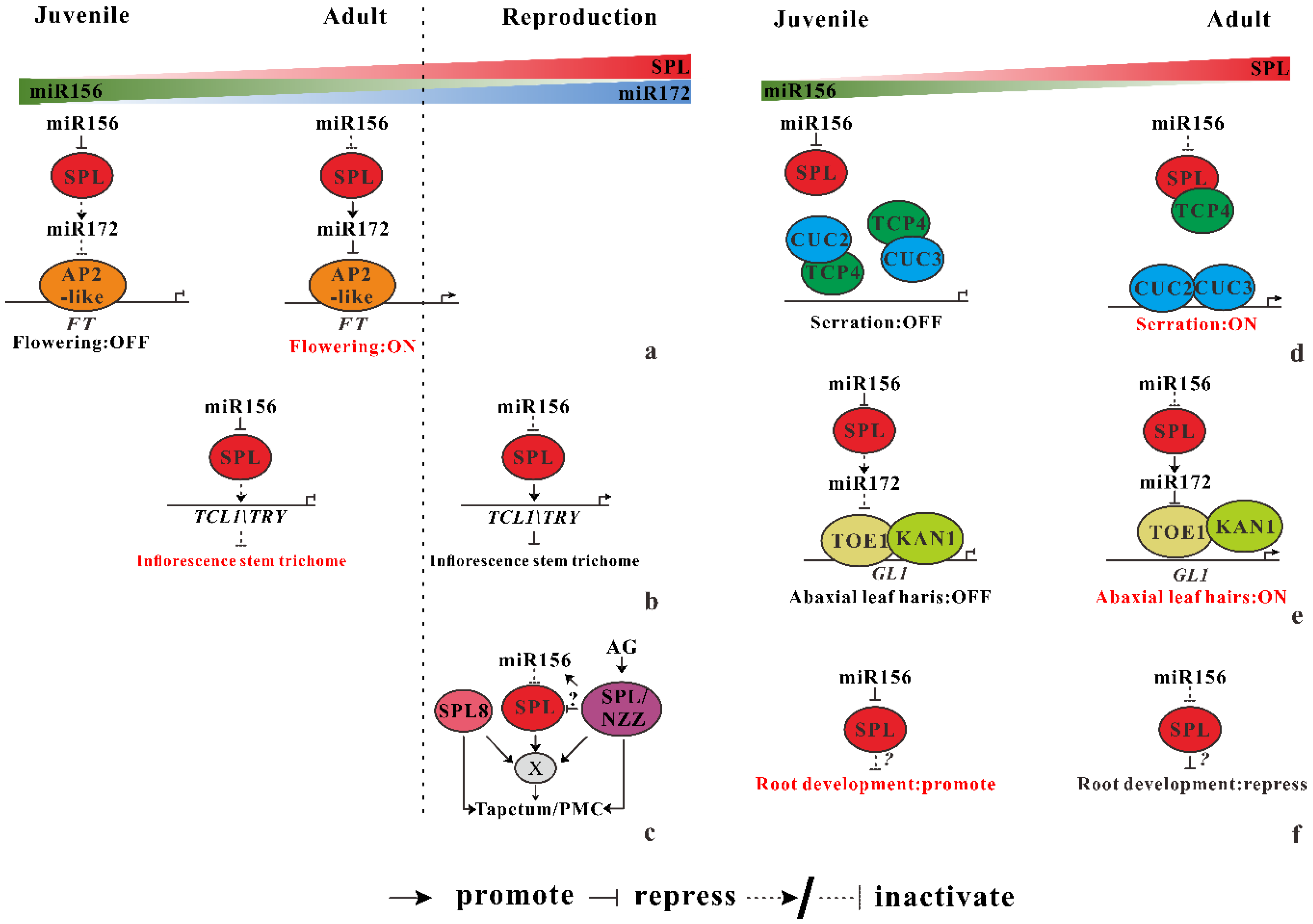
3.1.1. miR156-SPL Regulates Flowering Time and Reproductive Organ Development
miR156-SPL Regulates Flowering Time
miR156 Regulate the Distribution of Trichomes on the Inflorescence Stem
miR156 Secure Male Fertility
3.1.2. miR156 Mediate Heteroblastic Change of Leaf Morphology
miR156-miR319-miR164 Coregulate Leaf Morphological Change
miR156 Mediate the Leaf Hairs Initiation
3.1.3. miR156 Regulate Root Development
3.2. miR156-SPLs Regulates Response to Environment Stress
3.2.1. Inducing of miR156 in Response to Abiotic Stress
3.2.2. miR156 Participate in Defense against Invading Pathogens
4. A Big Portrayal of miR156 Regulation by Deep Sequencing
4.1. The miR156-SPL Module Linking Multiple Pathways
4.2. Inference of miR156-miRNAs Regulate Network in Arabidopsis
4.3. Limitation of HTS for Idenftied Function of miR156
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Poethig, R.S. Vegetative phase change and shoot maturation in plants. In Current Topics in Developmental Biology; Academic Press Inc.: New York, NY, USA, 2013; Volume 105, pp. 125–152. [Google Scholar]
- Fouracre, J.P.; Poethig, R.S. The role of small RNAs in vegetative shoot development. Curr. Opin. Plant Biol. 2016, 29, 64–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Lian, H.; Wang, J.W. Plant developmental transitions: The role of microRNAs and sugars. Curr. Opin. Plant Biol. 2015, 27, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shikata, M.; Yamaguchi, H.; Sasaki, K.; Ohtsubo, N. Overexpression of Arabidopsis miR157b induces bushy architecture and delayed phase transition in Torenia fournieri. Planta 2012, 236, 1027–1035. [Google Scholar] [CrossRef]
- Wang, J.-W.; Park, M.Y.; Wang, L.-J.; Koo, Y.; Chen, X.-Y.; Weigel, D.; Poethig, R.S. MiRNA control of vegetative phase change in trees. PLoS Genet. 2011, 7, e1002012. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [Green Version]
- Fu, C.; Sunkar, R.; Zhou, C.; Shen, H.; Zhang, J.-Y.; Matts, J.; Wolf, J.; Mann, D.G.J.; Stewart, C.N.; Tang, Y.; et al. Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 2012, 10, 443–452. [Google Scholar] [CrossRef] [Green Version]
- Pegler, J.; Oultram, J.; Grof, C.; Eamens, A. Profiling the abiotic stress responsive microRNA Landscape of Arabidopsis thaliana. Plants 2019, 8, 58. [Google Scholar] [CrossRef] [Green Version]
- Ishka, M.R.; Brown, E.; Weigand, C.; Tillett, R.L.; Schlauch, K.A.; Miller, G.; Harper, J.F. A comparison of heat-stress transcriptome changes between wild-type Arabidopsis pollen and a heat-sensitive mutant harboring a knockout of cyclic nucleotide-gated cation channel 16 (cngc16). BMC Genom. 2018, 19, 549. [Google Scholar]
- Wu, C.; Li, X.; Guo, S.; Wong, S.M. Analyses of RNA-Seq and sRNA-Seq data reveal a complex network of anti-viral defense in TCV-infected Arabidopsis thaliana. Sci. Rep. 2016, 6, 36007. [Google Scholar] [CrossRef]
- May, P.; Liao, W.; Wu, Y.; Shuai, B.; McCombie, W.R.; Zhang, M.Q.; Liu, Q.A. The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat. Commun. 2013, 4, 2145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y. MIR156 Participates in Iron Homeostasis Through Targeting SPL9/SPL15 in Arabidopsis Thaliana. Master’s Thesis, The University of Western Ontario, London, ON, Canada, 2015. [Google Scholar]
- Zhang, L.; Hu, Y.-B.; Wang, H.-S.; Feng, S.-J.; Zhang, Y.-T. Involvement of miR156 in the regulation of vegetative phase change in plants. J. Am. Soc. Hortic. Sci. 2015, 140, 387–395. [Google Scholar] [CrossRef] [Green Version]
- Pant, B.D.; Musialak-Lange, M.; Nuc, P.; May, P.; Buhtz, A.; Kehr, J.; Walther, D.; Scheible, W.R. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009, 150, 1541–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Street, N.R.; Scofield, D.G.; Ingvarsson, P.K. Variation in linked selection and recombination drive genomic divergence during allopatric speciation of European and American aspens. Mol. Biol. Evol. 2016, 33, 1754–1767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, L.-C.; Lin, S.-I.; Shih, A.C.-C.; Chen, J.-W.; Lin, W.-Y.; Tseng, C.-Y.; Li, W.-H.; Chiou, T.-J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armenta-Medina, A.; Lepe-Soltero, D.; Xiang, D.; Datla, R.; Abreu-Goodger, C.; Gillmor, C.S. Arabidopsis thaliana miRNAs promote embryo pattern formation beginning in the zygote. Dev. Biol. 2017, 431, 145–151. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miRr156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Li, C.; Zhou, C.M.; Zhang, T.Q.; Lian, H.; Sun, Y.; Wu, J.; Huang, J.; Wang, G.; Wang, J.W. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. Elife 2013, 2, e00269. [Google Scholar] [CrossRef]
- Yang, L.; Xu, M.; Koo, Y.; He, J.; Poethig, R.S. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife 2013, 2, e00260. [Google Scholar] [CrossRef]
- Gillmor, C.S.; Silva-Ortega, C.O.; Willmann, M.R.; Buendía-Monreal, M.; Poethig, R.S. The Arabidopsis Mediator CDK8 module genes CCT (MED12) and GCT (MED13) are global regulators of developmental phase transitions. Development 2014, 141, 4580–4589. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Hu, T.; Smith, M.R.; Poethig, R.S. Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 2016, 28, 28–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Guo, C.; Zhou, B.; Li, C.; Wang, H.; Zheng, B.; Ding, H.; Zhu, Z.; Peragine, A.; Cui, Y.; et al. Regulation of vegetative phase change by SWI2/SNF2 chromatin remodeling ATPase BRAHMA. Plant Physiol. 2016, 172, 2416–2428. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Leichty, A.R.; Hu, T.; Poethig, R.S. H2A.Z promotes the transcription of MIR156A and MIR156C in Arabidopsis by facilitating the deposition of h3k4me3. Development 2018, 145, dev152868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, K.; Kim, J.; Müller, S.Y.; Oh, M.; Underwood, C.; Henderson, I.; Lee, I. Regulation of microRNA-mediated developmental changes by the SWR1 chromatin remodeling complex. Plant Physiol. 2016, 171, 1128–1143. [Google Scholar] [PubMed] [Green Version]
- Weigel, D.; Meyerowitz, E.M. Activation of floral homeotic genes in Arabidopsis. Science 1993, 261, 1723–1726. [Google Scholar] [CrossRef]
- Huijser, P.; Schmid, M. The control of developmental phase transitions in plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.W. The multifaceted roles of miR156-targeted SPL transcription factors in plant developmental transitions. In Plant Transcription Factors: Evolutionary, Structural and Functional Aspects; Elsevier Inc.: Amsterdam, The Netherlands, 2015; pp. 281–293. ISBN 9780128011270. [Google Scholar]
- Yu, N.; Cai, W.J.; Wang, S.; Shan, C.M.; Wang, L.J.; Chena, X.Y. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 2010, 22, 2322–2335. [Google Scholar] [CrossRef] [Green Version]
- Xing, S.; Salinas, M.; Höhmann, S.; Berndtgen, R.; Huijser, P. miR156-targeted and non-targeted SBP-Box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef] [Green Version]
- Rubio-Somoza, I.; Zhou, C.M.; Confraria, A.; Martinho, C.; von Born, P.; Baena-Gonzalez, E.; Wang, J.W.; Weigel, D. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 2014, 24, 2714–2719. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Qian, Z.; Zhou, B.; Wu, G. Age-dependent heteroblastic development of leaf hairs in Arabidopsis. New Phytol. 2019, 224, 741–748. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, C.; Mai, Y.; Li, L.; Gao, J.; Shang, G.; Lian, H.; Han, L.; Zhang, T.; Tang, H.; et al. A spatiotemporally regulated transcriptional complex underlies heteroblastic development of leaf hairs in Arabidopsis thaliana. EMBO J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.S.; Xie, Q.; Fei, J.F.; Chua, N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chuaa, N.H. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, N.; Niu, Q.-W.; Ng, K.-H.; Chua, N.-H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015, 83, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.B.; Liu, Y.Q.; Chen, D.Y.; Chen, F.Y.; Fang, X.; Hong, G.J.; Wang, L.J.; Wang, J.W.; Chen, X.Y. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 2017, 8, 13925. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.-G.; Shan, J.-X.; Shi, M.; Gao, J.-P.; Lin, H.-X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Price, P.W. The plant vigor hypothesis and herbivore attack. Oikos 1991, 62, 244. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.P.; Li, J.; Deng, X.W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [Green Version]
- Schiltz, S.; Munier-Jolain, N.; Jeudy, C.; Burstin, J.; Salon, C. Dynamics of exogenous nitrogen partitioning and nitrogen remobilization from vegetative organs in pea revealed by 15N in vivo labeling throughout seed filling. Proc. Plant Physiol. 2005, 137, 1463–1473. [Google Scholar] [CrossRef] [Green Version]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, G.; He, H.; Yu, D. Identification of nitrogen starvation-responsive MicroRNAs in Arabidopsis thaliana. PLoS ONE 2012, 7, e48951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nodine, M.D.; Bartel, D.P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010, 24, 2678–2692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thatcher, S.R.; Burd, S.; Wright, C.; Lers, A.; Green, P.J. Differential expression of miRNAs and their target genes in senescing leaves and siliques: Insights from deep sequencing of small RNAs and cleaved target RNAs. Plant Cell Environ. 2015, 38, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, A.; Kellner, M.J.; Schon, M.A.; Mosiolek, M.; Nodine, M.D. MicroRNA dynamics and functions during Arabidopsis embryogenesis. Plant Cell 2019. [Google Scholar] [CrossRef]
- Mizzotti, C.; Rotasperti, L.; Moretto, M.; Tadini, L.; Resentini, F.; Galliani, B.M.; Galbiati, M.; Engelen, K.; Pesaresi, P.; Masiero, S. Time-course transcriptome analysis of Arabidopsis siliques discloses genes essential for fruit development and maturation. Plant Physiol. 2018, 178, 1249–1268. [Google Scholar] [CrossRef] [Green Version]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef]



| Biological Process | Conserved miRNAs | Novel miRNAs | Expression Pattern | Target Genes | Correlated miRNAs | References |
|---|---|---|---|---|---|---|
| Drought stress | 123 | NA | Up | SPL9 | 2 | [9] |
| Heat stress | 6 (5 families) | NA | Up | NA | 4 | [10] |
| Viral defense | 30 (17 families) | 29 | Down | SPLs * | 15 | [11] |
| Elevated CO2 content | 18 (8 families) | 1 | Down | SPL10 | 4 | [12] |
| Elevated temperature | 36 (14 families) | 4 | Up | SPL2 SPL3 SPL11 SPL13 SPL15 | 6 | [12] |
| Phosphaste deficiency | 55 (23 families) | NA | Up (root) | SPLs * GAI | 9 | [17] |
| Nitrogen starvation | 41 (34 families) | 9 | Up | SPLs * GAI | 10 | [45] |
| 20 (8 families) | NA | Up | NA | 6 | [15] | |
| Iron homeostasis | NA | NA | Up | SPL9 SPL15 | NA | [13] |
| Embryogenisis | 59 (47 families) | NA | Down | SPL2 SPL10 SPL15 EMB140 PMEI | 4 | [48] |
| Embryo pattern formation | 39 (13 families) | NA | Up | SPL2 SPL3 SPL10 SPL11 | 12 | [18] |
| 15 (10 families) | NA | Up | SPL10 SPL11 | 9 | [46] | |
| Siliques development | 15 (9 families) | 5 | Up | SPL15 | 8 | [47,49] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, C.; Ye, M.; Sang, M.; Wu, R. A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6166. https://doi.org/10.3390/ijms20246166
Zheng C, Ye M, Sang M, Wu R. A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana. International Journal of Molecular Sciences. 2019; 20(24):6166. https://doi.org/10.3390/ijms20246166
Chicago/Turabian StyleZheng, Chenfei, Meixia Ye, Mengmeng Sang, and Rongling Wu. 2019. "A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana" International Journal of Molecular Sciences 20, no. 24: 6166. https://doi.org/10.3390/ijms20246166
APA StyleZheng, C., Ye, M., Sang, M., & Wu, R. (2019). A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana. International Journal of Molecular Sciences, 20(24), 6166. https://doi.org/10.3390/ijms20246166





