Intra- and Inter-Tumor BRAF Heterogeneity in Acral Melanoma: An Immunohistochemical Analysis
Abstract
1. Introduction
2. Results
2.1. Patient Data
2.2. Consistency between IHC and RT-PCR in BRAFV600E Status in Acral Melanoma
2.3. Results of IHC Staining
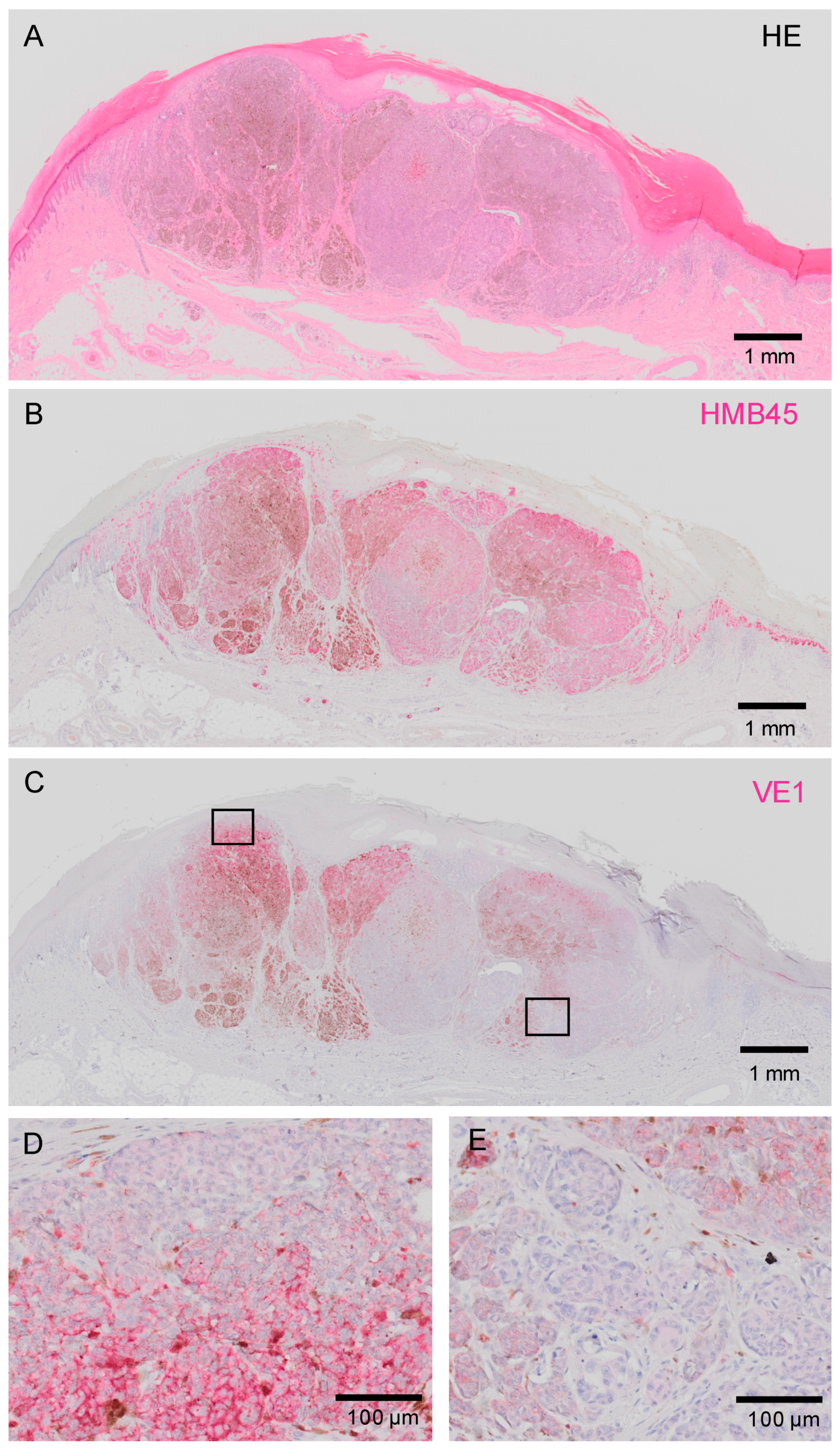
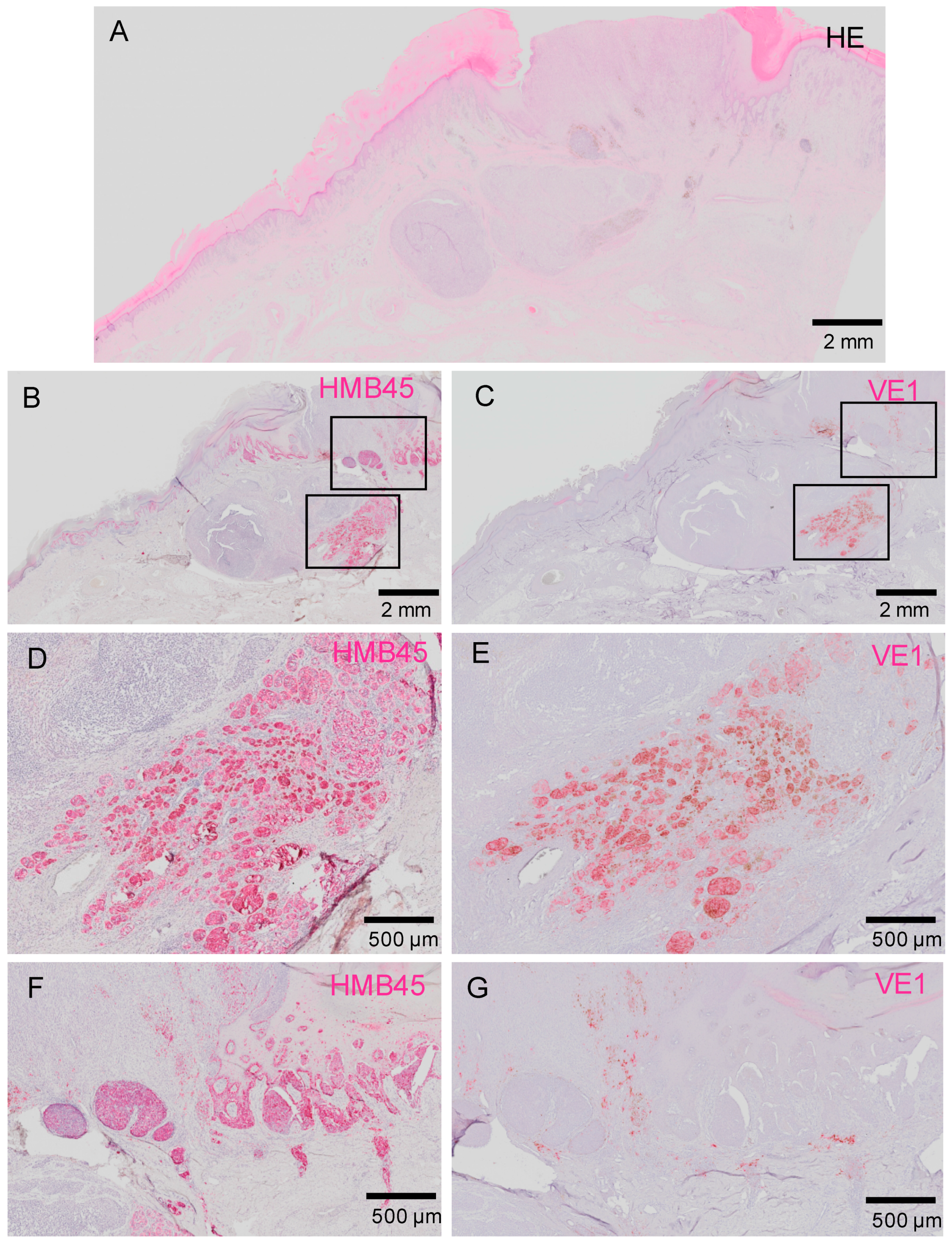
2.3.1. Intra-Tumor Heterogeneity in BRAFV600E Status in Acral Melanoma
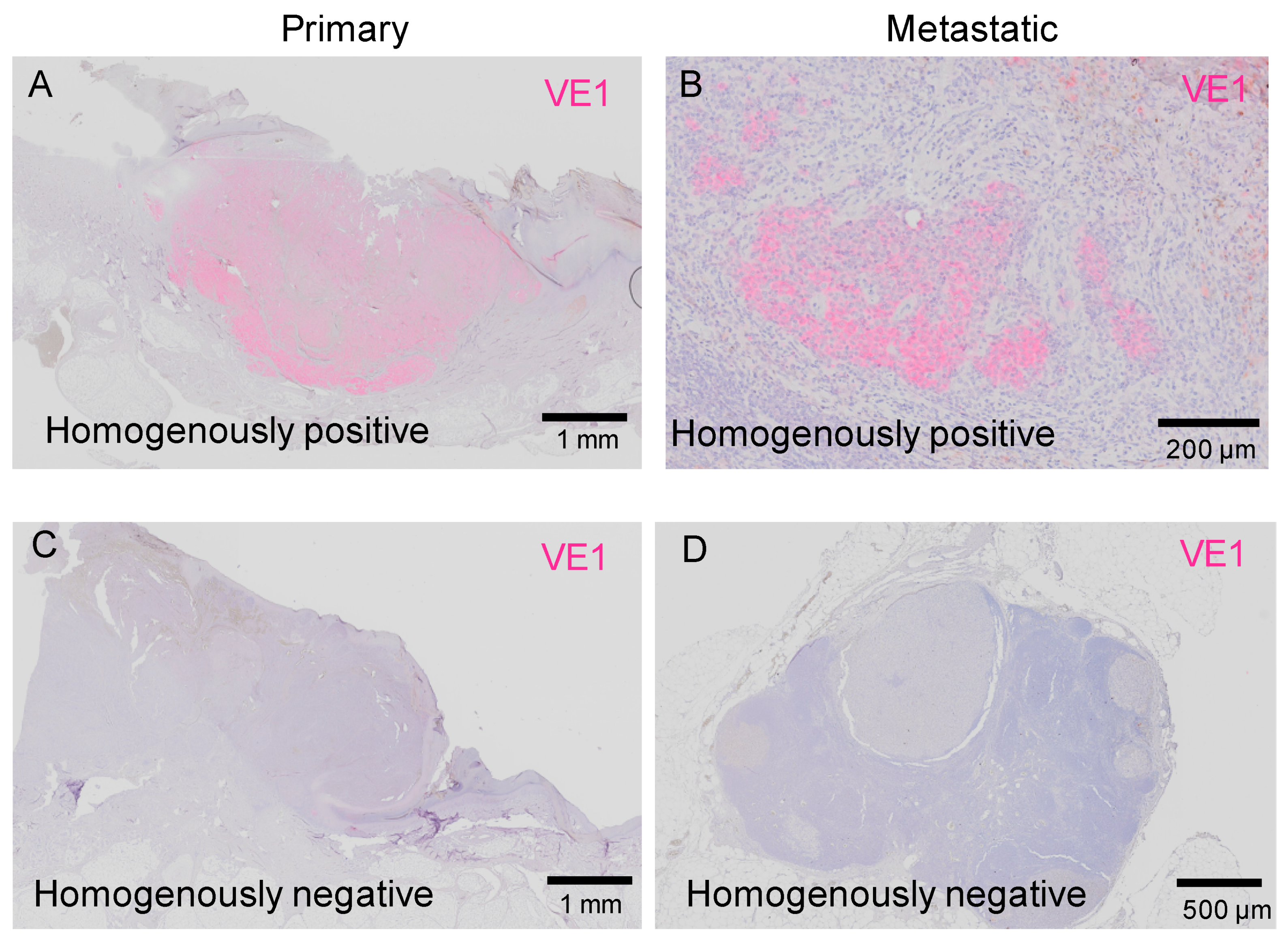
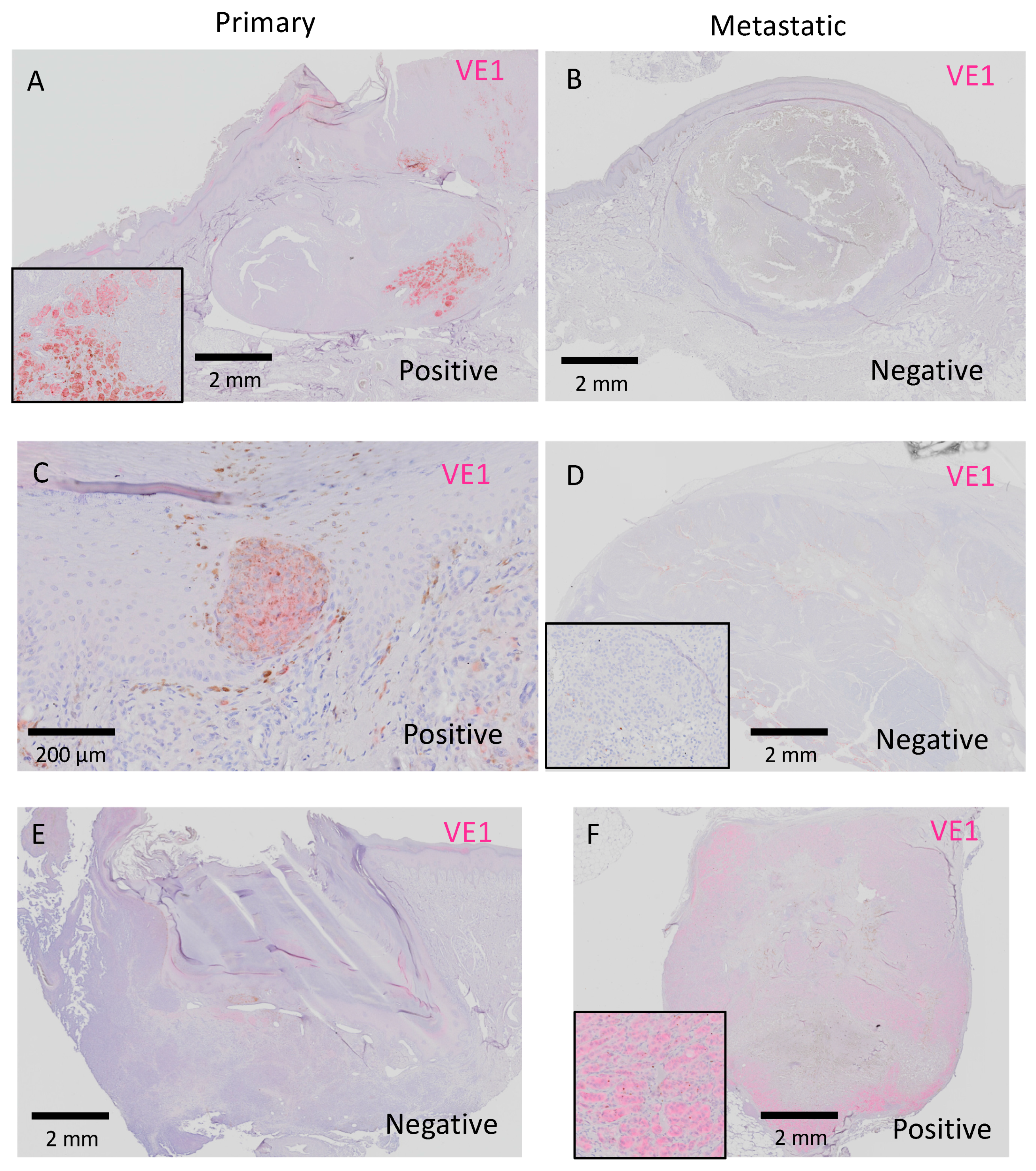
2.3.2. Inter-Tumor Heterogeneity of BRAFV600E in Acral Melanoma
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Patients and Tissue Samples
4.3. Immunohistochemistry
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MAPK | mitogen-activated protein-kinase |
| RT-PCR | real-time PCR |
| IHC | immunohistochemistry |
| FFPE | formalin-fixed paraffin-embedded |
References
- Siegel, R.; Ward, E.; Brawley, O.; Jemal, A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011, 61, 212–236. [Google Scholar] [CrossRef] [PubMed]
- Kohler, B.A.; Ward, E.; McCarthy, B.J.; Schymura, M.J.; Ries, L.A.; Eheman, C.; Jemal, A.; Anderson, R.N.; Ajani, U.A.; Edwards, B.K. Annual report to the nation on the status of cancer, 1975–2007, featuring tumors of the brain and other nervous system. J. Natl. Cancer Inst. 2011, 103, 714–736. [Google Scholar] [CrossRef] [PubMed]
- Sosman, J.A.; Kim, K.B.; Schuchter, L.; Gonzalez, R.; Pavlick, A.C.; Weber, J.S.; McArthur, G.A.; Hutson, T.E.; Moschos, S.J.; Flaherty, K.T.; et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 2012, 366, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.T.; Robert, C.; Hersey, P.; Nathan, P.; Garbe, C.; Milhem, M.; Demidov, L.V.; Hassel, J.C.; Rutkowski, P.; Mohr, P.; et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 2012, 367, 107–114. [Google Scholar] [CrossRef]
- Hauschild, A.; Grob, J.J.; Demidov, L.V.; Jouary, T.; Gutzmer, R.; Millward, M.; Rutkowski, P.; Blank, C.U.; Miller, W.H., Jr.; Kaempgen, E.; et al. Dabrafenib in BRAF-mutated metastatic melanoma: A multicentre, open-label, phase 3 randomised controlled trial. Lancet 2012, 380, 358–365. [Google Scholar] [CrossRef]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Fecher, L.A.; Amaravadi, R.K.; Flaherty, K.T. The MAPK pathway in melanoma. Curr. Opin. Oncol. 2008, 20, 183–189. [Google Scholar] [CrossRef]
- Coit, D.G.; Thompson, J.A.; Albertini, M.R.; Barker, C.; Carson, W.E.; Contreras, C.P.; Daniels, G.A.; DiMaio, D.; Fields, R.C.; Fleming, M.D.; et al. Cutaneous melanoma, version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2019, 17, 367–402. [Google Scholar] [CrossRef]
- Curry, J.L.; Torres-Cabala, C.A.; Tetzlaff, M.T.; Bowman, C.; Prieto, V.G. Molecular platforms utilized to detect BRAF V600E mutation in melanoma. Semin. Cutan. Med. Surg. 2012, 31, 267–273. [Google Scholar] [CrossRef]
- Ihle, M.A.; Fassunke, J.; König, K.; Grünewald, I.; Schlaak, M.; Kreuzberg, N.; Tietze, L.; Schildhaus, H.U.; Büttner, R.; Merkelbach-Bruse, S. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer 2014, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Skorokhod, A.; Capper, D.; von Deimling, A.; Enk, A.; Helmbold, P. Detection of BRAF V600E mutations in skin metastases of malignant melanoma by monoclonal antibody VE1. J. Am. Acad. Dermatol. 2012, 67, 488–491. [Google Scholar] [CrossRef] [PubMed]
- Capper, D.; Preusser, M.; Habel, A.; Sahm, F.; Ackermann, U.; Schindler, G.; Pusch, S.; Mechtersheimer, G.; Zentgraf, H.; von Deimling, A. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011, 122, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Yancovitz, M.; Litterman, A.; Yoon, J.; Ng, E.; Shapiro, R.L.; Berman, R.S.; Pavlick, A.C.; Darvishian, F.; Christos, P.; Mazumdar, M.; et al. Intra- and inter-tumor heterogeneity of BRAF(V600E) mutations [sic] in primary and metastatic melanoma. PLoS ONE 2012, 7, e29336. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Goto, Y.; Murata, H.; Sakaizawa, K.; Uchiyama, A.; Saida, T.; Takata, M. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. Br. J. Cancer 2011, 104, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Valachis, A.; Ullenhag, G.J. Discrepancy in BRAF status among patients with metastatic malignant melanoma: A meta-analysis. Eur. J. Cancer 2017, 81, 106–115. [Google Scholar] [CrossRef]
- Verlinden, I.; van den Hurk, K.; Clarijs, R.; Willig, A.P.; Stallinga, C.M.; Roemen, G.M.; van den Oord, J.J.; zur Hausen, A.; Speel, E.J.; Winnepenninckx, V.J. BRAFV600E immunopositive melanomas show low frequency of heterogeneity and association with epithelioid tumor cells: A STROBE-compliant article. Medicine (Baltimore) 2014, 93, e285. [Google Scholar] [CrossRef]
- Wada, M.; Ito, T.; Tsuji, G.; Nakahara, T.; Hagihara, A.; Furue, M.; Uchi, H. Acral lentiginous melanoma versus other melanoma: A single-center analysis in Japan. J. Dermatol. 2017, 44, 932–938. [Google Scholar] [CrossRef]
- Ito, T.; Wada, M.; Nagae, K.; Nakano-Nakamura, M.; Nakahara, T.; Hagihara, A.; Furue, M.; Uchi, H. Acral lentiginous melanoma: Who benefits from sentinel lymph node biopsy? J. Am. Acad. Dermatol. 2015, 72, 71–77. [Google Scholar] [CrossRef]
- Ito, T.; Wada, M.; Nagae, K.; Nakano-Nakamura, M.; Nakahara, T.; Hagihara, A.; Furue, M.; Uchi, H. Triple-marker PCR assay of sentinel lymph node as a prognostic factor in melanoma. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Sakaizawa, K.; Ashida, A.; Uchiyama, A.; Ito, T.; Fujisawa, Y.; Ogata, D.; Matsushita, S.; Fujii, K.; Fukushima, S.; Shibayama, Y.; et al. Clinical characteristics associated with BRAF, NRAS and KIT mutations in Japanese melanoma patients. J. Dermatol. Sci. 2015, 80, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Minagawa, A.; Omodaka, T.; Okuyama, R. Melanomas and mechanical stress points on the plantar surface of the foot. N. Engl. J. Med. 2016, 374, 2404–2406. [Google Scholar] [CrossRef] [PubMed]
- Wada-Ohno, M.; Ito, T.; Furue, M. Adjuvant herapy for nelanoma. Curr. Treat. Options Oncol. 2019, 20, 63. [Google Scholar] [CrossRef]
- Furue, M.; Ito, T.; Wada, N.; Wada, M.; Kadono, T.; Uchi, H. Melanoma and immune checkpoint inhibitors. Curr. Oncol. Rep. 2018, 20, 29. [Google Scholar] [CrossRef]
- Yaman, B.; Kandiloğlu, G.; Akalin, T. BRAF-V600 mutation heterogeneity in primary and metastatic melanoma: A study with pyrosequencing and immunohistochemistry. Am. J. Dermatopathol. 2016, 2, 113–120. [Google Scholar] [CrossRef]
- Manfredi, L.; Meyer, N.; Tournier, E.; Grand, D.; Uro-Coste, E.; Rochaix, P.; Brousset, P.; Lamant, L. Highly concordant results between immunohistochemistry and molecular testing of mutated V600E BRAF in primary and metastatic melanoma. Acta Derm. Venereol. 2016, 96, 630–634. [Google Scholar] [CrossRef]
- Eriksson, H.; Zebary, A.; Vassilaki, I.; Omholt, K.; Ghaderi, M.; Hansson, J. BRAFV600E protein expression in primary cutaneous malignant melanomas and paired metastases. JAMA Dermatol. 2015, 151, 410–416. [Google Scholar] [CrossRef]
- Riveiro-Falkenbach, E.; Villanueva, C.A.; Garrido, M.C.; Ruano, Y.; García-Martín, R.M.; Godoy, E.; Ortiz-Romero, P.L.; Ríos-Martín, J.J.; Santos-Briz, A.; Rodríguez-Peralto, J.L. Intra- and inter-tumoral homogeneity of BRAF(V600E) mutations in melanoma tumors. J. Investig. Dermatol. 2015, 135, 3078–3085. [Google Scholar] [CrossRef]
- Thiel, A.; Moza, M.; Kytölä, S.; Orpana, A.; Jahkola, T.; Hernberg, M.; Virolainen, S.; Ristimäki, A. Prospective immunohistochemical analysis of BRAF V600E mutation in melanoma. Hum. Pathol. 2015, 46, 169–175. [Google Scholar] [CrossRef]
- Fisher, K.E.; Cohen, C.; Siddiqui, M.T.; Palma, J.F.; Lipford, E.H., 3rd; Longshore, J.W. Accurate detection of BRAF p.V600E mutations in challenging melanoma specimens requires stringent immunohistochemistry scoring criteria or sensitive molecular assays. Hum. Pathol. 2014, 45, 2281–2293. [Google Scholar] [CrossRef] [PubMed]
- Saroufim, M.; Habib, R.H.; Gerges, R.; Saab, J.; Loya, A.; Amr, S.S.; Sheikh, S.; Satti, M.; Oberkanins, C.; Khalifeh, I. Comparing BRAF mutation status in matched primary and metastatic cutaneous melanomas: Implications on optimized targeted therapy. Exp. Mol. Pathol. 2014, 97, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Menzies, A.M.; Lum, T.; Wilmott, J.S.; Hyman, J.; Kefford, R.F.; Thompson, J.F.; O’Toole, S.; Long, G.V.; Scolyer, R.A. Intrapatient homogeneity of BRAFV600E expression in melanoma. Am. J. Surg. Pathol. 2014, 38, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Boursault, L.; Haddad, V.; Vergier, B.; Cappellen, D.; Verdon, S.; Bellocq, J.P.; Jouary, T.; Merlio, J.P. Tumor homogeneity between primary and metastatic sites for BRAF status in metastatic melanoma determined by immunohistochemical and molecular testing. PLoS ONE. 2013, 8, e70826. [Google Scholar] [CrossRef]
- Kaji, T.; Yamasaki, O.; Takata, M.; Otsuka, M.; Hamada, T.; Morizane, S.; Asagoe, K.; Yanai, H.; Hirai, Y.; Umemura, H.; et al. Comparative study on driver mutations in primary and metastatic melanomas at a single Japanese institute: A clue for intra- and inter-tumor heterogeneity. J. Dermatol. Sci. 2017, 85, 51–57. [Google Scholar] [CrossRef]
- Zebary, A.; Omholt, K.; Vassilaki, I.; Höiom, V.; Lindén, D.; Viberg, L.; Kanter-Lewensohn, L.; Johansson, C.H.; Hansson, J. KIT, NRAS, BRAF and PTEN mutations in a sample of Swedish patients with acral lentiginous melanoma. J. Dermatol. Sci. 2013, 72, 284–289. [Google Scholar] [CrossRef]
- Busam, K.J.; Hedvat, C.; Pulitzer, M.; von Deimling, A.; Jungbluth, A.A. Immunohistochemical analysis of BRAF(V600E) expression of primary and metastatic melanoma and comparison with mutation status and melanocyte differentiation antigens of metastatic lesions. Am. J. Surg. Pathol. 2013, 37, 413–420. [Google Scholar] [CrossRef]
- Long, G.V.; Wilmott, J.S.; Capper, D.; Preusser, M.; Zhang, Y.E.; Thompson, J.F.; Kefford, R.F.; von Deimling, A.; Scolyer, R.A. Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. Am. J. Surg. Pathol. 2013, 37, 61–65. [Google Scholar] [CrossRef]
- Colomba, E.; Hélias-Rodzewicz, Z.; Von Deimling, A.; Marin, C.; Terrones, N.; Pechaud, D.; Surel, S.; Côté, J.F.; Peschaud, F.; Capper, D.; et al. Detection of BRAF p.V600E mutations in melanomas: Comparison of four methods argues for sequential use of immunohistochemistry and pyrosequencing. J. Mol. Diagn. 2013, 15, 94–100. [Google Scholar] [CrossRef]
- Ito, T.; Kohashi, K.; Yamada, Y.; Iwasaki, T.; Maekawa, A.; Kuda, M.; Hoshina, D.; Abe, R.; Furue, M.; Oda, Y. Prognostic significance of forkhead box M1 (FOXM1) expression and antitumor effect of FOXM1 inhibition in angiosarcoma. J. Cancer 2016, 7, 823–830. [Google Scholar] [CrossRef]
- Ito, T.; Kohashi, K.; Yamada, Y.; Maekawa, A.; Kuda, M.; Furue, M.; Oda, Y. Prognostic significance of forkhead box M1 (FoxM1) expression and antitumour effect of FoxM1 inhibition in melanoma. Histopathology 2016, 69, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Tsuji, G.; Ohno, F.; Uchi, H.; Nakahara, T.; Hashimoto-Hachiya, A.; Yoshida, Y.; Yamamoto, O.; Oda, Y.; Furue, M. Activation of the OVOL1-OVOL2 axis in the hair bulb and in pilomatricoma. Am. J. Pathol. 2016, 186, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Tsuji, G.; Ohno, F.; Nakahara, T.; Uchi, H.; Furue, M. Potential role of the OVOL1-OVOL2 axis and c-Myc in the progression of cutaneous squamous cell carcinoma. Mod. Pathol. 2017, 30, 919–927. [Google Scholar] [CrossRef] [PubMed]

| Parameters | Number (%) |
|---|---|
| Age in years | |
| Range (mean ± SD) | 32–88 (69.8 ± 12.3) |
| Sex | |
| Male | 15 (48.4) |
| Female | 16 (51.6) |
| Race/ethnicity | |
| Japanese | 31 (100.0) |
| Type of melanoma | |
| Acral melanoma | 31 (100.0) |
| Primary tumor site | |
| Hand | 8 (25.8) |
| Foot | 23 (74.2) |
| Site of metastasis | |
| Lymph node | 24 (77.4) |
| Skin | 6 (19.4) |
| Lung | 1 (3.3) |
| Detection method for BRAFV600E | |
| IHC | 21 (67.7) |
| IHC + real-time PCR | 10 (32.3) |
| Total | 31 (100) |
| Real-Time PCR | IHC (Proportion, Intensity) |
|---|---|
| Positive | Positive (100%, 3+) |
| Negative | Negative (3%, 3+) |
| Negative | Negative (2%, 2+) |
| Negative | Negative (3%, 1+) |
| Negative | Negative (0%, –) |
| Negative | Negative (0%, –) |
| Negative | Negative (0%, –) |
| Negative | Negative (0%, –) |
| Negative | Negative (0%, –) |
| Negative | Negative (0%, –) |
| Sensitivity, 100%; specificity, 100% | |
| Mutation Status | BRAFV600E Mutation | ||
|---|---|---|---|
| Primary Melanoma | Metastatic Melanoma | Number (%) | |
| Concordance | Positive | Positive | 7 (22.6) |
| Negative | Negative | 21 (67.7) | |
| Discordance | Positive | Negative | 2 (6.5) |
| Negative | Positive | 1 (3.2) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ito, T.; Kaku-Ito, Y.; Murata, M.; Ichiki, T.; Kuma, Y.; Tanaka, Y.; Ide, T.; Ohno, F.; Wada-Ohno, M.; Yamada, Y.; et al. Intra- and Inter-Tumor BRAF Heterogeneity in Acral Melanoma: An Immunohistochemical Analysis. Int. J. Mol. Sci. 2019, 20, 6191. https://doi.org/10.3390/ijms20246191
Ito T, Kaku-Ito Y, Murata M, Ichiki T, Kuma Y, Tanaka Y, Ide T, Ohno F, Wada-Ohno M, Yamada Y, et al. Intra- and Inter-Tumor BRAF Heterogeneity in Acral Melanoma: An Immunohistochemical Analysis. International Journal of Molecular Sciences. 2019; 20(24):6191. https://doi.org/10.3390/ijms20246191
Chicago/Turabian StyleIto, Takamichi, Yumiko Kaku-Ito, Maho Murata, Toshio Ichiki, Yuki Kuma, Yuka Tanaka, Taketoshi Ide, Fumitaka Ohno, Maiko Wada-Ohno, Yuichi Yamada, and et al. 2019. "Intra- and Inter-Tumor BRAF Heterogeneity in Acral Melanoma: An Immunohistochemical Analysis" International Journal of Molecular Sciences 20, no. 24: 6191. https://doi.org/10.3390/ijms20246191
APA StyleIto, T., Kaku-Ito, Y., Murata, M., Ichiki, T., Kuma, Y., Tanaka, Y., Ide, T., Ohno, F., Wada-Ohno, M., Yamada, Y., Oda, Y., & Furue, M. (2019). Intra- and Inter-Tumor BRAF Heterogeneity in Acral Melanoma: An Immunohistochemical Analysis. International Journal of Molecular Sciences, 20(24), 6191. https://doi.org/10.3390/ijms20246191





