Translational Regulation of Pmt1 and Pmt2 by Bfr1 Affects Unfolded Protein O-Mannosylation
Abstract
1. Introduction
2. Results
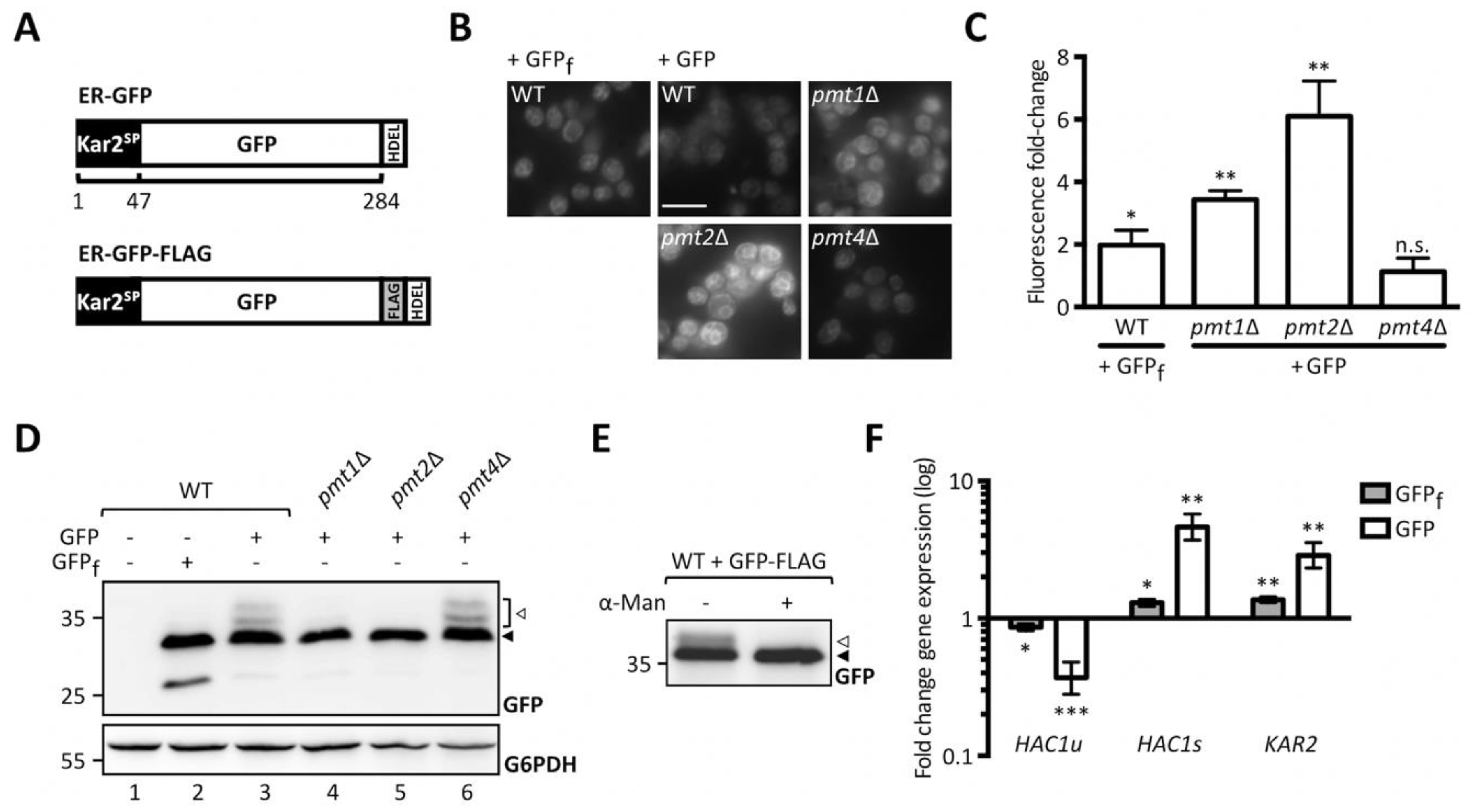
2.1. Genome-Wide Screen Reveals Bfr1 as a Factor Influencing UPOM
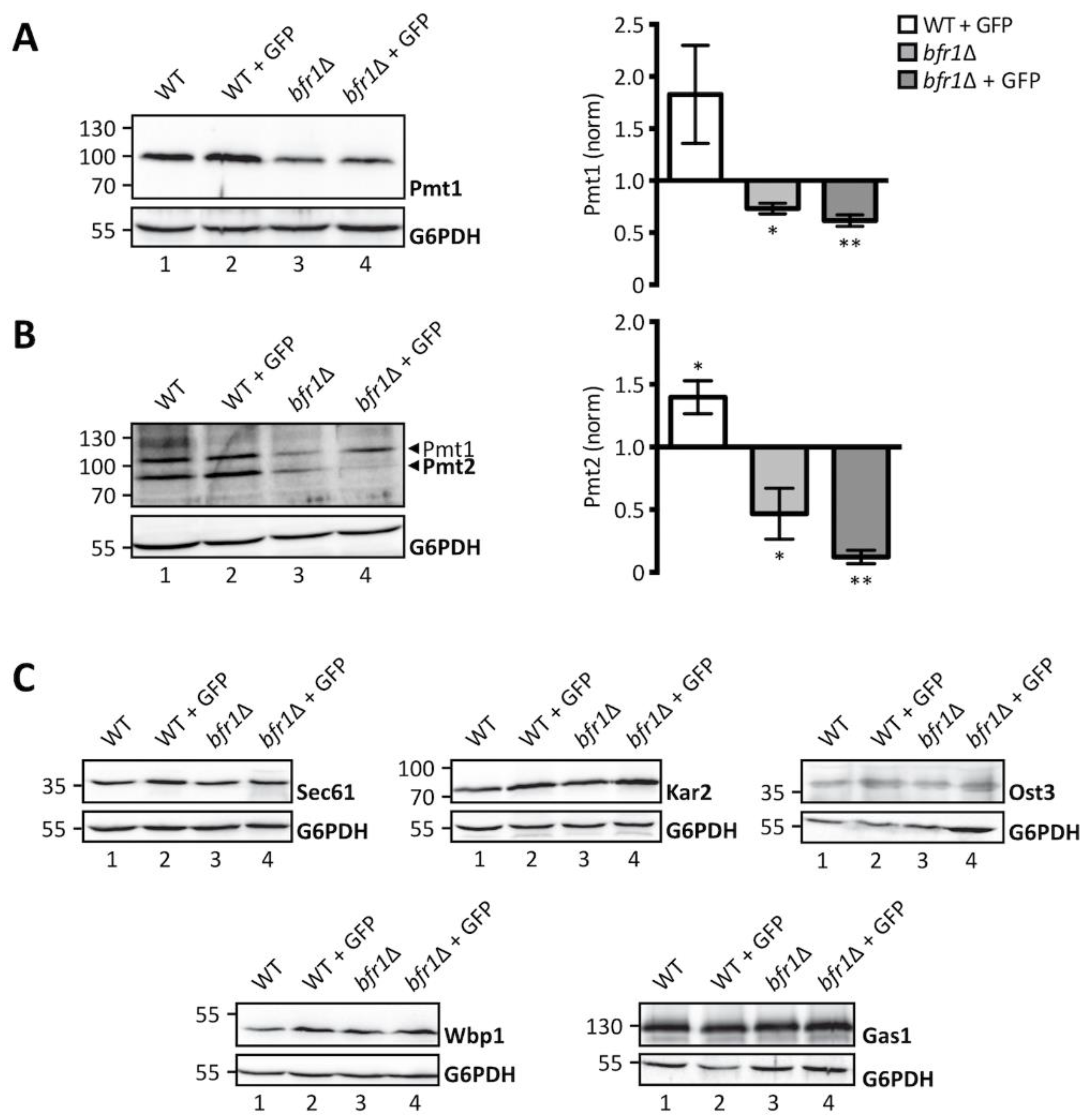
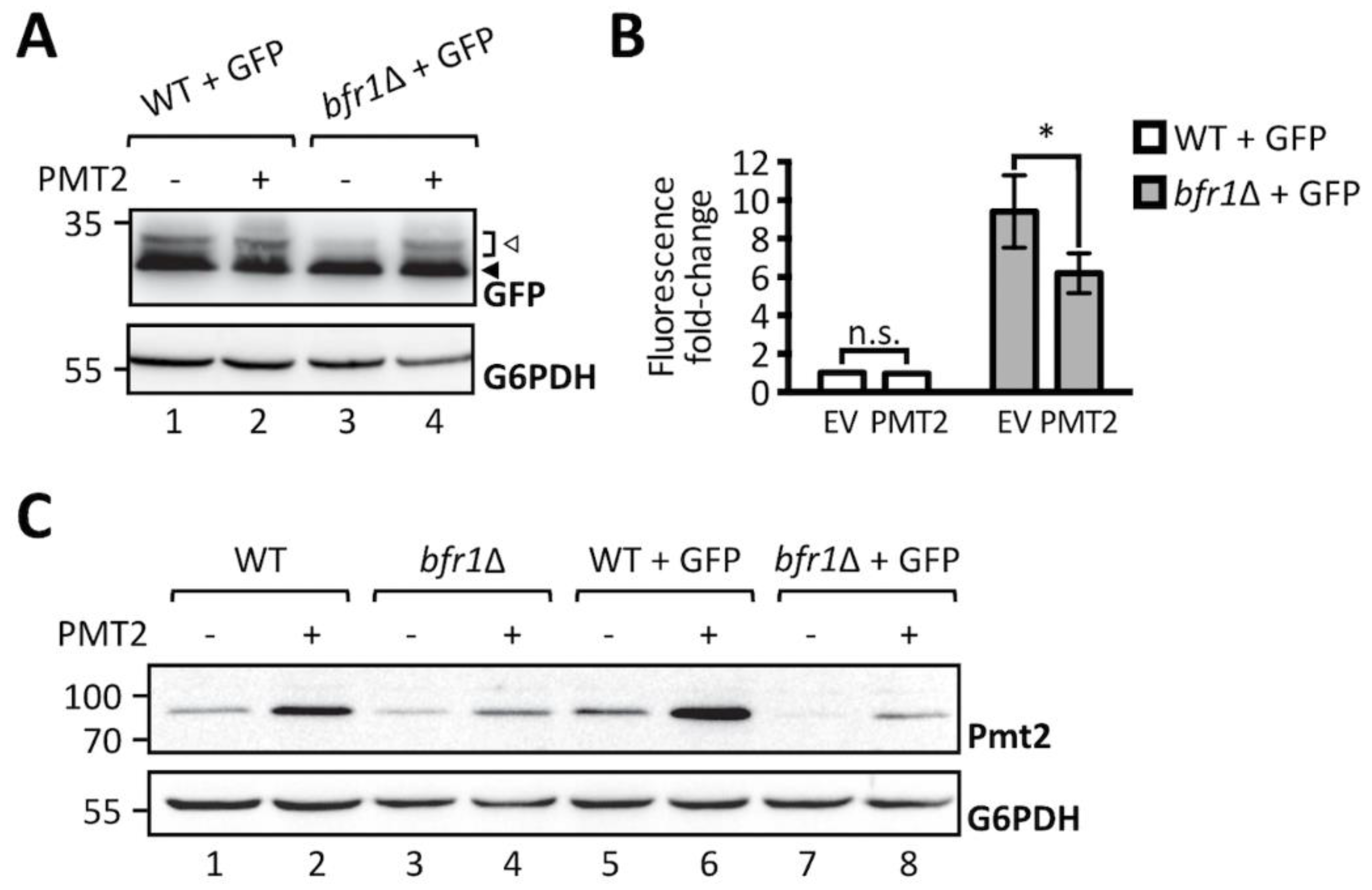
2.2. Bfr1 Affects UPOM by Modulating Pmt1 and Pmt2 Protein Levels
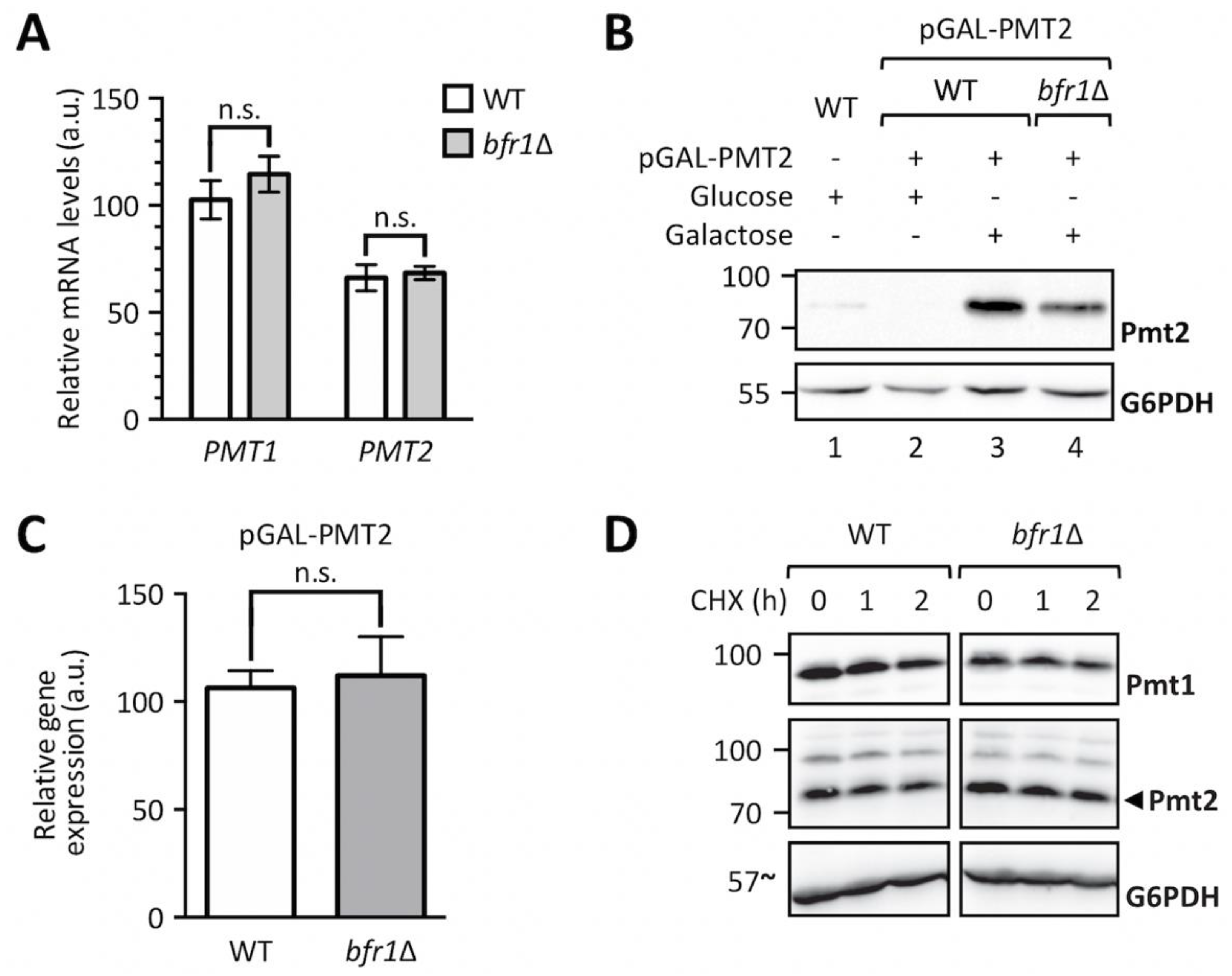
2.3. Bfr1 Affects Pmt1 and Pmt2 Protein Levels on a Posttranscriptional Level
2.4. BFR1 Deletion Does Not Affect PMT1 and PMT2 mRNA Localization to the ER
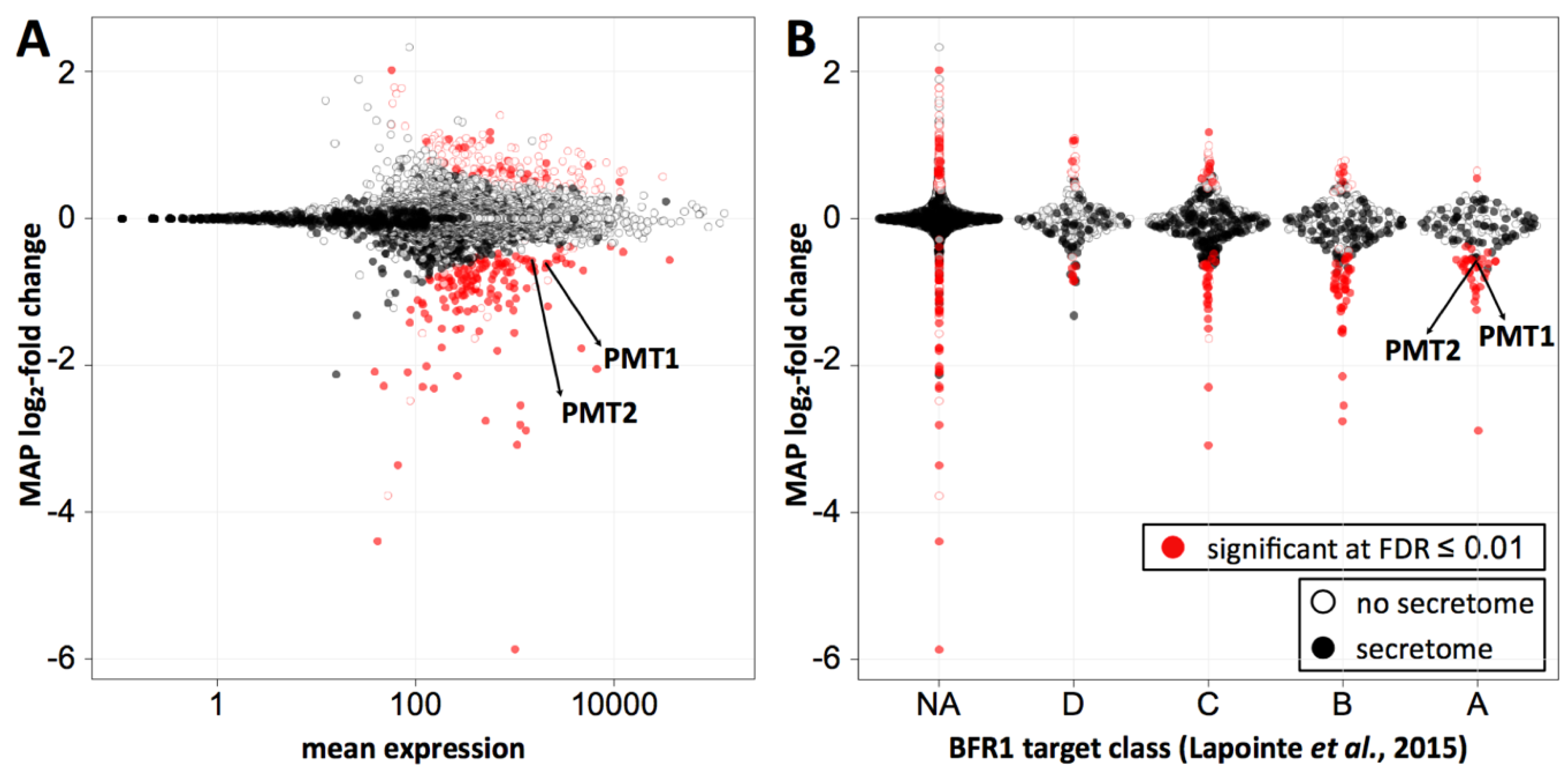
2.5. Bfr1 Affects Pmt1 and Pmt2 Translation
3. Discussion
4. Materials and Methods
4.1. Yeast Strains and Culture Conditions
4.2. Plasmids and Oligonucleotides
4.3. ER-GFP Screening
4.3.1. Automated Library Generation
4.3.2. High-Throughput Microscopy
4.3.3. Image Analysis
4.4. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.4.1. Total RNA Isolation
4.4.2. cDNA Synthesis
4.4.3. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.5. Preparation of Cell Extracts and Membranes
4.6. Flag-Tag Immunoprecipitation
4.7. Cycloheximide Chase Experiments
4.8. Western Blot Analysis
4.9. Cell Fractionation Coupled to RNA Preparation
4.9.1. Cell Fractionation by One Step Ultracentrifugation
4.9.2. Cell Fractionation by Sucrose Step Gradient Centrifugation
4.10. Flow Cytometry
4.11. Fluorescence Microscopy
4.12. Ribosome Profiling
4.12.1. Sample Preparation
4.12.2. Ribosome-Protected Footprint mRNA Extraction
4.12.3. Deep Sequencing Library Preparation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ER | endoplasmic reticulum |
| UPOM | unfolded protein O-mannosylation |
| GFP | green fluorescent protein |
| Bfr1 | brefeldin A resistance factor 1 |
| Dol-P-Man | Dol-P-mannose |
| PMT | protein O-mannosyltransferase |
| UPR | unfolded protein response |
| ERAD | ER associated degradation |
| MFI | median fluorescence intensity |
| HOG pathway | high osmolarity glycerol pathway |
| OST | oligosaccharyl transferase |
| RPM | reads per million mapped reads |
| FDR | false discovery rate |
| YPD medium | yeast extract-peptone-dextrose medium |
| SD medium | synthetic defined medium |
| RT-qPCR | real-time quantitative polymerase chain reaction |
| rep | replicate |
| TCL | total cell lysate |
| N.s. | not significant |
| NA | not assigned |
References
- Spiro, R.G. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002, 12, 43r–56r. [Google Scholar] [CrossRef] [PubMed]
- Neubert, P.; Strahl, S. Protein O-mannosylation in the early secretory pathway. Curr. Opin. Cell Biol. 2016, 41, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Strahl-Bolsinger, S.; Tanner, W. Protein O-glycosylation in Saccharomyces cerevisiae. Purification and characterization of the dolichyl-phosphate-D-mannose-protein O-D-mannosyltransferase. Eur. J. Biochem. 1991, 196, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Lussier, M.; Gentzsch, M.; Sdicu, A.M.; Bussey, H.; Tanner, W. Protein O-glycosylation in yeast. The PMT2 gene specifies a second protein O-mannosyltransferase that functions in addition to the PMT1-encoded activity. J. Biol. Chem. 1995, 270, 2770–2775. [Google Scholar] [CrossRef] [PubMed]
- Immervoll, T.; Gentzsch, M.; Tanner, W. PMT3 and PMT4, two new members of the protein-O-mannosyltransferase gene family of Saccharomyces cerevisiae. Yeast 1995, 11, 1345–1351. [Google Scholar] [CrossRef]
- Jurado, L.A.; Coloma, A.; Cruces, J. Identification of a human homolog of the Drosophila rotated abdomen gene (POMT1) encoding a putative protein O-mannosyl-transferase, and assignment to human chromosome 9q34.1. Genomics 1999, 58, 171–180. [Google Scholar] [CrossRef]
- Willer, T.; Amselgruber, W.; Deutzmann, R.; Strahl, S. Characterization of POMT2, a novel member of the PMT protein O-mannosyltransferase family specifically localized to the acrosome of mammalian spermatids. Glycobiology 2002, 12, 771–783. [Google Scholar] [CrossRef]
- Brancaccio, A. A molecular overview of the primary dystroglycanopathies. J. Cell. Mol. Med. 2019, 23, 3058–3062. [Google Scholar] [CrossRef]
- Carvalho, S.; Reis, C.A.; Pinho, S.S. Cadherins glycans in cancer: Sweet players in a bitter process. Trends Cancer 2016, 2, 519–531. [Google Scholar] [CrossRef]
- Kumari, K.; Das, B.; Adhya, A.K.; Rath, A.K.; Mishra, S.K. Genome-wide expression analysis reveals six contravened targets of EZH2 associated with breast cancer patient survival. Sci. Rep. 2019, 9, 1974. [Google Scholar] [CrossRef]
- Loibl, M.; Wunderle, L.; Hutzler, J.; Schulz, B.L.; Aebi, M.; Strahl, S. Protein O-mannosyltransferases associate with the translocon to modify translocating polypeptide chains. J. Biol. Chem. 2014, 289, 8599–8611. [Google Scholar] [CrossRef] [PubMed]
- Loibl, M.; Strahl, S. Protein O-mannosylation: What we have learned from baker’s yeast. Biochim. Biophys. Acta 2013, 1833, 2438–2446. [Google Scholar] [CrossRef] [PubMed]
- Girrbach, V.; Strahl, S. Members of the evolutionarily conserved PMT family of protein O-mannosyltransferases form distinct protein complexes among themselves. J. Biol. Chem. 2003, 278, 12554–12562. [Google Scholar] [CrossRef] [PubMed]
- Neubert, P.; Halim, A.; Zauser, M.; Essig, A.; Joshi, H.J.; Zatorska, E.; Larsen, I.S.; Loibl, M.; Castells-Ballester, J.; Aebi, M.; et al. Mapping the O-mannose glycoproteome in saccharomyces cerevisiae. Mol. Cell. Proteom. MCP 2016, 15, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Travers, K.J.; Patil, C.K.; Wodicka, L.; Lockhart, D.J.; Weissman, J.S.; Walter, P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000, 101, 249–258. [Google Scholar] [CrossRef]
- Arroyo, J.; Hutzler, J.; Bermejo, C.; Ragni, E.; Garcia-Cantalejo, J.; Botias, P.; Piberger, H.; Schott, A.; Sanz, A.B.; Strahl, S. Functional and genomic analyses of blocked protein O-mannosylation in baker’s yeast. Mol. Microbiol. 2011, 79, 1529–1546. [Google Scholar] [CrossRef]
- Hetz, C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102. [Google Scholar] [CrossRef]
- Xu, C.; Ng, D.T. Glycosylation-directed quality control of protein folding. Nat. Rev. Mol. Cell Biol. 2015, 16, 742–752. [Google Scholar] [CrossRef]
- Xu, C.; Wang, S.; Thibault, G.; Ng, D.T. Futile protein folding cycles in the ER are terminated by the unfolded protein O-mannosylation pathway. Science 2013, 340, 978–981. [Google Scholar] [CrossRef]
- Hirayama, H.; Fujita, M.; Yoko-o, T.; Jigami, Y. O-mannosylation is required for degradation of the endoplasmic reticulum-associated degradation substrate Gas1*p via the ubiquitin/proteasome pathway in Saccharomyces cerevisiae. J. Biochem. 2008, 143, 555–567. [Google Scholar] [CrossRef]
- Coughlan, C.M.; Walker, J.L.; Cochran, J.C.; Wittrup, K.D.; Brodsky, J.L. Degradation of mutated bovine pancreatic trypsin inhibitor in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J. Biol. Chem. 2004, 279, 15289–15297. [Google Scholar] [CrossRef] [PubMed]
- Nakatsukasa, K.; Okada, S.; Umebayashi, K.; Fukuda, R.; Nishikawa, S.; Endo, T. Roles of O-mannosylation of aberrant proteins in reduction of the load for endoplasmic reticulum chaperones in yeast. J. Biol. Chem. 2004, 279, 49762–49772. [Google Scholar] [CrossRef] [PubMed]
- Harty, C.; Strahl, S.; Romisch, K. O-mannosylation protects mutant alpha-factor precursor from endoplasmic reticulum-associated degradation. Mol. Biol. Cell 2001, 12, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Vashist, S.; Kim, W.; Belden, W.J.; Spear, E.D.; Barlowe, C.; Ng, D.T. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 2001, 155, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Murakami-Sekimata, A.; Sato, K.; Sato, K.; Takashima, A.; Nakano, A. O-Mannosylation is required for the solubilization of heterologously expressed human beta-amyloid precursor protein in Saccharomyces cerevisiae. Genes Cells Devoted Mol. Cell. Mech. 2009, 14, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Goder, V.; Melero, A. Protein O-mannosyltransferases participate in ER protein quality control. J. Cell Sci. 2011, 124, 144–153. [Google Scholar] [CrossRef]
- Fisher, A.C.; DeLisa, M.P. Laboratory evolution of fast-folding green fluorescent protein using secretory pathway quality control. PLoS ONE 2008, 3, e2351. [Google Scholar] [CrossRef]
- Winterhalter, P.R.; Lommel, M.; Ruppert, T.; Strahl, S. O-glycosylation of the non-canonical T-cadherin from rabbit skeletal muscle by single mannose residues. Febs Lett. 2013, 587, 3715–3721. [Google Scholar] [CrossRef]
- Cronin, S.R.; Rao, R.; Hampton, R.Y. Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J. Cell Biol. 2002, 157, 1017–1028. [Google Scholar] [CrossRef]
- Krumpe, K.; Frumkin, I.; Herzig, Y.; Rimon, N.; Ozbalci, C.; Brugger, B.; Rapaport, D.; Schuldiner, M. Ergosterol content specifies targeting of tail-anchored proteins to mitochondrial outer membranes. Mol. Biol. Cell 2012, 23, 3927–3935. [Google Scholar] [CrossRef]
- Stevens, K.L.P.; Black, A.L.; Wells, K.M.; Yeo, K.Y.B.; Steuart, R.F.L.; Stirling, C.J.; Schulz, B.L.; Mousley, C.J. Diminished Ost3-dependent N-glycosylation of the BiP nucleotide exchange factor Sil1 is an adaptive response to reductive ER stress. Proc. Natl. Acad. Sci. USA 2017, 114, 12489–12494. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Preissler, S.; Reuther, J.; Koch, M.; Scior, A.; Bruderek, M.; Frickey, T.; Deuerling, E. Not4-dependent translational repression is important for cellular protein homeostasis in yeast. Embo J. 2015, 34, 1905–1924. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Sakakibara, A.; Yamasaki, M.; Yoda, K. Saccharomyces cerevisiae VIG9 encodes GDP-mannose pyrophosphorylase, which is essential for protein glycosylation. J. Biol. Chem. 1997, 272, 16308–16314. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Breslow, D.K.; Cameron, D.M.; Collins, S.R.; Schuldiner, M.; Stewart-Ornstein, J.; Newman, H.W.; Braun, S.; Madhani, H.D.; Krogan, N.J.; Weissman, J.S. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 2008, 5, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Breker, M.; Gymrek, M.; Schuldiner, M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J. Cell Biol. 2013, 200, 839–850. [Google Scholar] [CrossRef]
- Jackson, C.L.; Kepes, F. BFR1, a multicopy suppressor of brefeldin A-induced lethality, is implicated in secretion and nuclear segregation in Saccharomyces cerevisiae. Genetics 1994, 137, 423–437. [Google Scholar]
- Lang, B.D.; Li, A.; Black-Brewster, H.D.; Fridovich-Keil, J.L. The brefeldin A resistance protein Bfr1p is a component of polyribosome-associated mRNP complexes in yeast. Nucleic Acids Res. 2001, 29, 2567–2574. [Google Scholar] [CrossRef][Green Version]
- Simpson, C.E.; Lui, J.; Kershaw, C.J.; Sims, P.F.; Ashe, M.P. mRNA localization to P-bodies in yeast is bi-phasic with many mRNAs captured in a late Bfr1p-dependent wave. J. Cell Sci. 2014, 127, 1254–1262. [Google Scholar] [CrossRef]
- Weidner, J.; Wang, C.; Prescianotto-Baschong, C.; Estrada, A.F.; Spang, A. The polysome-associated proteins Scp160 and Bfr1 prevent P body formation under normal growth conditions. J. Cell Sci. 2014, 127, 1992–2004. [Google Scholar] [CrossRef]
- Hogan, D.J.; Riordan, D.P.; Gerber, A.P.; Herschlag, D.; Brown, P.O. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008, 6, e255. [Google Scholar] [CrossRef] [PubMed]
- Lapointe, C.P.; Wilinski, D.; Saunders, H.A.; Wickens, M. Protein-RNA networks revealed through covalent RNA marks. Nat. Methods 2015, 12, 1163–1170. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Schekman, R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J. Cell Biol. 1987, 105, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.D.; Misra, L.M.; Vogel, J.P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 1989, 57, 1211–1221. [Google Scholar] [CrossRef]
- Karaoglu, D.; Kelleher, D.J.; Gilmore, R. Functional characterization of Ost3p. Loss of the 34-kD subunit of the Saccharomyces cerevisiae oligosaccharyltransferase results in biased underglycosylation of acceptor substrates. J. Cell Biol. 1995, 130, 567–577. [Google Scholar] [CrossRef]
- Te Heesen, S.; Janetzky, B.; Lehle, L.; Aebi, M. The yeast WBP1 is essential for oligosaccharyl transferase activity in vivo and in vitro. Embo J. 1992, 11, 2071–2075. [Google Scholar] [CrossRef]
- Nuoffer, C.; Jeno, P.; Conzelmann, A.; Riezman, H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 1991, 11, 27–37. [Google Scholar] [CrossRef]
- Aviram, N.; Schuldiner, M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 2017, 130, 4079–4085. [Google Scholar] [CrossRef]
- Gilmore, R.; Blobel, G.; Walter, P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982, 95, 463–469. [Google Scholar] [CrossRef]
- Meyer, D.I.; Krause, E.; Dobberstein, B. Secretory protein translocation across membranes-the role of the “docking protein”. Nature 1982, 297, 647–650. [Google Scholar] [CrossRef]
- Singer-Kruger, B.; Jansen, R.P. Here, there, everywhere. mRNA localization in budding yeast. Rna Biol. 2014, 11, 1031–1039. [Google Scholar] [CrossRef]
- Kraut-Cohen, J.; Afanasieva, E.; Haim-Vilmovsky, L.; Slobodin, B.; Yosef, I.; Bibi, E.; Gerst, J.E. Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2013, 24, 3069–3084. [Google Scholar] [CrossRef]
- Pyhtila, B.; Zheng, T.; Lager, P.J.; Keene, J.D.; Reedy, M.C.; Nicchitta, C.V. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA 2008, 14, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Brar, G.A.; Weissman, J.S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 2015, 16, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Ast, T.; Cohen, G.; Schuldiner, M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 2013, 152, 1134–1145. [Google Scholar] [CrossRef]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef]
- Castells-Ballester, J.; Zatorska, E.; Meurer, M.; Neubert, P.; Metschies, A.; Knop, M.; Strahl, S. Monitoring protein dynamics in protein o-mannosyltransferase mutants in vivo by tandem fluorescent protein timers. Molecules 2018, 23, 2622. [Google Scholar] [CrossRef]
- Xu, C.; Ng, D.T. O-mannosylation: The other glycan player of ER quality control. Semin. Cell Dev. Biol. 2015, 41, 129–134. [Google Scholar] [CrossRef]
- Ambroziak, J.; Henry, S.A. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 1994, 269, 15344–15349. [Google Scholar]
- Shyu, P., Jr.; Ng, B.S.H.; Ho, N.; Chaw, R.; Seah, Y.L.; Marvalim, C.; Thibault, G. Membrane phospholipid alteration causes chronic ER stress through early degradation of homeostatic ER-resident proteins. Sci. Rep. 2019, 9, 8637. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, D.M.; Holen, H.W.; Pedersen, J.T.; Martens, H.J.; Silvestro, D.; Stanchev, L.D.; Costa, S.R.; Gunther Pomorski, T.; Lopez-Marques, R.L.; Palmgren, M. The P5A ATPase Spf1p is stimulated by phosphatidylinositol 4-phosphate and influences cellular sterol homeostasis. Mol. Biol. Cell 2019, 30, 1069–1084. [Google Scholar] [CrossRef] [PubMed]
- Torres-Quiroz, F.; Garcia-Marques, S.; Coria, R.; Randez-Gil, F.; Prieto, J.A. The activity of yeast Hog1 MAPK is required during endoplasmic reticulum stress induced by tunicamycin exposure. J. Biol. Chem. 2010, 285, 20088–20096. [Google Scholar] [CrossRef] [PubMed]
- Burg, M.B.; Ferraris, J.D. Intracellular organic osmolytes: Function and regulation. J. Biol. Chem. 2008, 283, 7309–7313. [Google Scholar] [CrossRef]
- Guerra-Moreno, A.; Ang, J.; Welsch, H.; Jochem, M.; Hanna, J. Regulation of the unfolded protein response in yeast by oxidative stress. Febs Lett. 2019, 593, 1080–1088. [Google Scholar] [CrossRef]
- Carla Fama, M.; Raden, D.; Zacchi, N.; Lemos, D.R.; Robinson, A.S.; Silberstein, S. The Saccharomyces cerevisiae YFR041C/ERJ5 gene encoding a type I membrane protein with a J domain is required to preserve the folding capacity of the endoplasmic reticulum. Biochim. Biophys. Acta 2007, 1773, 232–242. [Google Scholar] [CrossRef]
- Copic, A.; Dorrington, M.; Pagant, S.; Barry, J.; Lee, M.C.; Singh, I.; Hartman, J.L.t.; Miller, E.A. Genomewide analysis reveals novel pathways affecting endoplasmic reticulum homeostasis, protein modification and quality control. Genetics 2009, 182, 757–769. [Google Scholar] [CrossRef]
- Bays, N.W.; Gardner, R.G.; Seelig, L.P.; Joazeiro, C.A.; Hampton, R.Y. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 2001, 3, 24–29. [Google Scholar] [CrossRef]
- Kostova, Z.; Wolf, D.H. For whom the bell tolls: Protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. Embo J. 2003, 22, 2309–2317. [Google Scholar] [CrossRef]
- Schwarz, M.; Knauer, R.; Lehle, L. Yeast oligosaccharyltransferase consists of two functionally distinct sub-complexes, specified by either the Ost3p or Ost6p subunit. Febs Lett. 2005, 579, 6564–6568. [Google Scholar] [CrossRef]
- Zatorska, E.; Gal, L.; Schmitt, J.; Bausewein, D.; Schuldiner, M.; Strahl, S. Cellular consequences of diminished protein o-mannosyltransferase activity in baker’s yeast. Int. J. Mol. Sci. 2017, 18, 1226. [Google Scholar] [CrossRef] [PubMed]
- Janik, A.; Sosnowska, M.; Kruszewska, J.; Krotkiewski, H.; Lehle, L.; Palamarczyk, G. Overexpression of GDP-mannose pyrophosphorylase in Saccharomyces cerevisiae corrects defects in dolichol-linked saccharide formation and protein glycosylation. Biochim. Biophys. Acta 2003, 1621, 22–30. [Google Scholar] [CrossRef]
- Zakrzewska, A.; Palamarczyk, G.; Krotkiewski, H.; Zdebska, E.; Saloheimo, M.; Penttila, M.; Kruszewska, J.S. Overexpression of the gene encoding GTP:mannose-1-phosphate guanyltransferase, mpg1, increases cellular GDP-mannose levels and protein mannosylation in Trichoderma reesei. Appl. Environ. Microbiol. 2003, 69, 4383–4389. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luo, Y.; Na, Z.; Slavoff, S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry 2018, 57, 2424–2431. [Google Scholar] [CrossRef]
- Albihlal, W.S.; Gerber, A.P. Unconventional RNA-binding proteins: An uncharted zone in RNA biology. Febs Lett. 2018, 592, 2917–2931. [Google Scholar] [CrossRef]
- Mitchell, S.F.; Jain, S.; She, M.; Parker, R. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2013, 20, 127–133. [Google Scholar] [CrossRef]
- Zweytick, D.; Hrastnik, C.; Kohlwein, S.D.; Daum, G. Biochemical characterization and subcellular localization of the sterol C-24(28) reductase, erg4p, from the yeast saccharomyces cerevisiae. Febs Lett. 2000, 470, 83–87. [Google Scholar] [CrossRef]
- Manchalu, S.; Mittal, N.; Spang, A.; Jansen, R.P. Local translation of yeast ERG4 mRNA at the endoplasmic reticulum requires the brefeldin A resistance protein Bfr1. RNA 2019, 25, 1661–1672. [Google Scholar] [CrossRef]
- Mueller, S.; Wahlander, A.; Selevsek, N.; Otto, C.; Ngwa, E.M.; Poljak, K.; Frey, A.D.; Aebi, M.; Gauss, R. Protein degradation corrects for imbalanced subunit stoichiometry in OST complex assembly. Mol. Biol. Cell 2015, 26, 2596–2608. [Google Scholar] [CrossRef]
- Cohen, N.; Breker, M.; Bakunts, A.; Pesek, K.; Chas, A.; Argemi, J.; Orsi, A.; Gal, L.; Chuartzman, S.; Wigelman, Y.; et al. Iron affects Ire1 clustering propensity and the amplitude of endoplasmic reticulum stress signaling. J. Cell Sci. 2017, 130, 3222–3233. [Google Scholar] [CrossRef]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Robinson, J.S.; Klionsky, D.J.; Banta, L.M.; Emr, S.D. Protein sorting in Saccharomyces cerevisiae: Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988, 8, 4936–4948. [Google Scholar] [CrossRef] [PubMed]
- Papic, D.; Elbaz-Alon, Y.; Koerdt, S.N.; Leopold, K.; Worm, D.; Jung, M.; Schuldiner, M.; Rapaport, D. The role of Djp1 in import of the mitochondrial protein Mim1 demonstrates specificity between a cochaperone and its substrate protein. Mol. Cell. Biol. 2013, 33, 4083–4094. [Google Scholar] [CrossRef] [PubMed]
- Taxis, C.; Knop, M. System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques 2006, 40, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Christianson, T.W.; Sikorski, R.S.; Dante, M.; Shero, J.H.; Hieter, P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992, 110, 119–122. [Google Scholar] [CrossRef]
- Guldener, U.; Heck, S.; Fielder, T.; Beinhauer, J.; Hegemann, J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996, 24, 2519–2524. [Google Scholar] [CrossRef]
- Hutzler, F.; Gerstl, R.; Lommel, M.; Strahl, S. Protein N-glycosylation determines functionality of the Saccharomyces cerevisiae cell wall integrity sensor Mid2p. Mol. Microbiol. 2008, 68, 1438–1449. [Google Scholar] [CrossRef]
- Tong, A.H.; Boone, C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 2006, 313, 171–192. [Google Scholar]
- Cohen, Y.; Schuldiner, M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol. Biol. 2011, 781, 127–159. [Google Scholar]
- Gentzsch, M.; Immervoll, T.; Tanner, W. Protein O-glycosylation in Saccharomyces cerevisiae: The protein O-mannosyltransferases Pmt1p and Pmt2p function as heterodimer. Febs Lett. 1995, 377, 128–130. [Google Scholar] [CrossRef]
- Stirling, C.J.; Rothblatt, J.; Hosobuchi, M.; Deshaies, R.; Schekman, R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell 1992, 3, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Popolo, L.; Grandori, R.; Vai, M.; Lacana, E.; Alberghina, L. Immunochemical characterization of gp115, a yeast glycoprotein modulated by the cell cycle. Eur. J. Cell Biol. 1988, 47, 173–180. [Google Scholar] [PubMed]
- Te Heesen, S.; Knauer, R.; Lehle, L.; Aebi, M. Yeast Wbp1p and Swp1p form a protein complex essential for oligosaccharyl transferase activity. Embo J. 1993, 12, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Aronov, S.; Dover-Biterman, S.; Suss-Toby, E.; Shmoish, M.; Duek, L.; Choder, M. Pheromone-encoding mRNA is transported to the yeast mating projection by specific RNP granules. J. Cell Biol. 2015, 209, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Doring, K.; Ahmed, N.; Riemer, T.; Suresh, H.G.; Vainshtein, Y.; Habich, M.; Riemer, J.; Mayer, M.P.; O’Brien, E.P.; Kramer, G.; et al. Profiling Ssb-nascent chain interactions reveals principles of Hsp70-assisted folding. Cell 2017, 170, 298–311.e220. [Google Scholar] [CrossRef] [PubMed]
- Galmozzi, C.V.; Merker, D.; Friedrich, U.A.; Doring, K.; Kramer, G. Selective ribosome profiling to study interactions of translating ribosomes in yeast. Nat. Protoc. 2019, 14, 2279–2317. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ignatiadis, N.; Klaus, B.; Zaugg, J.B.; Huber, W. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 2016, 13, 577–580. [Google Scholar] [CrossRef]


| Strain | Genotype | Reference/Source |
|---|---|---|
| BY4741 (wild type) | MATa met15-Δ0 his3-Δ1 leu2-Δ0 ura3-Δ0 | [81] |
| SEY6210 | MATα lys2-801 his3-Δ200 leu2-3,112 trp1-Δ901 ura3-52 suc2-Δ9 | [82] |
| YMS721 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::STE2pr-spHIS5 lyp1Δ::STE3pr-LEU2 | [83] |
| JEY05 | YMS721 hoΔ::ER-GFPf-URA3 | This study |
| JEY06 | YMS721 hoΔ::ER-GFP-URA3 | This study |
| JCY010 | JEY06 except pmt1Δ::kanMX4 | This study |
| JCY011 | JEY06 except pmt2Δ::kanMX4 | This study |
| JCY012 | JEY06 except pmt4Δ::kanMX4 | This study |
| bfr1Δ | BY4741 except bfr1Δ::kanMX4 | Euroscarf |
| psa1DAmP ER-GFP | JEY06 except psa1Δ::psa1DAmP | This study |
| bfr1Δ ER-GFP | JEY06 except bfr1Δ::kanMX4 | This study |
| spf1Δ ER-GFP | JEY06 except spf1Δ::kanMX4 | This study |
| JCY015 | BY4741 except psa1Δ::psa1DAmP | This study |
| JCY016 | JEY06 except bfr1Δ::kanMX4 | This study |
| JCY017 | BY4741 except bfr1Δ::BFR1-3xHA | This study |
| MLY014 | SEY6210 except PMT2-GAGA-HA3-kanMX6 | This study |
| MLY098 | SEY6210 except klTRP1-PGAL1-UBI4-R-PMT2-GAGA-HA3-kanMX6-HA | This study |
| JCY034 | MLY098 except bfr1Δ::URA3 | This study |
| EZY70 | BY4741 except hoΔ::ER-GFP-URA3 | This study |
| EZY77 | ost3Δ except hoΔ::ER-GFP-URA3 | This study |
| EZY78 | ost6Δ except hoΔ::ER-GFP-URA3 | This study |
| pmt2Δ ER-GFP | pmt2Δ except hoΔ::ER-GFP-URA3 | This study |
| Plasmid | Description | Reference/Source |
|---|---|---|
| pPN014 | ori, CEN/ARS, PTDH3-ER-GFP-3xFLAG-HDEL | This study |
| pWX204 | ori, CEN/ARS, PTDH3-Kar2SS-ER-GFPf-HDEL, URA3 | [19] |
| pWX206 | ori, CEN/ARS, PTDH3-Kar2SS-ER-GFP-HDEL, URA3 | [19] |
| pJC01 | ori, bla, 2µ, PMT2, LEU2 | This study |
| pJC02 | ori, bla, 2µ, PMT2-3xHA, HIS3 | This study |
| pRS41N | ori, CEN/ARS, natNT2 | [84] |
| pJC09 | ori, CEN/ARS, PMT2, natNT2 | This study |
| pJC10 | ori, CEN/ARS, PMT2-3xHA, natNT2 | This study |
| pRS415 | ori, CEN/ARS, bla, LEU2 | [85] |
| pJC16 | ori, CEN/ARS, PTDH3-Kar2SS-ER -GFP-HDEL, LEU2 | This study |
| pUG6 | ori, bla, kanMX4 | [86] |
| pJH24 | ori, bla, 2µ, URA3, kanMX6 | [87] |
| Name | Description | Reference/Source |
|---|---|---|
| αPmt1 | rabbit; 1:2500 | [3] |
| αPmt2 | rabbit; 1:2500 | [90] |
| αPmt4 | rabbit; 1:250 | [13] |
| αSec61 | rabbit; 1:2500 | [91] |
| αHA | mouse; 1:10,000 | #MMS-101R; Covance; Princeton, NJ, USA |
| αGas1 | rabbit; 1:2500 | [92] |
| αWbp1 | rabbit; 1:2500 | [93] |
| αKar2 | rabbit; 1:500 | |
| αOst3 | rabbit; 1:1000 | Gift from M. Aebi |
| αG6PDH | rabbit; 1:5000 | #A9521; Sigma-Aldrich Chemie; Munich, Germany |
| αGFP | rabbit; 1:2500 | #A6455; Thermo Fisher Scientific; Waltham, MA, USA |
| αRpl5 | rabbit; 1:7000 | Gift from E. Hurt |
| αmouseHRP | rabbit; 1:10,000 | #A9044; Sigma-Aldrich; Munich, Germany |
| αrabbitHRP | goat; 1:10,000 | #A6154; Sigma-Aldrich; Munich, Germany |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castells-Ballester, J.; Rinis, N.; Kotan, I.; Gal, L.; Bausewein, D.; Kats, I.; Zatorska, E.; Kramer, G.; Bukau, B.; Schuldiner, M.; et al. Translational Regulation of Pmt1 and Pmt2 by Bfr1 Affects Unfolded Protein O-Mannosylation. Int. J. Mol. Sci. 2019, 20, 6220. https://doi.org/10.3390/ijms20246220
Castells-Ballester J, Rinis N, Kotan I, Gal L, Bausewein D, Kats I, Zatorska E, Kramer G, Bukau B, Schuldiner M, et al. Translational Regulation of Pmt1 and Pmt2 by Bfr1 Affects Unfolded Protein O-Mannosylation. International Journal of Molecular Sciences. 2019; 20(24):6220. https://doi.org/10.3390/ijms20246220
Chicago/Turabian StyleCastells-Ballester, Joan, Natalie Rinis, Ilgin Kotan, Lihi Gal, Daniela Bausewein, Ilia Kats, Ewa Zatorska, Günter Kramer, Bernd Bukau, Maya Schuldiner, and et al. 2019. "Translational Regulation of Pmt1 and Pmt2 by Bfr1 Affects Unfolded Protein O-Mannosylation" International Journal of Molecular Sciences 20, no. 24: 6220. https://doi.org/10.3390/ijms20246220
APA StyleCastells-Ballester, J., Rinis, N., Kotan, I., Gal, L., Bausewein, D., Kats, I., Zatorska, E., Kramer, G., Bukau, B., Schuldiner, M., & Strahl, S. (2019). Translational Regulation of Pmt1 and Pmt2 by Bfr1 Affects Unfolded Protein O-Mannosylation. International Journal of Molecular Sciences, 20(24), 6220. https://doi.org/10.3390/ijms20246220





