Bioactive Polyphenols Modulate Enzymes Involved in Grapevine Pathogenesis and Chitinase Activity at Increasing Complexity Levels
Abstract
:1. Introduction
2. Results
2.1. HPLC-MS Quantitative Analysis of the Assayed PEs
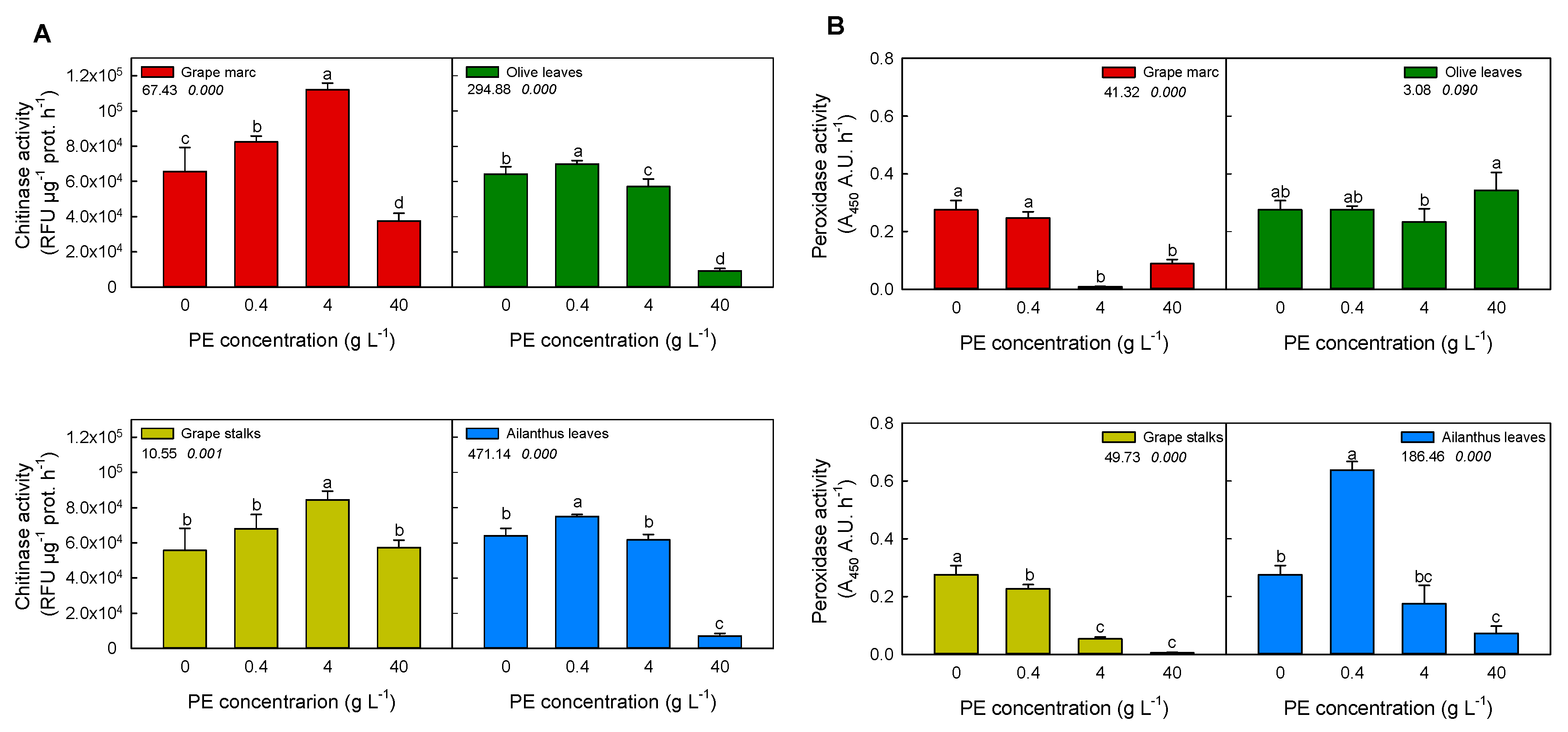
2.2. In Vitro Modulation of Plant Defense-Related Enzyme Activities: Chitinase and Peroxidase
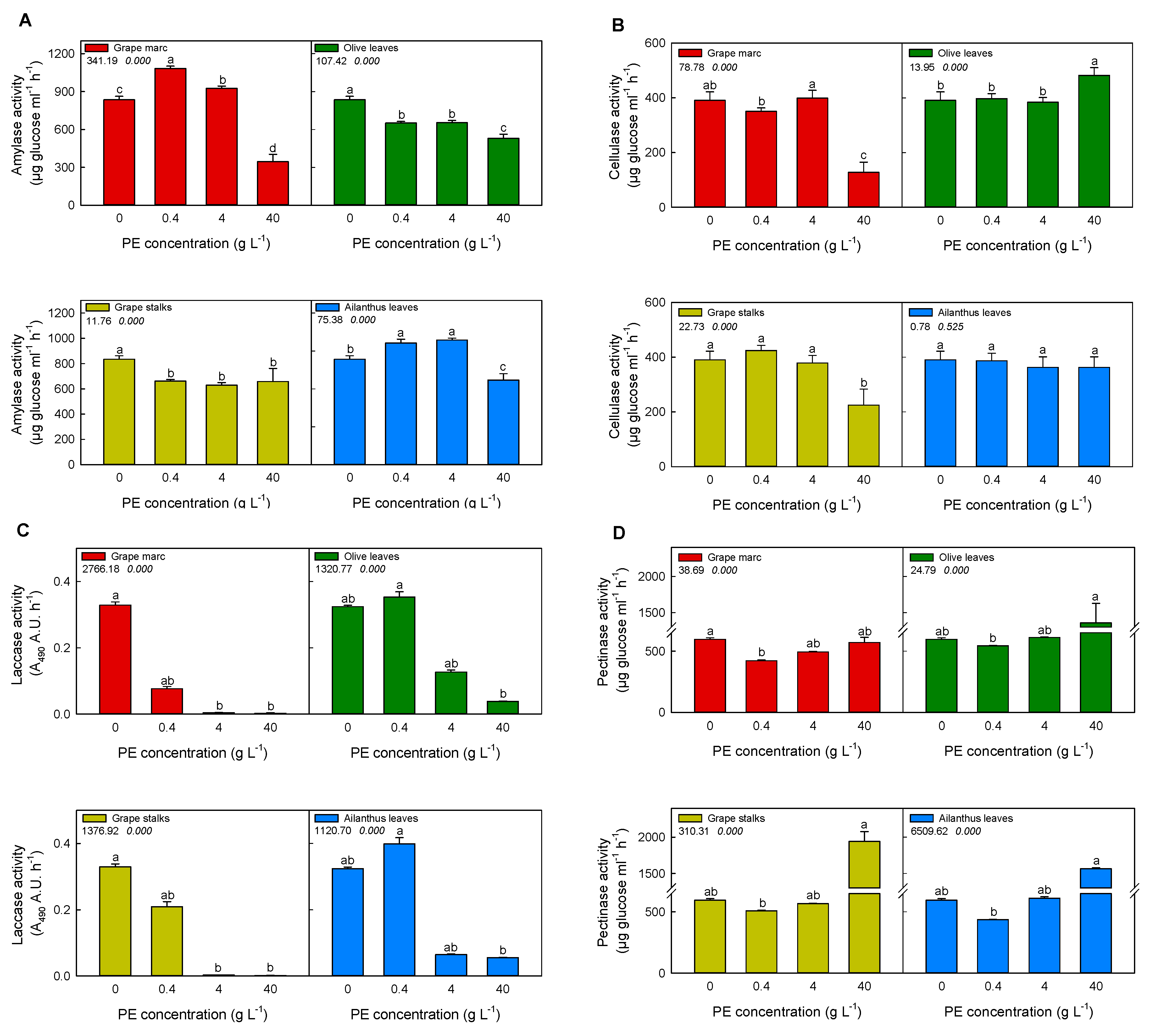
2.3. In Vitro Modulation of Purified Pathogen-Related Enzymes Activities
2.4. Ex Vivo Modulation of Chitinase Activity in V. vinifera cv. Cabernet Sauvignon Cell Suspension Cultures
2.5. Cell Death of V. vinifera cv. Cabernet Sauvignon Cell Suspension Cultures
2.6. In Vivo Time-Course Modulation of Chitinase Activity Stimulation
2.7. A Synthetic Assessment of the In Vitro Effectiveness of the Considered Plant Extracts
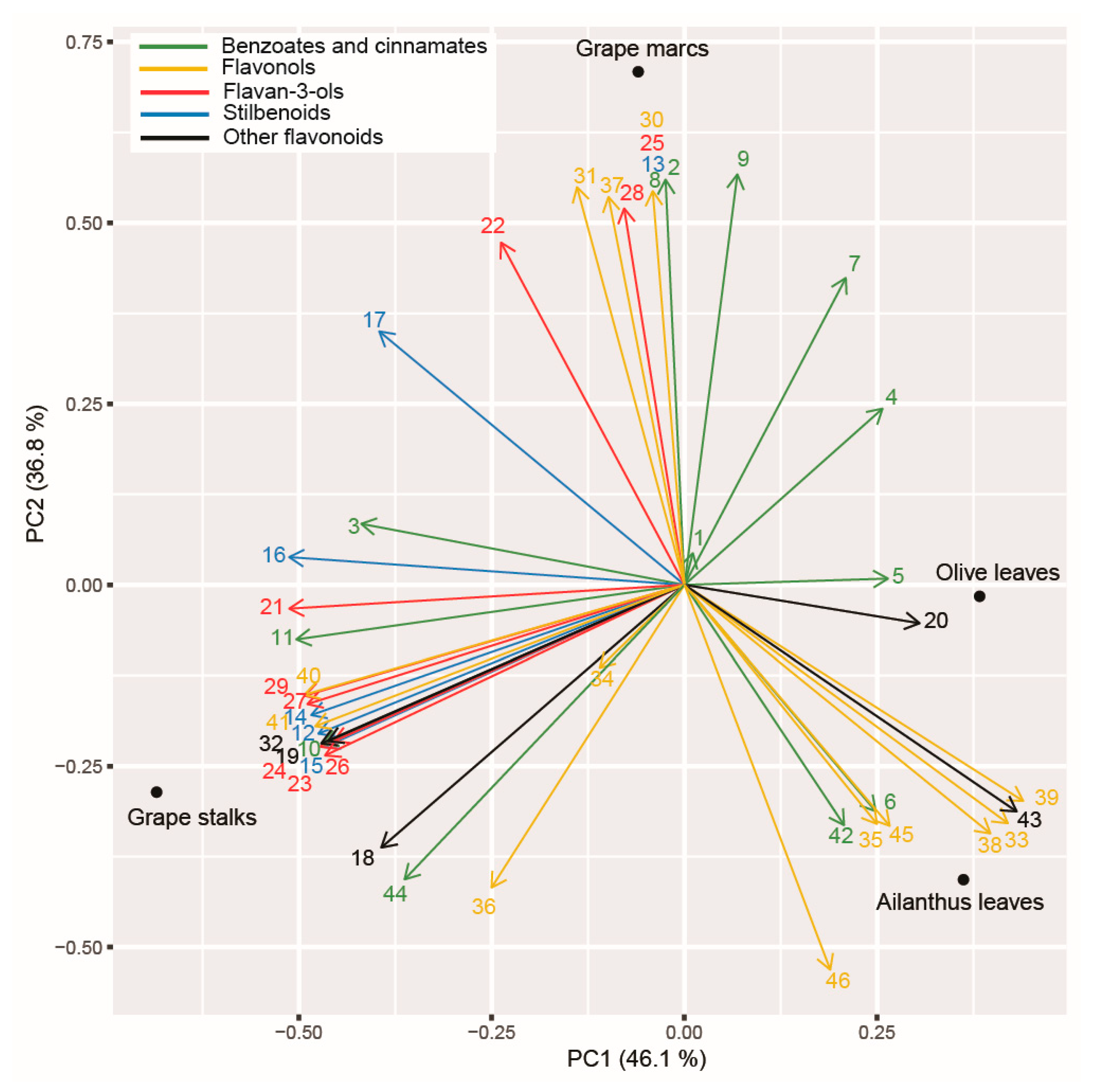
2.8. Principal Component Analysis (PCA) Applied on Polyphenolic Profile of the Tested PEs and Assayed Enzymatic Activities
3. Discussion
4. Materials and Methods
4.1. Bioactive Compound Extraction
4.2. Analysis of Polyphenols by Means of UHPLC-MS
4.3. Cell Culture Suspension and Plant Material: Growth and Maintenance
4.4. Cell Suspension Cultures (Ex Vivo) Treatment by Bioactive Compounds
4.5. Plant (In Vivo) Treatment by Bioactive Compounds
4.6. Cell Death Determination in Cell Suspension Cultures
4.7. Modulation of In Vitro Pathogen-Related and Plant Defense-Related Enzyme Activities
4.7.1. Purified Enzymes; In Vitro Modulation
4.7.2. Chitinase Activity; Ex Vivo Modulation in V. vinifera cv. Cabernet Sauvignon Suspension Cultures
4.7.3. Chitinase Activity: In Vivo Modulation in V. vinifera cv. Verduzzo Friulano Plants
4.8. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fermaud, M.; Smits, N.; Merot, A.; Roudet, J.; Thiéry, D.; Wery, J.; Delbac, L. New multipest damage indicator to assess protection strategies in grapevine cropping systems. Aust. J. Grape Wine Res. 2016, 22, 450–461. [Google Scholar] [CrossRef]
- Martínez, J.A. Natural fungicides obtained from plants. Fungic. Plant Anim. Dis. 2012. [Google Scholar]
- Enoki, S.; Suzuki, S. Pathogenesis-related proteins in grape. Grape Wine Biotechnol. 2016, 43. [Google Scholar]
- McSpadden Gardener, B.B.; Fravel, D.R. Biological control of plant pathogens: research, commercialization, and application in the USA. Plant Health Prog. 2002, 3, 17. [Google Scholar] [CrossRef] [Green Version]
- Akula, R.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [CrossRef] [PubMed]
- Galanakis, C.M. Nutraceutical and Functional Food Components: Effects of Innovative Processing Techniques; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Arunkumar, K.; Sivakumar, S.R.; Rengasamy, R. Review on bioactive potential in seaweeds (marine macroalgae): A special emphasis on bioactivity of seaweeds against plant pathogens. Asian J. Plant Sci. 2010, 9, 227–240. [Google Scholar] [CrossRef] [Green Version]
- De Bona, G.S.; Adrian, M.; Negrel, J.; Chiltz, A.; Klinguer, A.; Poinssot, B.; Héloir, M.-C.; Angelini, E.; Vincenzi, S.; Bertazzon, N. Dual mode of action of grape cane extracts against Botrytis cinerea. J. Agric. Food Chem. 2019, 67, 5512–5520. [Google Scholar] [CrossRef]
- Krzyzaniak, Y.; Trouvelot, S.; Negrel, J.; Cluzet, S.; Valls, J.; Richard, T.; Bougaud, A.; Jacquens, L.; Klinguer, A.; Chiltz, A.; et al. A plant extract acts both as a resistance inducer and an oomycide against grapevine downy mildew. Front. Plant Sci. 2018, 9, 1085. [Google Scholar] [CrossRef]
- Goupil, P.; Benouaret, R.; Charrier, O.; ter Halle, A.; Richard, C.; Eyheraguibel, B.; Thiery, D.; Ledoigt, G. Grape marc extract acts as elicitor of plant defence responses. Ecotoxicology 2012, 21, 1541–1549. [Google Scholar] [CrossRef] [Green Version]
- Wiesel, L.; Newton, A.C.; Elliott, I.; Booty, D.; Gilroy, E.M.; Birch, P.R.J.; Hein, I. Molecular effects of resistance elicitors from biological origin and their potential for crop protection. Front. Plant Sci. 2014, 5, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Dana, M.; de las, M.; Pintor-Toro, J.A.; Cubero, B. Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol. 2006, 142, 722–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filippi, A.; Petrussa, E.; Rajcevic, U.; Čurin Šerbec, V.; Passamonti, S.; Renzone, G.; Scaloni, A.; Zancani, M.; Vianello, A.; Braidot, E. Flavonoid Interaction with a chitinase from grape berry skin: protein identification and modulation of the enzymatic activity. Molecules 2016, 21, 1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karthik, N.; Binod, P.; Pandey, A. 15—Chitinases. In Current Developments in Biotechnology and Bioengineering; Pandey, A., Negi, S., Soccol, C.R., Eds.; Elsevier: Amsterdam, the Netherlands, 2017; pp. 335–368. [Google Scholar]
- Collinge, D.B.; Kragh, K.M.; Mikkelsen, J.D.; Nielsen, K.K.; Rasmussen, U.; Vad1, K. Plant chitinases. Plant J. 1993, 3, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Petrasch, S.; Knapp, S.J.; van Kan, J.A.L.; Blanco-Ulate, B. Grey mould of strawberry, a devastating disease caused by the ubiquitous necrotrophic fungal pathogen Botrytis cinerea. Mol. Plant Pathol. 2019, 20, 877–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteiro, S.; Piçarra-Pereira, M.A.; Loureiro, V.B.; Teixeira, A.R.; Ferreira, R.B. The diversity of pathogenesis-related proteins decreases during grape maturation. Phytochemistry 2007, 68, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Kortekamp, A. Expression analysis of defence-related genes in grapevine leaves after inoculation with a host and a non-host pathogen. Plant Physiol. Biochem. PPB 2006, 44, 58–67. [Google Scholar] [CrossRef]
- Figueiredo, A.; Fortes, A.M.; Ferreira, S.; Sebastiana, M.; Choi, Y.H.; Sousa, L.; Acioli-Santos, B.; Pessoa, F.; Verpoorte, R.; Pais, M.S. Transcriptional and metabolic profiling of grape (Vitis vinifera L.) leaves unravel possible innate resistance against pathogenic fungi. J. Exp. Bot. 2008, 59, 3371–3381. [Google Scholar] [CrossRef] [Green Version]
- Ali, K.; Maltese, F.; Choi, Y.H.; Verpoorte, R. Metabolic constituents of grapevine and grape-derived products. Phytochem. Rev. 2010, 9, 357–378. [Google Scholar] [CrossRef] [Green Version]
- Aziz, A.; Poinssot, B.; Daire, X.; Adrian, M.; Bézier, A.; Lambert, B.; Joubert, J.-M.; Pugin, A. Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola. Mol. Plant. Microbe Interact. 2003, 16, 1118–1128. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Gamboa, G.; Romanazzi, G.; Garde-Cerdán, T.; Pérez-Álvarez, E.P. A review of the use of biostimulants in the vineyard for improved grape and wine quality: effects on prevention of grapevine diseases: use of biostimulants in the vineyard for improved grape and wine quality. J. Sci. Food Agric. 2019, 99, 1001–1009. [Google Scholar] [CrossRef]
- Ping, L.; Brosse, N.; Sannigrahi, P.; Ragauskas, A. Evaluation of grape stalks as a bioresource. Ind. Crops Prod. 2011, 33, 200–204. [Google Scholar] [CrossRef]
- Vincenzi, S.; Bierma, J.; Wickramasekara, S.I.; Curioni, A.; Gazzola, D.; Bakalinsky, A.T. Characterization of a grape class IV chitinase. J. Agric. Food Chem. 2014, 62, 5660–5668. [Google Scholar] [CrossRef] [PubMed]
- Batran, R.Z.; Khedr, M.A.; Abdel Latif, N.A.; Abd El Aty, A.A.; Shehata, A.N. Synthesis, homology modeling, molecular docking, dynamics, and antifungal screening of new 4-hydroxycoumarin derivatives as potential chitinase inhibitors. J. Mol. Struct. 2019, 1180, 260–271. [Google Scholar] [CrossRef]
- Tayel, A.A.; Shaban, S.M.; Moussa, S.H.; Elguindy, N.M.; Diab, A.M.; Mazrou, K.E.; Ghanem, R.A.; El-Sabbagh, S.M. Bioactivity and application of plant seeds’ extracts to fight resistant strains of Staphylococcus aureus. Ann. Agric. Sci. 2018, 63, 47–53. [Google Scholar] [CrossRef]
- Wink, M. Modes of action of herbal medicines and plant secondary metabolites. Medicines 2015, 2, 251–286. [Google Scholar] [CrossRef] [PubMed]
- Kulbat, K. The role of phenolic compounds in plant resistance. Biotechnol. Food Sci. 2016, 80, 97–108. [Google Scholar]
- Haslam, E. Natural polyphenols (vegetable tannins) as drugs: possible modes of action. J. Nat. Prod. 1996, 59, 205–215. [Google Scholar] [CrossRef]
- Wang, S.; Alseekh, S.; Fernie, A.R.; Luo, J. The structure and function of major plant metabolite modifications. Mol. Plant 2019, 12, 899–919. [Google Scholar] [CrossRef]
- Olmo-García, L.; Kessler, N.; Neuweger, H.; Wendt, K.; Olmo-Peinado, J.; Fernández-Gutiérrez, A.; Baessmann, C.; Carrasco-Pancorbo, A. Unravelling the distribution of secondary metabolites in Olea europaea L.: exhaustive characterization of eight olive-tree derived matrices by complementary platforms (LC-ESI/APCI-MS and GC-APCI-MS). Molecules 2018, 23, 2419. [Google Scholar] [CrossRef] [Green Version]
- Fontana, A.R.; Antoniolli, A.; Bottini, R. Grape pomace as a sustainable source of bioactive compounds: extraction, characterization, and biotechnological applications of phenolics. J. Agric. Food Chem. 2013, 61, 8987–9003. [Google Scholar] [CrossRef]
- Pereira, A.P.; Ferreira, I.C.F.R.; Marcelino, F.; Valentão, P.; Andrade, P.B.; Seabra, R.; Estevinho, L.; Bento, A.; Pereira, J.A. Phenolic compounds and antimicrobial activity of Olive (Olea europaea L. cv. Cobrançosa) Leaves. Mol. J. Synth. Chem. Nat. Prod. Chem. 2007, 12, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Straney, D.; Khan, R.; Tan, R.; Bagga, S. Host recognition by pathogenic fungi through plant flavonoids. In Flavonoids in Cell Function; Buslig, B.S., Manthey, J.A., Eds.; Advances in Experimental Medicine and Biology; Springer US: Boston, MA, USA, 2002; pp. 9–22. [Google Scholar]
- Connolly, M.S.; Williams, N.; Heckman, C.A.; Morris, P.F. Soybean isoflavones trigger a calcium influx in Phytophthora sojae. Fungal Genet. Biol. 1999, 28, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Casañal, A.; Zander, U.; Muñoz, C.; Dupeux, F.; Luque, I.; Botella, M.A.; Schwab, W.; Valpuesta, V.; Marquez, J.A. The strawberry Pathogenesis-related 10 (PR-10) Fra a proteins control flavonoid biosynthesis by binding to metabolic intermediates. J. Biol. Chem. 2013, 288, 35322–35332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, Y.-L.; Liang, H.-L.; Hung, C.-H.; Kuo, P.-L. Syringetin, a flavonoid derivative in grape and wine, induces human osteoblast differentiation through bone morphogenetic protein-2/extracellular signal-regulated kinase 1/2 pathway. Mol. Nutr. Food Res. 2009, 53, 1452–1461. [Google Scholar] [CrossRef] [PubMed]
- Ghareib, H.R.A.; Abdelhamed, M.S.; Ibrahim, O.H. Antioxidative effects of the acetone fraction and vanillic acid from Chenopodium murale on tomato plants. Weed Biol. Manag. 2010, 10, 64–72. [Google Scholar] [CrossRef]
- Quan, N.T.; Xuan, T.D. Foliar application of vanillic and p-hydroxybenzoic acids enhanced drought tolerance and formation of phytoalexin momilactones in rice. Arch. Agron. Soil Sci. 2018, 64, 1831–1846. [Google Scholar] [CrossRef]
- Repka, V. Elicitor-stimulated induction of defense mechanisms and defense gene activation in grapevine cell suspension cultures. Biol. Plant. 2001, 44, 555–565. [Google Scholar] [CrossRef]
- Faurie, B.; Cluzet, S.; Mérillon, J.-M. Implication of signaling pathways involving calcium, phosphorylation and active oxygen species in methyl jasmonate-induced defense responses in grapevine cell cultures. J. Plant Physiol. 2009, 166, 1863–1877. [Google Scholar] [CrossRef]
- Zamboni, A.; Gatto, P.; Cestaro, A.; Pilati, S.; Viola, R.; Mattivi, F.; Moser, C.; Velasco, R. Grapevine cell early activation of specific responses to DIMEB, a resveratrol elicitor. BMC Genom. 2009, 10, 363. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Tan, X. Comprehensive analysis of the chitinase family genes in tomato (Solanum lycopersicum). Plants 2019, 8, 52. [Google Scholar] [CrossRef] [Green Version]
- Passarinho, P.A.; Van Hengel, A.J.; Fransz, P.F.; de Vries, S.C. Expression pattern of the Arabidopsis thaliana AtEP3/AtchitIV endochitinase gene. Planta 2001, 212, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Kim, N.H.; Hwang, B.K. The Capsicum annuum class IV chitinase ChitIV interacts with receptor-like cytoplasmic protein kinase PIK1 to accelerate PIK1-triggered cell death and defence responses. J. Exp. Bot. 2015, 66, 1987–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, A.; Kim, E.L.; Kang, S.C. Antibacterial and antioxidant properties of Ailanthus Altissima Swingle leave extract to reduce foodborne pathogens and spoiling bacteria. J. Food Saf. 2009, 29, 499–510. [Google Scholar] [CrossRef]
- Liu, Z.; Shi, L.; Yang, S.; Lin, Y.; Weng, Y.; Li, X.; Hussain, A.; Noman, A.; He, S. Functional and promoter analysis of ChiIV3, a chitinase of pepper plant, in response to Phytophthora capsici infection. Int. J. Mol. Sci. 2017, 18, 1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vrhovsek, U.; Masuero, D.; Gasperotti, M.; Franceschi, P.; Caputi, L.; Viola, R.; Mattivi, F. A versatile targeted metabolomics method for the rapid quantification of multiple classes of phenolics in fruits and beverages. J. Agric. Food Chem. 2012, 60, 8831–8840. [Google Scholar] [CrossRef]
- Krisa, S.; Waffo Téguo, P.; Decendit, A.; Deffieux, G.; Vercauteren, J.; Mérillon, J.-M. Production of 13C-labelled anthocyanins by Vitis vinifera cell suspension cultures. Phytochemistry 1999, 51, 651–656. [Google Scholar] [CrossRef]
- Gusakov, A.V.; Kondratyeva, E.G.; Sinitsyn, A.P. Comparison of two methods for assaying reducing sugars in the determination of carbohydrase activities. Int. J. Anal. Chem. 2011. [Google Scholar] [CrossRef] [Green Version]
- Biz, A.; Farias, F.C.; Motter, F.A.; de Paula, D.H.; Richard, P.; Krieger, N.; Mitchell, D.A. Pectinase activity determination: an early deceleration in the release of reducing sugars throws a spanner in the works! PLoS ONE 2014, 9, e109529. [Google Scholar] [CrossRef] [Green Version]
- Keharom, S.; Mahachai, R.; Chanthai, S. The optimization study of α-amylase activity based on central composite design-response surface methodology by dinitrosalicylic acid method. Int. Food Res. J. 2016, 23, 10–17. [Google Scholar]
- Abd El Monssef, R.A.; Hassan, E.A.; Ramadan, E.M. Production of laccase enzyme for their potential application to decolorize fungal pigments on aging paper and parchment. Ann. Agric. Sci. 2016, 61, 145–154. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Chan, Z.; Yang, F. Comparative analyses of universal extraction buffers for assay of stress related biochemical and physiological parameters. Prep. Biochem. Biotechnol. 2015, 45, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-J.; Hwang, C.-L. Fuzzy multiple attribute decision making methods. In Fuzzy Multiple Attribute Decision Making: Methods and Applications; Chen, S.-J., Hwang, C.-L., Eds.; Lecture notes in economics and mathematical systems; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 1992; pp. 289–486. [Google Scholar]
- Yoon, K.P.; Hwang, C.-L. Multiple Attribute Decision Making: An Introduction; SAGE Publications: Thousand Oaks, CA, USA, 1995. [Google Scholar]
- R Core Team. Foundation for Statistical Computing, Vienna, Austria R: the R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 9 October 2019).


| Polyphenols | Grape Marc (mg L−1) | Grape Stalks (mg L−1) | Olive Leaves (mg L−1) | Ailanthus Leaves (mg L−1) | LOQ (pg) | Reference Number |
|---|---|---|---|---|---|---|
| Benzoates and cinnamates | ||||||
| 4-aminobenzoic acid | <L.O.Q. | <L.O.Q. | - | - | n.a. | - |
| p-hydroxybenzoic acid | 0.11 | 0.19 | 0.37 | - | 40 | [1] |
| vanillic acid | 0.81 | 0.02 | 0.09 | - | 20 | [2] |
| gallic acid | 4.07 | 4.79 | - | 2.58 | 160 | [3] |
| 3,5-dihydroxy-benzoic acid | 0.17 | - | 0.38 | - | 16 | [4] |
| protocatechuic acid | 0.11 | - | 2.89 | - | 80 | [5] |
| methyl gallate | - | <L.O.Q. | - | 1.21 | 8 | [6] |
| p-coumaric acid | 0.13 | - | 0.15 | - | 12 | [7] |
| caffeic acid | 0.16 | - | - | - | 12 | [8] |
| ferulic acid | 0.23 | 0.01 | 0.11 | - | 2 | [9] |
| caftaric acid | 0.36 | 33.41 | 0.80 | 0.63 | 120 | [10] |
| fertaric acid | 0.24 | 0.78 | 0.10 | 0.03 | 40 | [11] |
| ellagic acid | 1.65 | 5.51 | - | 53.79 | 800 | [24] |
| Stilbenoids | ||||||
| t-resveratrol | 0.07 | 2.36 | - | - | 224 | [12] |
| cis-resveratrol | 0.02 | - | - | - | 8 | [13] |
| piceatannol | 0.07 | 0.86 | - | - | 12 | [14] |
| t-piceid | 0.05 | 0.79 | - | 0.07 | 15 | [15] |
| cis-piceid | 0.16 | 0.34 | - | <L.O.Q. | 8 | [16] |
| isorhapontin | 0.05 | 0.04 | - | - | 8 | [17] |
| Chalcones | ||||||
| phlorizin | 0.03 | 0.36 | 0.03 | 0.16 | 4 | [18] |
| Flavones | ||||||
| sinensetin | - | 0.01 | - | - | 160 | [19] |
| luteolin | 0.02 | - | 10.94 | 1.40 | 200 | [20] |
| luteolin-7-O-Glc | 0.19 | 0.24 | 23.86 | 31.86 | 12 | [25] |
| Flavan-3-ols | ||||||
| catechin | 16.30 | 46.97 | <L.O.Q. | 0.04 | 20 | [21] |
| epicatechin | 13.35 | 5.50 | - | 0.91 | 400 | [22] |
| epigallocatechin | 1.05 | 3.44 | - | 1.40 | 16 | [23] |
| gallocatechin | 0.82 | 36.70 | - | 0.94 | 20 | [26] |
| catechin gallate | 0.07 | - | - | - | 20 | [27] |
| epicatechin gallate | 0.14 | 8.05 | - | 0.50 | 2 | [28] |
| procyanidin B1 | 7.71 | 68.93 | - | 0.10 | 120 | [29] |
| procyanidin B2 + B4 | 358.93 | 44.43 | - | 38.34 | 80 | [30] |
| procyanidin B3 (as B1) | 4.19 | 30.77 | - | 0.14 | 20 | [31] |
| Flavonols | ||||||
| kaempferol | 0.27 | - | - | - | 8 | [32] |
| quercetin | 3.46 | 1.36 | 1.46 | - | 8 | [33] |
| quercetin-3-Rha | 0.01 | 0.24 | 5.49 | 7.87 | 8 | [34] |
| myricitrin | 0.02 | 0.02 | - | 0.03 | 20 | [35] |
| quercetin-3-Glc + quercetin-3-Gal (as que-3-Glc) | 0.32 | 5.59 | 7.03 | 142.94 | 12 | [36] |
| isorhamnetin-3-Glc | 0.14 | 0.43 | 0.10 | 0.39 | 8 | [37] |
| syringetin-3-Glc + syringetin-3-Gal (as syr-3-Glc) | 2.68 | 0.31 | - | - | 8 | [38] |
| rutin | 0.08 | 2.41 | 39.42 | 71.42 | 16 | [39] |
| quercetin-3,4-diglucoside | 0.06 | 0.05 | 0.80 | 0.95 | 20 | [40] |
| quercetin-3-glucuronide | 2.27 | 16.11 | 0.02 | 0.12 | 40 | [41] |
| kaempferol-3-glucuronide | 0.03 | 0.59 | - | - | 8 | [42] |
| arbutin | - | 0.34 | 0.07 | 0.14 | 200 | [43] |
| kaempferol-3-rutinoside | - | 0.31 | 0.87 | 9.89 | 4 | [44] |
| isorhamnetin-3-rutinoside | - | 0.31 | 0.35 | 0.58 | 8 | [45] |
| Flavanonols | ||||||
| taxifolin | 0.30 | 6.19 | 0.71 | 0.16 | 12 | [46] |
| Agent | 0.4 (g L−1 FW) | 4 (g L−1 FW) | 40 (g L−1 FW) | Mean |
|---|---|---|---|---|
| Ailanthus leaves | 0.51 | 0.50 | 0.39 | 0.46 |
| Grape marc | 0.53 | 0.54 | 0.64 | 0.57 |
| Olive leaves | 0.49 | 0.54 | 0.45 | 0.49 |
| Grape stalks | 0.51 | 0.57 | 0.48 | 0.52 |
| Mean | 0.51 | 0.54 | 0.49 | 0.51 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filippi, A.; Petrussa, E.; Boscutti, F.; Vuerich, M.; Vrhovsek, U.; Rabiei, Z.; Braidot, E. Bioactive Polyphenols Modulate Enzymes Involved in Grapevine Pathogenesis and Chitinase Activity at Increasing Complexity Levels. Int. J. Mol. Sci. 2019, 20, 6357. https://doi.org/10.3390/ijms20246357
Filippi A, Petrussa E, Boscutti F, Vuerich M, Vrhovsek U, Rabiei Z, Braidot E. Bioactive Polyphenols Modulate Enzymes Involved in Grapevine Pathogenesis and Chitinase Activity at Increasing Complexity Levels. International Journal of Molecular Sciences. 2019; 20(24):6357. https://doi.org/10.3390/ijms20246357
Chicago/Turabian StyleFilippi, Antonio, Elisa Petrussa, Francesco Boscutti, Marco Vuerich, Urska Vrhovsek, Zohreh Rabiei, and Enrico Braidot. 2019. "Bioactive Polyphenols Modulate Enzymes Involved in Grapevine Pathogenesis and Chitinase Activity at Increasing Complexity Levels" International Journal of Molecular Sciences 20, no. 24: 6357. https://doi.org/10.3390/ijms20246357
APA StyleFilippi, A., Petrussa, E., Boscutti, F., Vuerich, M., Vrhovsek, U., Rabiei, Z., & Braidot, E. (2019). Bioactive Polyphenols Modulate Enzymes Involved in Grapevine Pathogenesis and Chitinase Activity at Increasing Complexity Levels. International Journal of Molecular Sciences, 20(24), 6357. https://doi.org/10.3390/ijms20246357






