Role of Immunogenetics in the Outcome of HCMV Infection: Implications for Ageing
Abstract
:1. Introduction
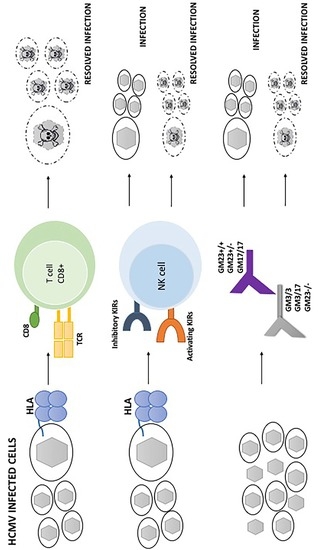
2. HLA and HCMV
3. KIR and HCMV
4. GM Allotypes and HCMV
5. Summary
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FcγR | fragment crystallisable γ receptor |
| gB | glycoprotein B |
| GWAS | genome-wide association study |
| GM | genetic marker |
| HCMV | human cytomegalovirus |
| HLA | human leukocyte antigens |
| IGHG | immunoglobulin heavy gamma |
| KIRs | killer cell immunoglobulin-like receptors |
| NK | natural killer |
References
- Burgner, D.; Jamieson, S.E.; Blackwell, J.M. Genetic susceptibility to infectious diseases: Big is beautiful, but will bigger be even better? Lancet Infect. Dis. 2006, 6, 653–663. [Google Scholar] [CrossRef]
- Lederberg, J.J.B.S. Haldane (1949) on infectious disease and evolution. Genetics 1999, 153, 1–3. [Google Scholar] [PubMed]
- Souquette, A.; Frere, J.; Smithey, M.; Sauce, D.; Thomas, P.G. A constant companion: Immune recognition and response to cytomegalovirus with aging and implications for immune fitness. Geroscience 2017, 39, 293–330. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, P.; Baraniak, I.; Reeves, M. The pathogenesis of human cytomegalovirus. J. Pathol. 2015, 235, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Hassan, J.; O’Neill, D.; Honari, B.; De Gascun, C.; Connell, J.; Keogan, M.; Hickey, D. Cytomegalovirus Infection in Ireland: Seroprevalence, HLA Class I Alleles, and Implications. Medicine (Baltimore) 2016, 95, e2735. [Google Scholar] [CrossRef] [PubMed]
- Futohi, F.; Saber, A.; Nemati, E.; Einollahi, B.; Rostami, Z. Human Leukocyte Antigen Alleles and Cytomegalovirus Infection after Renal Transplantation. Nephrourol. Mon. 2015, 7, e31635. [Google Scholar] [CrossRef]
- Roizman, B.; Pellet, P.E. The Family Herpesviridae: A Brief Introduction. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Raven Press: New York, NY, USA, 2001; pp. 2381–2397. [Google Scholar]
- Wreghitt, T.G.; Teare, E.L.; Sule, O.; Devi, R.; Rice, P. Cytomegalovirus infection in immunocompetent patients. Clin. Infect. Dis. 2003, 37, 1603–1606. [Google Scholar] [CrossRef]
- Cannon, M.J.; Schmid, D.S.; Hyde, T.B. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 2010, 20, 202–213. [Google Scholar] [CrossRef]
- Pawelec, G.; McElhaney, J.E.; Aiello, A.E.; Derhovanessian, E. The impact of CMV infection on survival in older humans. Curr. Opin. Immunol. 2012, 24, 507–511. [Google Scholar] [CrossRef]
- Cook, C.H. Cytomegalovirus reactivation in “immunocompetent” patients: A call for scientific prophylaxis. J. Infect. Dis. 2010, 196, 1273–1275. [Google Scholar] [CrossRef]
- Gandhi, M.K.; Khanna, R. Human cytomegalovirus: Clinical aspects, immune regulation, and emerging treatments. Lancet Infect. Dis. 2004, 4, 725–738. [Google Scholar] [CrossRef]
- Hummel, M.; Abecassis, M.M. A model for reactivation of CMV from latency. J. Clin. Virol. 2002, S2, S123–S136. [Google Scholar] [CrossRef]
- La Rosa, C.; Diamond, D.J. The immune response to human CMV. Future Virol. 2012, 7, 279–293. [Google Scholar] [CrossRef]
- Jackson, S.E.; Redeker, A.; Arens, R.; van Baarle, D.; van den Berg, S.P.H.; Benedict, C.A.; Čičin-Šain, L.; Hill, A.B.; Wills, M.R. CMV immune evasion and manipulation of the immune system with aging. Geroscience 2017, 39, 273–291. [Google Scholar] [CrossRef]
- Miller-Kittrell, M.; Sparer, T.E. Feeling manipulated: Cytomegalovirus immune manipulation. Virol. J. 2009, 6, 4. [Google Scholar] [CrossRef]
- Accardi, G.; Caruso, C. Immune-inflammatory responses in the elderly: An update. Immun. Ageing 2018, 15, 11. [Google Scholar] [CrossRef]
- Nikolich-Žugich, J.; van Lier, R.A.W. Cytomegalovirus (CMV) research in immune senescence comes of age: Overview of the 6th International Workshop on CMV and Immunosenescence. Geroscience 2017, 39, 245–249. [Google Scholar] [CrossRef]
- Parry, H.M.; Zuo, J.; Frumento, G.; Mirajkar, N.; Inman, C.; Edwards, E.; Griffiths, M.; Pratt, G.; Moss, P. Cytomegalovirus viral load within blood increases markedly in healthy people over the age of 70 years. Immun. Ageing 2016, 13, 1. [Google Scholar] [CrossRef]
- Broadley, I.; Pera, A.; Morrow, G.; Davies, K.A.; Kern, F. Expansions of Cytotoxic CD4(+)CD28(-) T Cells Drive Excess Cardiovascular Mortality in Rheumatoid Arthritis and Other Chronic Inflammatory Conditions and Are Triggered by CMV Infection. Front. Immunol. 2017, 8, 195. [Google Scholar] [CrossRef]
- Yang, F.J.; Shu, K.H.; Chen, H.Y.; Chen, I.Y.; Lay, F.Y.; Chuang, Y.F.; Wu, C.S.; Tsai, W.C.; Peng, Y.S.; Hsu, S.P.; et al. Anti-cytomegalovirus IgG antibody titer is positively associated with advanced T cell differentiation and coronary artery disease in end-stage renal disease. Immun. Ageing 2018, 15, 15. [Google Scholar] [CrossRef]
- Weinberger, B.; Keller, M.; Grubeck-Loebenstein, B. Long-term maintenance of diphtheria-specific antibodies after booster vaccination is hampered by latent infection with Cytomegalovirus. Immun. Ageing 2017, 14, 16. [Google Scholar] [CrossRef]
- Kilgour, A.H.; Firth, C.; Harrison, R.; Moss, P.; Bastin, M.E.; Wardlaw, J.M.; Deary, I.J.; Starr, J.M. Seropositivity for CMV and IL-6 levels are associated with grip strength and muscle size in the elderly. Immun. Ageing 2013, 10, 33. [Google Scholar] [CrossRef]
- Caruso, C.; Candore, G.; Romano, G.C.; Lio, D.; Bonafè, M.; Valensin, S.; Franceschi, C. Immunogenetics of longevity. Is major histocompatibility complex polymorphism relevant to the control of human longevity? A review of literature data. Mech. Ageing Dev. 2001, 122, 445–462. [Google Scholar] [CrossRef]
- Caruso, C.; Candore, G.; Colonna Romano, G.; Lio, D.; Bonafè, M.; Valensin, S.; Franceschi, C. HLA, aging, and longevity: A critical reappraisal. Hum. Immunol. 2000, 61, 942–949. [Google Scholar] [CrossRef]
- Rizzo, C.; Accardi, G.; Caruso, C. Genetic variation in human leukocyte antigen and susceptibility to acute myeloid leukemia. Acta Haematol. 2015, 133, 162–163. [Google Scholar] [CrossRef]
- Blancho, G.; Josien, R.; Douillard, D.; Bignon, J.D.; Cesbron, A.; Soulillou, J.P. The influence of HLA A-B-DR matching on cytomegalovirus disease after renal transplantation. Evidence that HLA-DR7-matched recipients are more susceptible to cytomegalovirus disease. Transplantation 1992, 54, 871–874. [Google Scholar] [CrossRef]
- Kraat, Y.J.; Christiaans, M.H.; Nieman, F.H.; van den Berg-Loonen, P.M.; van Hooff, J.P.; Bruggeman, C. A Increased frequency of CMV infection in HLA-DR7 matched renal allograft recipients. Lancet 1993, 341, 494–495. [Google Scholar] [CrossRef]
- Kraat, Y.J.; Christiaans, M.H.; Nieman, F.H.; van den Berg-Loonen, P.M.; van Hooff, J.P.; Bruggeman, C.A. Risk factors for cytomegalovirus infection and disease in renal transplant recipients: HLA-DR7 and triple therapy. Transpl. Int. 1994, 7, 362–367. [Google Scholar] [CrossRef]
- Gambino, C.M.; Aiello, A.; Accardi, G.; Caruso, C.; Candore, G. Autoimmune diseases and 8.1 ancestral haplotype: An update. HLA 2018, 92, 137–143. [Google Scholar] [CrossRef]
- Aiello, A.; Candore, G.; Accardi, G.; Caruso, C.; Colomba, C.; Duro, G.; Gambino, C.M.; Ligotti, M.E.; Di Bona, D. Translation of Basic Research into Clinics: Killer Immunoglobulin-like Receptors Genes in Autoimmune and Infectious Diseases. Curr. Pharm. Des. 2018, 24, 3113–3122. [Google Scholar] [CrossRef]
- Misra, M.K.; Augusto, D.G.; Martin, G.M.; Nemat-Gorgani, N.; Sauter, J.; Hofmann, J.A.; Traherne, J.A.; González-Quezada, B.; Gorodezky, C.; Bultitude, W.P.; et al. Report from the Killer-cell Immunoglobulin-like Receptors (KIR) component of the 17thInternational HLA and Immunogenetics Workshop. Hum. Immunol. 2018. [Google Scholar] [CrossRef]
- Shilling, H.G.; Young, N.; Guethlein, L.A.; Cheng, N.W.; Gardiner, C.M.; Tyan, D.; Parham, P. Genetic control of human NK cell repertoire. J. Immunol. 2002, 169, 239–247. [Google Scholar] [CrossRef]
- Aiello, A.; Accardi, G.; Candore, G.; Gambino, C.M.; Caruso, C.; Di Bona, D. The importance of interactions between KIRs and HLA ligands in the development of autoimmune and viral diseases. In Causality and Chance in Ageing, Age-Related Diseases and Longevity, Proceedings of the Symposium Updated in Pathobiology, Palermo, Italy, 24 March 2017; Caruso, C., Accardi, G., Eds.; Unipapress: Palermo, Italy, 2017; pp. 91–110. [Google Scholar]
- Lanier, L.L. NKG2D Receptor and Its Ligands in Host Defense. Cancer Immunol. Res. 2015, 3, 575–582. [Google Scholar] [CrossRef]
- Gazit, R.; Garty, B.Z.; Monselise, Y.; Hoffer, V.; Finkelstein, Y.; Markel, G.; Katz, G.; Hanna, J.; Achdout, H.; Gruda, R.; et al. Expression of KIR2DL1 on the entire NK cell population: A possible novel immunodeficiency syndrome. Blood 2004, 103, 1965–1966. [Google Scholar]
- Stern, M.; Elsässer, H.; Hönger, G.; Steiger, J.; Schaub, S.; Hess, C. The number of activating KIR genes inversely correlates with the rate of CMV infection/reactivation in kidney transplant recipients. Am. J. Transpl. 2008, 8, 1312–1317. [Google Scholar] [CrossRef]
- Hadaya, K.; de Rham, C.; Bandelier, C.; Bandelier, C.; Ferrari-Lacraz, S.; Jendly, S.; Berney, T.; Buhler, L.; Kaiser, L.; Seebach, J.D.; et al. Natural killer cell receptor repertoire and their ligands, and the risk of CMV infection after kidney transplantation. Am. J. Transpl. 2008, 8, 2674–2683. [Google Scholar] [CrossRef]
- Zaia, J.A.; Sun, J.Y.; Gallez-Hawkins, G.M.; Thao, L.; Oki, A.; Lacey, S.F.; Dagis, A.; Palmer, J.; Diamond, D.J.; Forman, S.J.; et al. The effect of single and combined activating killer immunoglobulin-like receptor genotypes on cytomegalovirus infection and immunity after hematopoietic cell transplantation. Biol. Blood Marrow Transpl. 2009, 15, 315–325. [Google Scholar] [CrossRef]
- Cook, M.; Briggs, D.; Craddock, C.; Mahendra, P.; Milligan, D.; Fegan, C.; Darbyshire, P.; Lawson, S.; Boxall, E.; Moss, P. Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell replete stem cell transplantation. Blood 2006, 107, 1230–1232. [Google Scholar] [CrossRef]
- Di Bona, D.; Scafidi, V.; Plaia, A.; Colomba, C.; Nuzzo, D.; Occhino, C.; Tuttolomondo, A.; Giammanco, G.; De Grazia, S.; Montalto, G.; et al. HLA and killer cell immunoglobulin-like receptors influence the natural course of CMV infection. J. Infect. Dis. 2014, 210, 1083–1089. [Google Scholar] [CrossRef]
- Piazza, A.; Olivetti, E.; Griffo, R.M.; Rendine, S.; Amoroso, A.; Barbanti, M.; Caruso, C.; Conighi, C.; Conte, R.; Favoino, B.; et al. The distribution of HLA antigens in Italy. Gene Geogr. 1989, 3, 141–164. [Google Scholar]
- Kruskall, M.S.; Eynon, E.E.; Awdeh, Z.; Alper, C.A.; Yunis, E.J. Identification of HLA-B44 subtypes associated with extended MHC haplotypes. Immunogenetics 1987, 26, 216–219. [Google Scholar] [CrossRef]
- Naumova, E.; Ivanova, M.; Pawelec, G.; Constantinescu, I.; Bogunia-Kubik, K.; Lange, A.; Qguz, F.; Carin, M.; Franceschi, C.; Caruso, C.; et al. ‘Immunogenetics of Aging’: Report on the activities of the 15th International HLA and Immunogenetics Working Group and 15th International HLA and Immunogenetics Workshop. Tissue Antigens 2011, 77, 187–189. [Google Scholar] [CrossRef]
- Naumova, E.; Ivanova, M.; Pawelec, G.; Constantinescu, I.; Bogunia-Kubik, K.; Lange, A.; Oguz, F.; Ozdilli, K.; Franceschi, C.; Caruso, C.; et al. 16(th) IHIW: Immunogenetics of aging. Int. J. Immunogenet. 2013, 40, 77–81. [Google Scholar] [CrossRef]
- Lefranc, M.P.; Lefranc, G. Human Gm, Km, and Am allotypes and their molecular characterization: A remarkable demonstration of polymorphism. Methods Mol. Biol. 2012, 882, 635–680. [Google Scholar]
- Oxelius, V.A.; Pandey, J.P. Human immunoglobulin constant heavy G chain (IGHG) (Fcγ) (GM) genes, defining innate variants of IgG molecules and B cells, have impact on disease and therapy. Clin. Immunol. 2013, 149, 475–486. [Google Scholar] [CrossRef]
- de Vries, R.R.; Meera Khan, P.; Bernini, L.F.; van Loghem, E.; van Rood, J.J. Genetic control of survival in epidemics. J. Immunogenet. 1979, 6, 271–287. [Google Scholar] [CrossRef]
- Caruso, C.; Pandey, J.P.; Puca, A.A. Genetics of exceptional longevity: Possible role of GM allotypes. Immun. Ageing 2018, 15, 25. [Google Scholar] [CrossRef]
- Pandey, J.P.; Kistner-Griffin, E.; Radwan, F.F.; Kaur, N.; Namboodiri, A.M.; Black, L.; Butler, M.A.; Carreon, T.; Ruder, A.M. Immunoglobulin genes influence the magnitude of humoral immunity to cytomegalovirus glycoprotein B. J. Infect. Dis. 2014, 210, 1823–1826. [Google Scholar] [CrossRef]
- Kuparinen, T.; Seppälä, I.; Jylhävä, J.; Marttila, S.; Aittoniemi, J.; Kettunen, J.; Viikari, J.; Kähönen, M.; Raitakari, O.; Lehtimäki, T.; et al. Genome-wide association study does not reveal major genetic determinants for anti-cytomegalovirus antibody response. Genes Immun. 2012, 13, 184–190. [Google Scholar] [CrossRef]
- Pandey, J.P. Candidate gene approach’s missing link. Science 2010, 329, 1148. [Google Scholar] [CrossRef]
- Pandey, J.P. Genome wide association studies and assessment of the risk of disease. N. Engl. J. Med. 2010, 363, 2076–2077. [Google Scholar]
- Casadevall, A.; Pirofski, L.A. A new synthesis for antibody-mediated immunity. Nat. Immunol. 2012, 13, 21–28. [Google Scholar] [CrossRef]
- Corrales-Aguilar, E.; Hoffmann, K.; Hengel, H. CMV-encoded Fcγ receptors: Modulators at the interface of innate and adaptive immunity. Semin. Immunopathol. 2014, 36, 627–640. [Google Scholar] [CrossRef]
- Namboodiri, A.M.; Pandey, J.P. The human cytomegalovirus TRL11/IRL11-encoded FcγR binds differentially to allelic variants of immunoglobulin G1. Arch. Virol. 2011, 156, 907–910. [Google Scholar] [CrossRef]
- Pandey, J.P.; Namboodiri, A.M.; Radwan, F.F.; Nietert, P.J. The decoy Fcγ receptor encoded by the cytomegalovirus UL119-UL118 gene has differential affinity to IgG proteins expressing different GM allotypes. Hum. Immunol. 2015, 76, 591–594. [Google Scholar]
- Pandey, J.P.; Namboodiri, A.M.; Mohan, S.; Nietert, P.J.; Peterson, L. Genetic markers of immunoglobulin G and immunity to cytomegalovirus in patients with breast cancer. Cell. Immunol. 2017, 312, 67–70. [Google Scholar] [CrossRef]
- Di Bona, D.; Accardi, G.; Aiello, A.; Bilancia, M.; Candore, G.; Colomba, C.; Caruso, C.; Duro, G.; Gambino, C.M.; Macchia, L.; et al. Association between γ marker, human leucocyte antigens and killer immunoglobulin-like receptors and the natural course of human cytomegalovirus infection: A pilot study performed in a Sicilian population. Immunology 2018, 153, 523–531. [Google Scholar] [CrossRef]
- Puca, A.A.; Ferrario, A.; Maciag, A.; Accardi, G.; Aiello, A.; Gambino, C.M.; Candore, G.; Caruso, C.; Namboodiri, A.M.; Pandey, J.P. Association of immunoglobulin GM allotypes with longevity in long-living individuals from Southern Italy. Immun. Ageing 2018, 15, 26. [Google Scholar] [CrossRef]

| HLA-Bw4 Groups | HCMV Patients (N = 31) | |
|---|---|---|
| N | % | |
| HLA-Bw4T | ||
| B*44 | 11 | 33.30 |
| B*13 | 4 | 12.12 |
| B*35 | 3 | 9.09 |
| B*14 | 1 | 3.03 |
| B*40 | 1 | 3.03 |
| B*27 | 2 | 6.06 |
| B*73 | 1 | 3.03 |
| B*50 | 1 | 3.03 |
| B*07 | 0 | 0 |
| B*18 | 0 | 0 |
| HLA-Bw4I | ||
| B*27 | 1 | 3.03 |
| B*14 | 1 | 3.03 |
| B*39 | 1 | 3.03 |
| B*57 | 3 | 9.09 |
| B*35 | 5 | 15.15 |
| B*38 | 2 | 6.06 |
| B*40 | 1 | 3.03 |
| B*49 | 3 | 9.09 |
| B*50 | 2 | 6.06 |
| B*08 | 1 | 3.03 |
| B*15 | 1 | 3.03 |
| B*51 | 4 | 12.12 |
| B*67 | 1 | 3.03 |
| B*18 | 0 | 0 |
| B*53 | 1 | 3.03 |
| B*58 | 3 | 9.09 |
| B*44 | 1 | 3.03 |
| B*52 | 1 | 3.03 |
| HLA-Bw4T/HLA-Bw4I | ||
| B*44 | 4 | 12.12 |
| B*13 | 0 | 0 |
| B*49 | 1 | 3.03 |
| B*53 | 1 | 3.03 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aiello, A.; Accardi, G.; Candore, G.; Caruso, C.; Colomba, C.; Di Bona, D.; Duro, G.; Gambino, C.M.; Ligotti, M.E.; Pandey, J.P. Role of Immunogenetics in the Outcome of HCMV Infection: Implications for Ageing. Int. J. Mol. Sci. 2019, 20, 685. https://doi.org/10.3390/ijms20030685
Aiello A, Accardi G, Candore G, Caruso C, Colomba C, Di Bona D, Duro G, Gambino CM, Ligotti ME, Pandey JP. Role of Immunogenetics in the Outcome of HCMV Infection: Implications for Ageing. International Journal of Molecular Sciences. 2019; 20(3):685. https://doi.org/10.3390/ijms20030685
Chicago/Turabian StyleAiello, Anna, Giulia Accardi, Giuseppina Candore, Calogero Caruso, Claudia Colomba, Danilo Di Bona, Giovanni Duro, Caterina Maria Gambino, Mattia Emanuela Ligotti, and Janardan P. Pandey. 2019. "Role of Immunogenetics in the Outcome of HCMV Infection: Implications for Ageing" International Journal of Molecular Sciences 20, no. 3: 685. https://doi.org/10.3390/ijms20030685
APA StyleAiello, A., Accardi, G., Candore, G., Caruso, C., Colomba, C., Di Bona, D., Duro, G., Gambino, C. M., Ligotti, M. E., & Pandey, J. P. (2019). Role of Immunogenetics in the Outcome of HCMV Infection: Implications for Ageing. International Journal of Molecular Sciences, 20(3), 685. https://doi.org/10.3390/ijms20030685