Cancer-Specifically Re-Spliced TSG101 mRNA Promotes Invasion and Metastasis of Nasopharyngeal Carcinoma
Abstract
1. Introduction
2. Results
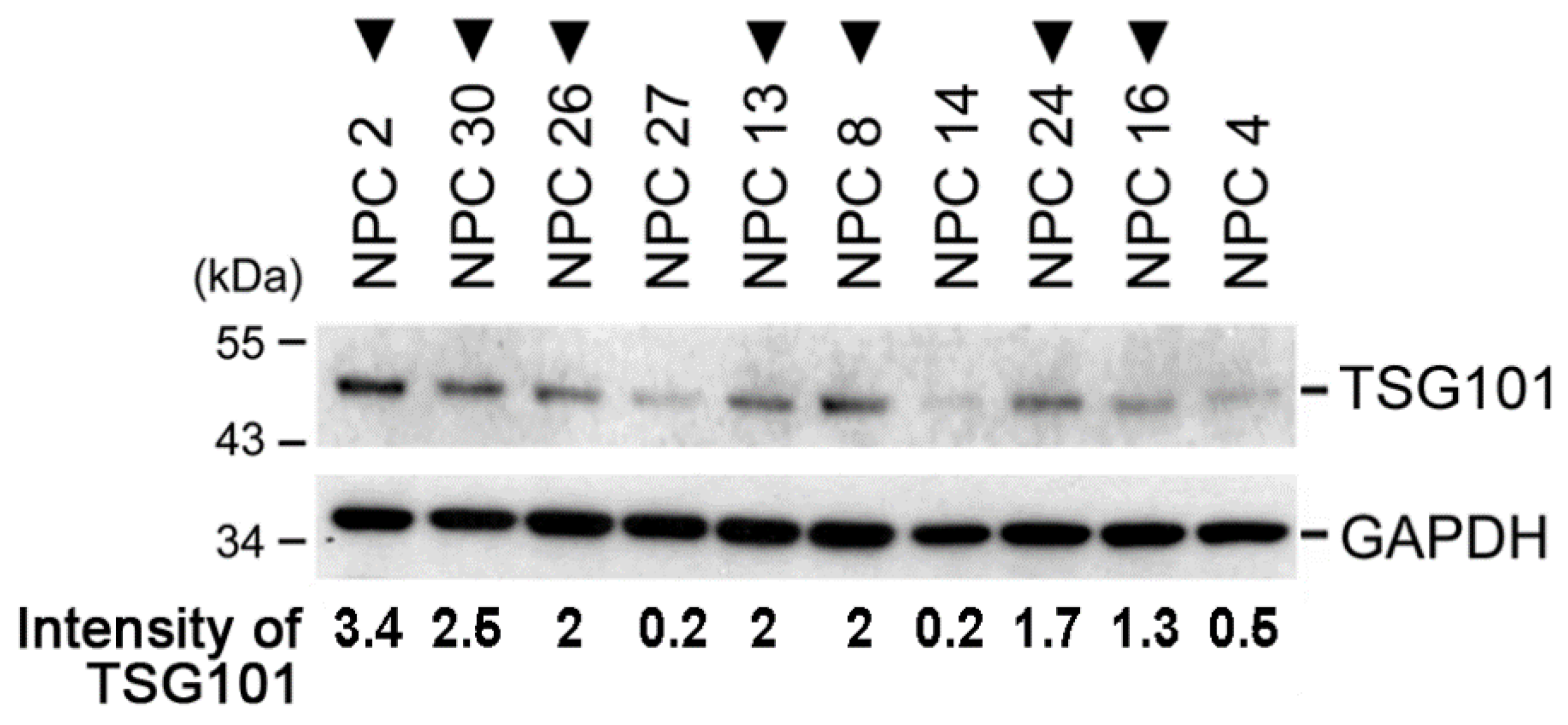
2.1. TSG101 Pre-mRNA Is Aberrantly Spliced in Nasopharyngeal Carcinoma Tissues from Patients
2.2. TSG∆154-1054 Expression Augments the Protein Levels of TSG101
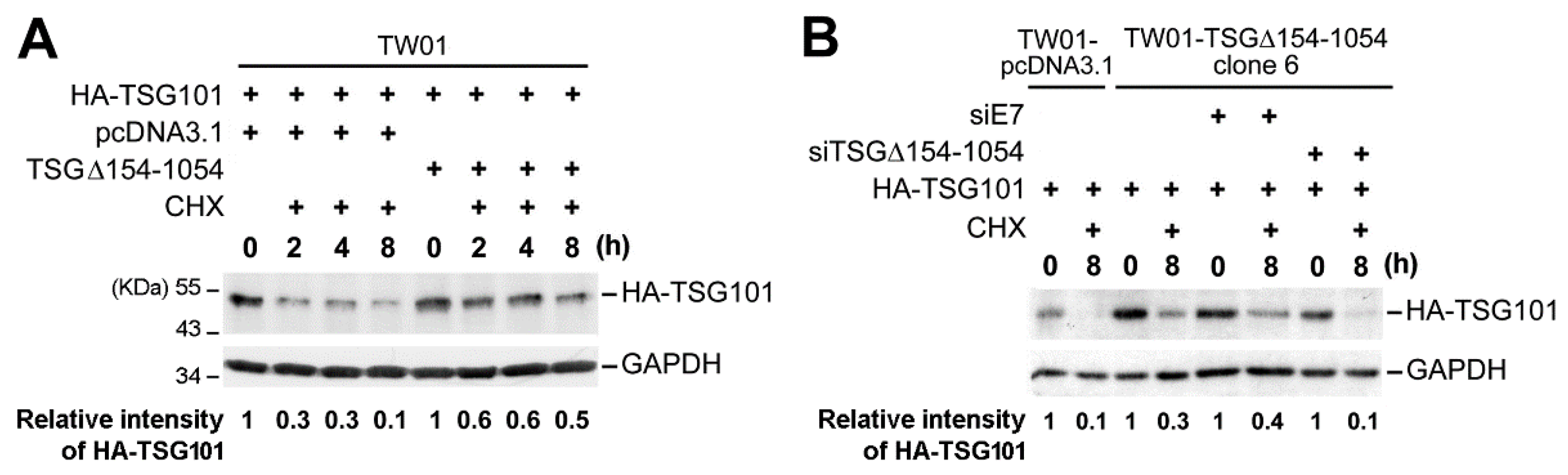
2.3. TSG∆154-1054 Induces TSG101 Protein Stabilization
2.4. TSG∆154-1054 Increases the TSG101-Dependent Metastatic Activity of Tumor Cells
2.5. TSG∆154-1054 Expression Correlates with Tumor Metastasis in Patients of NPC
3. Discussion
4. Materials and Methods
4.1. Primary Specimens from NPC Patients and Cell Lines
4.2. Construction of Plasmids and siRNAs
4.3. Transfection of Cells and siRNA Knockdown Experiments
4.4. RNA Extraction, Reverse Transcription, and PCR Analysis
4.5. Immunoblot Analysis
4.6. Cycloheximide Chase Assay for TSG101 Protein Stability
4.7. Scratch-Wound Assay for Cell Migration Ability
4.8. Transwell Cell Invation Assay
4.9. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TSG101 | Tumor susceptibility 101 gene |
| NPC | Nasopharyngeal carcinoma |
| ESCRT | Endosomal sorting complex required for transport |
| PCR | Polymerase chain reaction |
| DMEM | Dulbecco’s modified Eagle’s medium |
| FCS | Fetal calf serum |
References
- Tanaka, N.; Kyuuma, M.; Sugamura, K. Endosomal sorting complex required for transport proteins in cancer pathogenesis, vesicular transport, and non-endosomal functions. Cancer Sci. 2008, 99, 1293–1303. [Google Scholar] [CrossRef] [PubMed]
- Scourfield, E.J.; Martin-Serrano, J. Growing functions of the ESCRT machinery in cell biology and viral replication. Biochem. Soc. Trans. 2017, 45, 613–634. [Google Scholar] [CrossRef]
- Brodsky, F.M.; Sosa, R.T.; Ybe, J.A.; O’Halloran, T.J. Unconventional functions for clathrin, ESCRTs, and other endocytic regulators in the cytoskeleton, cell cycle, nucleus, and beyond: Links to human disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a017004. [Google Scholar] [CrossRef] [PubMed]
- Alfred, V.; Vaccari, T. When membranes need an ESCRT: Endosomal sorting and membrane remodelling in health and disease. Swiss Med. Wkly. 2016, 146, w14347. [Google Scholar] [CrossRef] [PubMed]
- Slagsvold, T.; Pattni, K.; Malerod, L.; Stenmark, H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006, 16, 317–326. [Google Scholar] [CrossRef]
- Jiang, Y.; Ou, Y.; Cheng, X. Role of TSG101 in cancer. Front. Biosci. 2013, 18, 279–288. [Google Scholar]
- Li, L.; Cohen, S.N. Tsg101: A novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell 1996, 85, 319–329. [Google Scholar] [CrossRef]
- Ruland, J.; Sirard, C.; Elia, A.; MacPherson, D.; Wakeham, A.; Li, L.; de la Pompa, J.L.; Cohen, S.N.; Mak, T.W. p53 accumulation, defective cell proliferation, and early embryonic lethality in mice lacking tsg101. Proc. Natl. Acad. Sci. USA 2001, 98, 1859–1864. [Google Scholar] [CrossRef]
- Wagner, K.U.; Krempler, A.; Qi, Y.; Park, K.; Henry, M.D.; Triplett, A.A.; Riedlinger, G.; Rucker, I.E.; Hennighausen, L. Tsg101 is essential for cell growth, proliferation, and cell survival of embryonic and adult tissues. Mol. Cell. Biol. 2003, 23, 150–162. [Google Scholar] [CrossRef]
- Krempler, A.; Henry, M.D.; Triplett, A.A.; Wagner, K.U. Targeted deletion of the Tsg101 gene results in cell cycle arrest at G1/S and p53–independent cell death. J. Biol. Chem. 2002, 277, 43216–43223. [Google Scholar] [CrossRef]
- Zhu, G.; Gilchrist, R.; Borley, N.; Chng, H.W.; Morgan, M.; Marshall, J.F.; Camplejohn, R.S.; Muir, G.H.; Hart, I.R. Reduction of TSG101 protein has a negative impact on tumor cell growth. Int. J. Cancer 2004, 109, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Pan, Y.; Han, Z.; Hong, L.; Liu, N.; Han, S.; Yao, L.; Xie, H.; Zhaxi, C.; Shi, Y.; et al. Reversal of multidrug resistance of gastric cancer cells by downregulation of TSG101 with TSG101siRNA. Cancer Biol. Ther. 2004, 3, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Chua, H.H.; Lee, H.H.; Chang, S.S.; Lu, C.C.; Yeh, T.H.; Hsu, T.Y.; Cheng, T.H.; Cheng, J.T.; Chen, M.R.; Tsai, C.H. Role of the TSG101 gene in Epstein-Barr virus late gene transcription. J. Virol. 2007, 81, 2459–2471. [Google Scholar] [CrossRef] [PubMed]
- Venables, J.P. Unbalanced alternative splicing and its significance in cancer. Bioessays 2006, 28, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Venables, J.P. Aberrant and alternative splicing in cancer. Cancer Res. 2004, 64, 7647–7654. [Google Scholar] [CrossRef] [PubMed]
- Kalnina, Z.; Zayakin, P.; Silina, K.; Line, A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer 2005, 42, 342–357. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, X.; Francke, U.; Cohen, S.N. The TSG101 tumor susceptibility gene is located in chromosome 11 band p15 and is mutated in human breast cancer. Cell 1997, 88, 143–154. [Google Scholar] [CrossRef]
- Sun, Z.; Pan, J.; Bubley, G.; Balk, S.P. Frequent abnormalities of TSG101 transcripts in human prostate cancer. Oncogene 1997, 15, 3121–3125. [Google Scholar] [CrossRef]
- Gayther, S.A.; Barski, P.; Batley, S.J.; Li, L.; de Foy, K.A.; Cohen, S.N.; Ponder, B.A.; Caldas, C. Aberrant splicing of the TSG101 and FHIT genes occurs frequently in multiple malignancies and in normal tissues and mimics alterations previously described in tumours. Oncogene 1997, 15, 2119–2126. [Google Scholar] [CrossRef]
- Oh, Y.; Proctor, M.L.; Fan, Y.H.; Su, L.K.; Hong, W.K.; Fong, K.M.; Sekido, Y.S.; Gazdar, A.F.; Minna, J.D.; Mao, L. TSG101 is not mutated in lung cancer but a shortened transcript is frequently expressed in small cell lung cancer. Oncogene 1998, 17, 1141–1148. [Google Scholar] [CrossRef]
- Lin, P.M.; Liu, T.C.; Chang, J.G.; Chen, T.P.; Lin, S.F. Aberrant TSG101 transcripts in acute myeloid leukaemia. Br. J. Haematol. 1998, 102, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Chen, P.H.; Lin, S.Y.; Chang, J.G. Analysis of aberrant transcription of TSG101 in hepatocellular carcinomas. Eur. J. Cancer 1999, 35, 302–308. [Google Scholar] [CrossRef]
- Ferrer, M.; Lopez-Borges, S.; Lazo, P.A. Expression of a new isoform of the tumor susceptibility TSG101 protein lacking a leucine zipper domain in Burkitt lymphoma cell lines. Oncogene 1999, 18, 2253–2259. [Google Scholar] [CrossRef] [PubMed]
- Klaes, R.; Kloor, M.; Willeke, F.; Melsheimer, P.; von Knebel Doeberitz, M.; Ridder, R. Significant increase of a specific variant TSG101 transcript during the progression of cervical neoplasia. Eur. J. Cancer 1999, 35, 733–737. [Google Scholar] [CrossRef]
- McIver, B.; Grebe, S.K.; Wang, L.; Hay, I.D.; Yokomizo, A.; Liu, W.; Goellner, J.R.; Grant, C.S.; Smith, D.I.; Eberhardt, N.L. FHIT and TSG101 in thyroid tumours: Aberrant transcripts reflect rare abnormal RNA processing events of uncertain pathogenetic or clinical significance. Clin. Endocrinol. 2000, 52, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.F.; Lin, P.M.; Liu, T.C.; Chang, J.G.; Sue, Y.C.; Chen, T.P. Clinical implications of aberrant TSG101 transcripts in acute myeloblastic leukemia. Leuk. Lymphoma 2000, 36, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.P.; Feinberg, A.P. Aberrant splicing but not mutations of TSG101 in human breast cancer. Cancer Res. 1997, 57, 3131–3134. [Google Scholar] [PubMed]
- Moyret-Lalle, C.; Duriez, C.; Van Kerckhove, J.; Gilbert, C.; Wang, Q.; Puisieux, A. p53 induction prevents accumulation of aberrant transcripts in cancer cells. Cancer Res. 2001, 61, 486–488. [Google Scholar]
- Kameyama, T.; Suzuki, H.; Mayeda, A. Re-splicing of mature mRNA in cancer cells promotes activation of distant weak alternative splice sites. Nucleic Acids Res. 2012, 40, 7896–7906. [Google Scholar] [CrossRef]
- Lo, Y.F.; Chen, T.C.; Chen, S.C.; Chao, C.C. Aberrant expression of TSG101 in Taiwan Chinese breast cancer. Breast Cancer Res. Treat. 2000, 60, 259–266. [Google Scholar] [CrossRef]
- Chua, H.H.; Huang, C.S.; Weng, P.L.; Yeh, T.H. TSG∆154-1054 splice variant increases TSG101 oncogenicity by inhibiting its E3-ligase-mediated proteasomal degradation. Oncotarget 2016, 7, 8240–8252. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liao, J.; Ruland, J.; Mak, T.W.; Cohen, S.N. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc. Natl. Acad. Sci. USA 2001, 98, 1619–1624. [Google Scholar] [CrossRef] [PubMed]
- McDonald, B.; Martin-Serrano, J. Regulation of Tsg101 expression by the steadiness box: A role of Tsg101-associated ligase. Mol. Biol. Cell 2008, 19, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Amit, I.; Yakir, L.; Katz, M.; Zwang, Y.; Marmor, M.D.; Citri, A.; Shtiegman, K.; Alroy, I.; Tuvia, S.; Reiss, Y.; et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004, 18, 1737–1752. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.H.; Lih, C.J.; Cohen, S.N. TSG101 protein steady-state level is regulated posttranslationally by an evolutionarily conserved COOH-terminal sequence. Cancer Res 2000, 60, 1736–1741. [Google Scholar] [PubMed]
- Liu, R.T.; Huang, C.C.; You, H.L.; Chou, F.F.; Hu, C.C.; Chao, F.P.; Chen, C.M.; Cheng, J.T. Overexpression of tumor susceptibility gene TSG101 in human papillary thyroid carcinomas. Oncogene 2002, 21, 4830–4837. [Google Scholar] [CrossRef] [PubMed]
- Young, T.W.; Rosen, D.G.; Mei, F.C.; Li, N.; Liu, J.; Wang, X.F.; Cheng, X. Up-regulation of tumor susceptibility gene 101 conveys poor prognosis through suppression of p21 expression in ovarian cancer. Clin. Cacer Res. 2007, 13, 3848–3854. [Google Scholar] [CrossRef]
- Oh, K.B.; Stanton, M.J.; West, W.W.; Todd, G.L.; Wagner, K.U. Tsg101 is upregulated in a subset of invasive human breast cancers and its targeted overexpression in transgenic mice reveals weak oncogenic properties for mammary cancer initiation. Oncogene 2007, 26, 5950–5959. [Google Scholar] [CrossRef]
- Carlton, J.G.; Martin-Serrano, J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science 2007, 316, 1908–1912. [Google Scholar] [CrossRef]
- Young, T.W.; Mei, F.C.; Rosen, D.G.; Yang, G.; Li, N.; Liu, J.; Cheng, X. Up-regulation of tumor susceptibility gene 101 protein in ovarian carcinomas revealed by proteomics analyses. Mol. Cell. Proteomics 2007, 6, 294–304. [Google Scholar] [CrossRef]
- Liu, Z.; Tian, Z.; Cao, K.; Zhang, B.; Wen, Q.; Zhou, X.; Yang, W.; Wang, T.; Shi, H.; Wang, R. TSG101 promotes the proliferation, migration and invasion of hepatocellular carcinoma cells by regulating the PEG10. J. Cell. Mol. Med. 2019, 23, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Saharat, K.; Lirdprapamongkol, K.; Chokchaichamnankit, D.; Srisomsap, C.; Svasti, J.; Paricharttanakul, N.M. Tumor Susceptibility Gene 101 Mediates Anoikis Resistance of Metastatic Thyroid Cancer Cells. Cancer Genom. Proteom. 2018, 15, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, Z.; Liu, D.; Li, D.; Zou, Q.; Yuan, Y.; Li, J.; Liang, L.; Chen, M.; Chen, S. TSG101 and PEG10 are prognostic markers in squamous cell/adenosquamous carcinomas and adenocarcinoma of the gallbladder. Oncol. Lett. 2014, 7, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Gupta, D.; Shankar, S.; Srivastava, R.K. Biomolecular characterization of exosomes released from cancer stem cells: Possible implications for biomarker and treatment of cancer. Oncotarget 2015, 6, 3280–3291. [Google Scholar] [CrossRef] [PubMed]



| Characteristics | No. of Patients | p Value ** | |
|---|---|---|---|
| TSG ∆154-1054 (+) * | TSG ∆154-1054 (−) | ||
| Age | |||
| Median (Extent) | 44.8 (32–62) | 47.3 (24–78) | |
| Gender | 0.825 | ||
| Male | 12 | 14 | |
| Female | 6 | 6 | |
| Pathology Subtype | 0.840 | ||
| Non-keratinizing | 4 | 5 | |
| Undifferentiated | 14 | 15 | |
| Primary Tumor Stage (T) | 0.165 | ||
| T1 | 2 | 2 | |
| T2 | 3 | 8 | |
| T3 | 3 | 1 | |
| T4 | 9 | 4 | |
| Nodal Stage (N) | 0.287 | ||
| N0 | 3 | 5 | |
| N1 | 4 | 6 | |
| N2 | 7 | 2 | |
| N3 | 3 | 2 | |
| Metastasis | 4 | 0 | 0.045 |
| Stage *** | 0.077 | ||
| Early (I + II) | 3 | 7 | |
| Late (III + IV) | 14 | 8 | |
| Total Number of Patients | 18 | 20 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chua, H.-H.; Kameyama, T.; Mayeda, A.; Yeh, T.-H. Cancer-Specifically Re-Spliced TSG101 mRNA Promotes Invasion and Metastasis of Nasopharyngeal Carcinoma. Int. J. Mol. Sci. 2019, 20, 773. https://doi.org/10.3390/ijms20030773
Chua H-H, Kameyama T, Mayeda A, Yeh T-H. Cancer-Specifically Re-Spliced TSG101 mRNA Promotes Invasion and Metastasis of Nasopharyngeal Carcinoma. International Journal of Molecular Sciences. 2019; 20(3):773. https://doi.org/10.3390/ijms20030773
Chicago/Turabian StyleChua, Huey-Huey, Toshiki Kameyama, Akila Mayeda, and Te-Huei Yeh. 2019. "Cancer-Specifically Re-Spliced TSG101 mRNA Promotes Invasion and Metastasis of Nasopharyngeal Carcinoma" International Journal of Molecular Sciences 20, no. 3: 773. https://doi.org/10.3390/ijms20030773
APA StyleChua, H.-H., Kameyama, T., Mayeda, A., & Yeh, T.-H. (2019). Cancer-Specifically Re-Spliced TSG101 mRNA Promotes Invasion and Metastasis of Nasopharyngeal Carcinoma. International Journal of Molecular Sciences, 20(3), 773. https://doi.org/10.3390/ijms20030773







