Classical Soybean (Glycine max (L.) Merr) Symbionts, Sinorhizobium fredii USDA191 and Bradyrhizobium diazoefficiens USDA110, Reveal Contrasting Symbiotic Phenotype on Pigeon Pea (Cajanus cajan (L.) Millsp)
Abstract
:1. Introduction
2. Results
2.1. Nodulation Outer Proteins (NOPs) of Sinorhizobium fredii USDA191, Bradyrhizobium diazoefficiens USDA110, and their T3SS Mutants
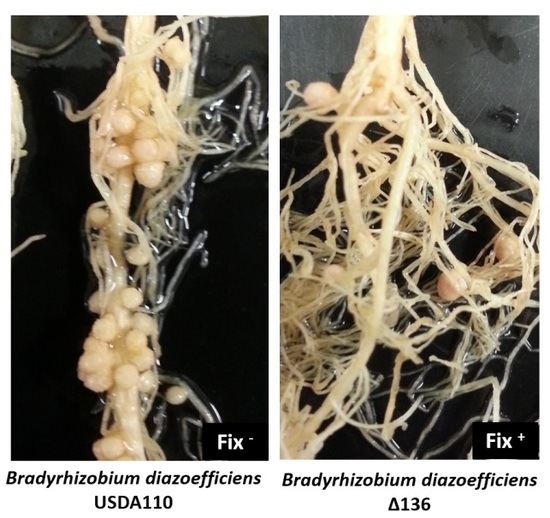
2.2. Sinorhizobium fredii USDA191 Exhibits Nitrogen Fixing Nodules While Bradyrhizobium diazoefficiens USDA110 Forms Ineffective Nodules on Pigeon Pea
2.3. Anatomy of Pigeon Pea Nodules Induced by Sinorhizobium fredii USDA191 and Bradyrhizobium diazoefficiens USDA110
2.4. Ultrastructural Observation of Pigeon Pea Nodules Produced by Sinorhizobium fredii USDA191 and Bradyrhizobium diazoefficiens USDA110 and their T3SS Mutants
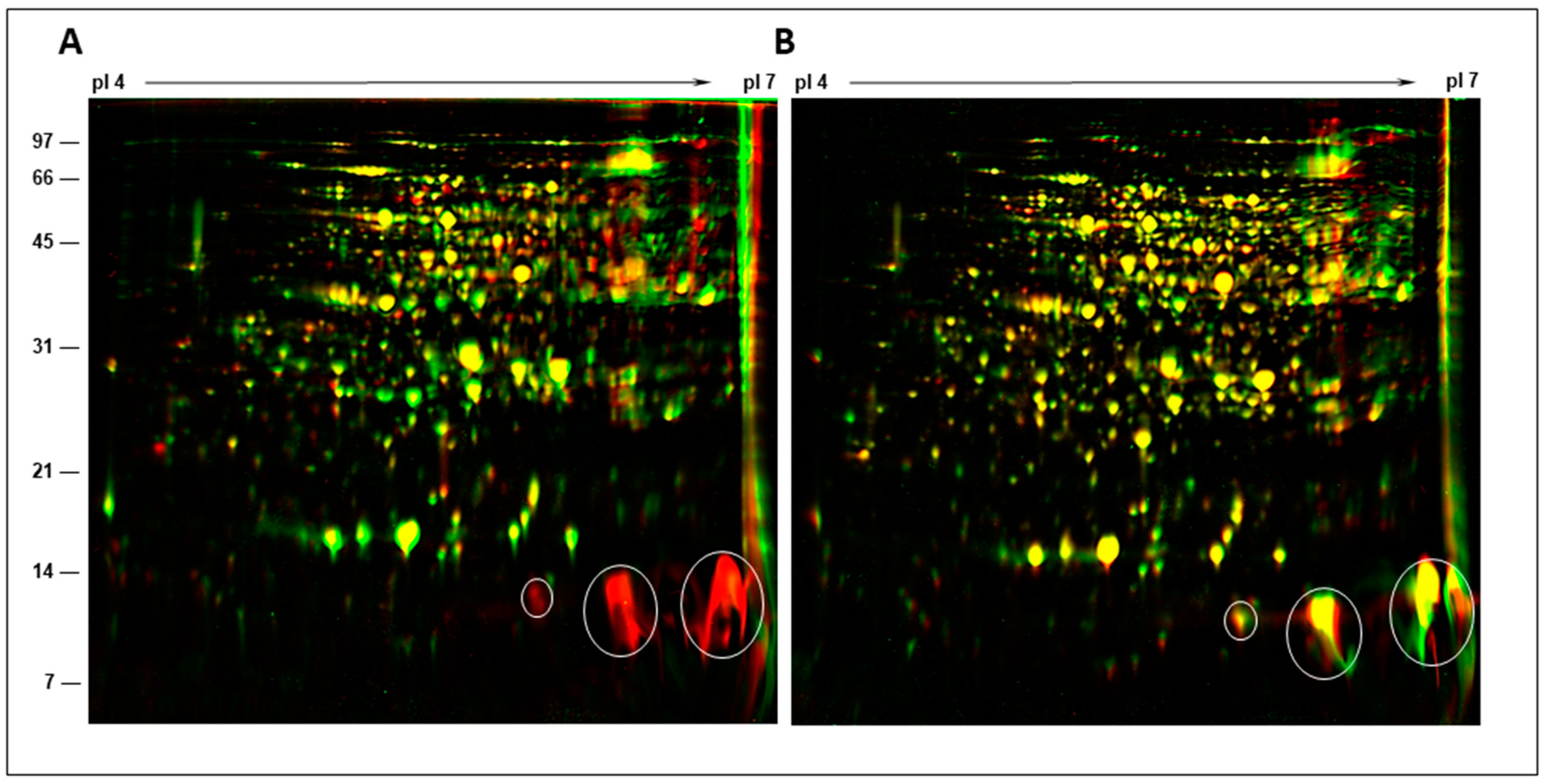
2.5. Two-Dimensional Gel Analysis of Nodule Proteins
2.6. Leghemoglobin and Nitrogenase Are Absent in Pigeon Pea Nodules Produced by Bradyrhizobium diazoefficiens USDA110
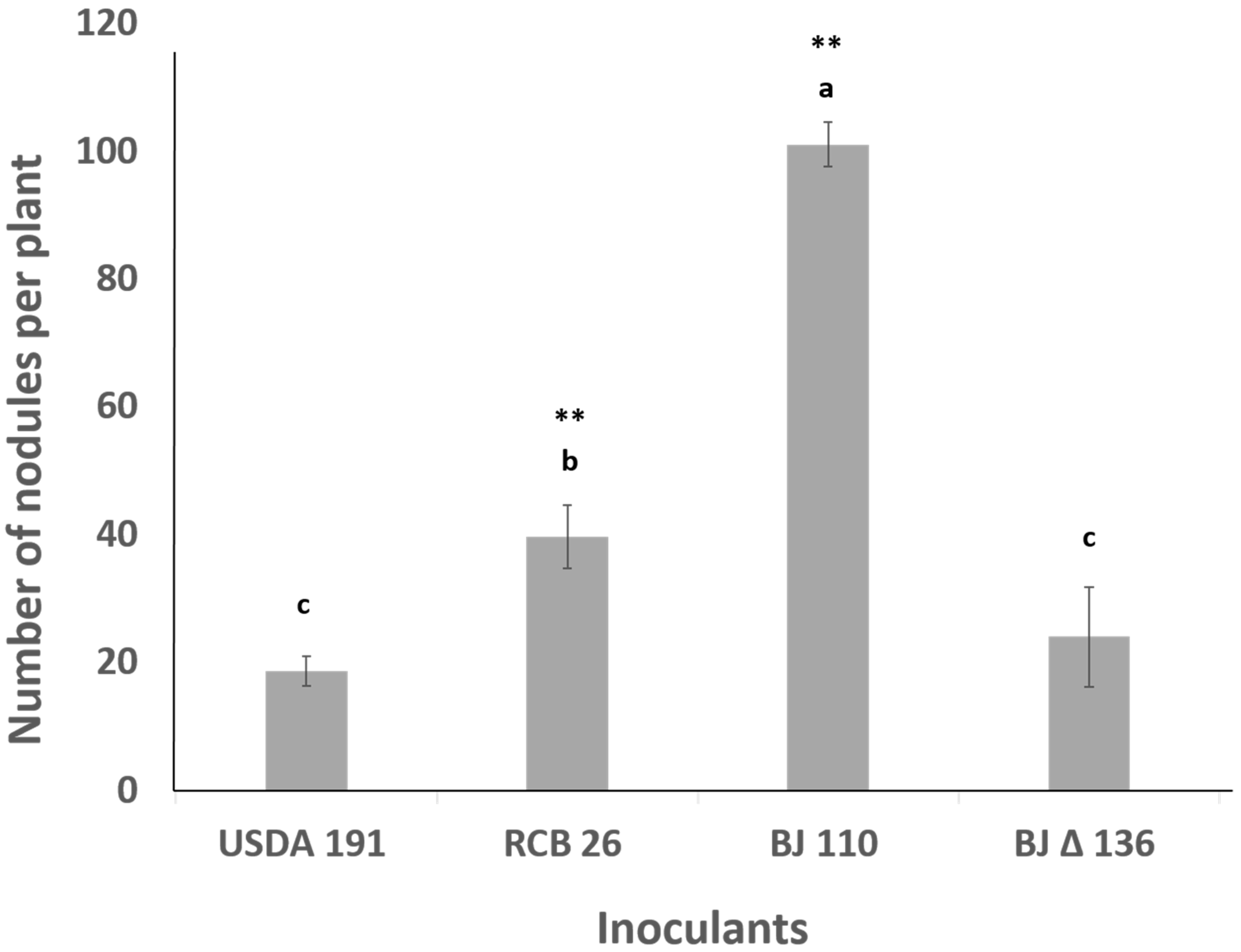
2.7. Coinoculation with Bradyrhizobium diazoefficiens T3SS Mutant Drastically Lowers the Number of Nodules Formed by the Wild Type Strain
3. Discussion
4. Materials and Methods
4.1. Bacteria Strains and Growth Condition
4.2. Nodulation Assay
4.3. Light and Electron Microscopy Images
4.4. Isolation of Extracellular Proteins
4.5. One-Dimensional Gel Electrophoresis
4.6. 2-D Electrophoresis
4.7. Western Blot Analysis
4.8. Competitive Nodulation Assay
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NF | Nod factor |
| T3SS | Type three secretion system |
| Nops | Nodulation outer proteins |
| S. fredii | Sinorhizobium fredii |
| B. diazoefficiens | Bradyrhizobium diazoefficiens |
| TEM | Transmission electron microscopy |
| PBM | Peribacteroid membrane |
| DAI | Days after inoculation |
| PHB | Poly β-hydroxybutyrate |
| EPS | Exopopolysaccharides |
| LPS | Lipopolysaccharides |
| PGPR | Plant growth-promoting rhizobacteria |
References
- Hassan, S.; Mathesius, U. The role of flavonoids in root–rhizosphere signalling: Opportunities and challenges for improving plant–microbe interactions. J. Exp. Bot. 2012, 63, 3429–3444. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Yang, S.; Tang, F.; Zhu, H. Symbiosis specificity in the legume–rhizobial mutualism. Cell. Microbiol. 2012, 14, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Perret, X.; Staehelin, C.; Broughton, W.J. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 2000, 64, 180–201. [Google Scholar] [CrossRef] [PubMed]
- López-Baena, F.J.; Ruiz-Sainz, J.E.; Rodríguez-Carvajal, M.A.; Vinardell, J.M. Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 2016, 17, 755. [Google Scholar] [CrossRef] [PubMed]
- Marczak, M.; Mazur, A.; Koper, P.; Żebracki, K.; Skorupska, A. Synthesis of rhizobial exopolysaccharides and their importance for symbiosis with legume plants. Genes 2017, 8, 360. [Google Scholar] [CrossRef] [PubMed]
- Kawaharada, Y.; Kelly, S.; Nielsen, M.W.; Hjuler, C.T.; Gysel, K.; Muszyński, A.; Carlson, R.; Thygesen, M.B.; Sandal, N.; Asmussen, M. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 2015, 523, 308. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Indrasumunar, A.; Hayashi, S.; Lin, M.H.; Lin, Y.H.; Reid, D.E.; Gresshoff, P.M. Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant. Biol. 2010, 52, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, M.L.C.d.O.; Moreira, M.; Ambrósio, L.A.; Cardoso, E.J.B.N. Occurence and host specificity of indigenous rhizobia from soils of São Paulo State, Brazil. Sci. Agric. 2009, 66, 543–548. [Google Scholar] [CrossRef]
- Gage, D.J. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 2004, 68, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.K.; Day, D.A. Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Biol. 1997, 48, 493–523. [Google Scholar] [CrossRef] [PubMed]
- Hueck, C.J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 1998, 62, 379–433. [Google Scholar] [PubMed]
- Galán, J.E.; Collmer, A. Type III secretion machines: Bacterial devices for protein delivery into host cells. Science 1999, 284, 1322–1328. [Google Scholar]
- Krishnan, H.; Kuo, C.-I.; Pueppke, S. Elaboration of flavonoid-induced proteins by the nitrogen-fixing soybean symbiont Rhizobium fredii is regulated by both nodD1 and nodD2, and is dependent on the cultivar-specificity locus, nolXWBTUV. Microbiology 1995, 141, 2245–2251. [Google Scholar] [CrossRef] [Green Version]
- Saad, M.M.; Crèvecoeur, M.; Masson-Boivin, C.; Perret, X. The T3SS of Cupriavidus taiwanensis strain LMG19424 compromizes symbiosis with Leucaena leucocephala. Appl. Environ. Microbiol. 2012, 78, 7476–7479. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Tampakaki, A.P. Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria. Front. Plant. Sci. 2014, 5, 114. [Google Scholar] [CrossRef] [PubMed]
- He, S.Y. Type III protein secretion systems in plant and animal pathogenic bacteria. Annu. Rev. Phytopathol. 1998, 36, 363–392. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, H.B.; Lorio, J.; Kim, W.S.; Jiang, G.; Kim, K.Y.; DeBoer, M.; Pueppke, S.G. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant-Microbe Interact. 2003, 16, 617–625. [Google Scholar] [CrossRef] [PubMed]
- de Lyra, M.d.C.; López-Baena, F.J.; Madinabeitia, N.; Vinardell, J.M.; Espuny, M.d.R.; Cubo, M.T.; Bellogín, R.A.; Ruiz-Sainz, J.E.; Ollero, F.J. Inactivation of the Sinorhizobium fredii HH103 rhcJ gene abolishes nodulation outer proteins (Nops) secretion and decreases the symbiotic capacity with soybean. Int. Microbiol. 2006, 9, 125–133. [Google Scholar]
- Viprey, V.; Del Greco, A.; Golinowski, W.; Broughton, W.J.; Perret, X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 1998, 28, 1381–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krause, A.; Doerfel, A.; Göttfert, M. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 2002, 15, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Zehner, S.; Hempel, J.; Lang, K.; Göttfert, M. Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol. Lett. 2009, 295, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okazaki, S.; Okabe, S.; Higashi, M.; Shimoda, Y.; Sato, S.; Tabata, S.; Hashiguchi, M.; Akashi, R.; Göttfert, M.; Saeki, K. Identification and functional analysis of type III effector proteins in Mesorhizobium loti. Mol. Plant-Microbe Interact. 2010, 23, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Heron, D.; Ersek, T.; Krishnan, H.; Pueppke, S. Nodulation mutants of Rhizobium fredii USDA257. Mol. Plant-Microbe Interact. 1989. [Google Scholar] [CrossRef]
- Krishnan, H.B.; Pueppke, S.G. Sequence and analysis of the nodABC region of Rhizobium fredii USDA257, a nitrogen-fixing symbiont of soybean and other legumes. Mol. Plant-Microbe Interact. 1991, 4, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, H.B.; Pueppke, S.G. Inactivation of nolC conditions developmental abnormalities in nodulation of Peking soybean by Rhizobium fredii USDA257. Mol. Plant-Microbe Interact. 1992, 5, 14–21. [Google Scholar] [CrossRef]
- Krishnan, H.; Pueppke, S. Flavonoid inducers of nodulation genes stimulate Rhizobium fredii USDA257 to export proteins into the environment. Mol. Plant-Microbe Interact. 1993, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.K.; Kumbhar, B.; Sarkar, B. Process parameter optimization for instant pigeonpea dhal using response surface methodology. J. Food Eng. 2007, 81, 171–178. [Google Scholar] [CrossRef]
- Salunkhe, D.; Chavan, J.; Kadam, S.; Reddy, N. Pigeonpea as an important food source. Crit. Rev. Food Sci. Nutr. 1986, 23, 103–145. [Google Scholar] [CrossRef] [PubMed]
- La Favre, J.S.; Focht, D.D. Comparison of N2 fixation and yields in Cajanus cajan between hydrogenase-positive and hydrogenase-negative rhizobia by in situ acetylene reduction assays and direct 15N partitioning. Plant Physiol. 1983, 72, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Sanginga, N.; Okogun, A.; Akobundu, I.; Carsky, R.; Tian, G.; Wirkom, L. Nodulation and estimation of symbiotic nitrogen fixation by herbaceous and shrub legumes in Guinea savanna in Nigeria. Biol. Fertility Soils 1996, 23, 441–448. [Google Scholar] [CrossRef]
- España, M.; Cabrera-Bisbal, E.; López, M. Study of Nitrogen Fixation by Tropical Legumes in Acid Soil from Venezuelan Savannas Using 15N. Interciencia 2006, 31, 197–201. [Google Scholar]
- Fossou, R.K.; Ziegler, D.; Zézé, A.; Barja, F.; Perret, X. Two Major Clades of Bradyrhizobia Dominate Symbiotic Interactions with Pigeonpea in Fields of Côte d’Ivoire. Front. Microbiol. 2016, 7, 1793. [Google Scholar] [CrossRef] [PubMed]
- Degefu, T.; Wolde-meskel, E.; Adem, M.; Fikre, A.; Amede, T.; Ojiewo, C. Morphophysiological diversity of rhizobia nodulating pigeon pea (Cajanus cajan L. Millsp.) growing in Ethiopia. Afr. J. Biotechnol. 2018, 17, 167–177. [Google Scholar]
- Rufini, M.; Oliveira, D.P.; Trochmann, A.; Soares, B.L.; Andrade, M.J.B.d.; Moreira, F.M.d.S. Bradyrhizobium spp. Strains in Symbiosis with Pigeon Pea cv. Fava-Larga under Greenhouse and Field Conditions. Rev. Bras. Cienc. Solo 2016, 40. [Google Scholar] [CrossRef]
- Süß, C.; Hempel, J.; Zehner, S.; Krause, A.; Patschkowski, T.; Göttfert, M. Identification of genistein-inducible and type III-secreted proteins of Bradyrhizobium japonicum. J. Biotechnol. 2006, 126, 69–77. [Google Scholar]
- Kanbe, M.; Yagasaki, J.; Zehner, S.; Göttfert, M.; Aizawa, S.-I. Characterization of two sets of subpolar flagella in Bradyrhizobium japonicum. J. Bacteriol. 2007, 189, 1083–1089. [Google Scholar] [CrossRef] [PubMed]
- Hempel, J.; Zehner, S.; Göttfert, M.; Patschkowski, T. Analysis of the secretome of the soybean symbiont Bradyrhizobium japonicum. J. Biotechnol. 2009, 140, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Bellato, C.; Krishnan, H.; Cubo, T.; Temprano, F.; Pueppke, S. The soybean cultivar specificity gene noIX is present, expressed in a nodD-dependent manner, and of symbiotic significance in cultivar-nonspecific strains of Rhizobium (Sinorhizobium) fredii. Microbiology 1997, 143, 1381–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, D.; Fortin, M.; Stanley, J.; Mauro, V.; Purohit, S.; Morrison, N. Nodulins and nodulin genes of Glycine max. Plant Mol. Biol. 1986, 7, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Appleby, C. The origin and functions of haemoglobin in plants. Sci. Prog. 1992, 365–398. [Google Scholar]
- Appleby, C.; Bergersen, F. Methods for evaluating biological nitrogen fixation. New. Age. Inter. 1980. [Google Scholar]
- Yang, S.; Tang, F.; Gao, M.; Krishnan, H.B.; Zhu, H. R gene-controlled host specificity in the legume–rhizobia symbiosis. Proc. Nati. Acad. Sci. USA 2010, 107, 18735–18740. [Google Scholar] [CrossRef] [PubMed]
- Vest, G.; Caldwell, B. Rj4—A gene conditioning ineffective nodulation in soybean 1. Crop Sci. 1972, 12, 692–693. [Google Scholar] [CrossRef]
- Pazdernik, D.L.; Vance, C.P.; Sadowsky, M.J.; Graham, P.H.; Orf, J.H. A host-controlled, serogroup-specific, ineffective-nodulation system in the Bradyrhizobium-soybean (Glycine max) symbiosis. Mol. Plant-Microbe Interact. 1997, 10, 994–1001. [Google Scholar] [CrossRef]
- Tirichine, L.; de Billy, F.; Huguet, T. Mtsym6, a Gene ConditioningSinorhizobium Strain-Specific Nitrogen Fixation inMedicago truncatula. Plant Physiol. 2000, 123, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, L.; Krishnan, H.; Balatti, P.; Pueppke, S. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 1993, 9, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Kimbrel, J.A.; Thomas, W.J.; Jiang, Y.; Creason, A.L.; Thireault, C.A.; Sachs, J.L.; Chang, J.H. Mutualistic co-evolution of type III effector genes in Sinorhizobium fredii and Bradyrhizobium japonicum. PLoS Path. 2013, 9, e1003204. [Google Scholar] [CrossRef] [PubMed]
- Keyser, H.H.; Bohlool, B.B.; Hu, T.; Weber, D.F. Fast-growing rhizobia isolated from root nodules of soybean. Science 1982, 215, 1631–1632. [Google Scholar] [CrossRef] [PubMed]
- Newcomb, W.; Sippell, D.; Peterson, R. The early morphogenesis of Glycine max and Pisum sativum root nodules. Cana. J. Bot. 1979, 57, 2603–2616. [Google Scholar] [CrossRef]
- Caldwell, B. Inheritance of a strain-specific ineffective nodulation in soybeans. Crop Sci. 1966, 6, 427–428. [Google Scholar] [CrossRef]
- Caldwell, B.; Hinson, K.; Johnson, H. A strain-specific ineffective nodulation reaction in the soybean Glycine max L. Merrill. Crop Sci. 1966, 6, 495–496. [Google Scholar] [CrossRef]
- Cregan, P.; Keyser, H. Host Restriction of Nodulation by Bradyrhizobium japonicum Strain USDA 123 in Soybean 1. Crop Sci. 1986, 26, 911–916. [Google Scholar] [CrossRef]
- Kneen, B.; LaRue, T. Peas (Pisum sativum L.) with strain specificity for Rhizobium leguminosarum. Heredity 1984, 52, 383–389. [Google Scholar] [CrossRef]
- Pankhurst, C. Ineffective Rhizobium trifolii Mutants Examined by Immunediffusion, Gel-electrophoresis and Electron Microscopy. Microbiology 1974, 82, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Bergersen, F. Formation and function of bacteroids. In The Biology of Nitrogen Fixation; North Holland Publising Co: Amsterdam, The Netherlands, 1974; pp. 473–498. [Google Scholar]
- Holl, F. Host plant control of the inheritance of dinitrogen fixation in the Pisum-Rhizobium symbiosis. Euphytica 1975, 24, 767–770. [Google Scholar] [CrossRef]
- Blauenfeldt, J.; Joshi, P.; Gresshoff, P.; Caetano-Anollés, G. Nodulation of white clover (Trifolium repens) in the absence ofRhizobium. Protoplasma 1994, 179, 106–110. [Google Scholar] [CrossRef]
- Joshi, P.A.; Caetano-Anolles, G.; Graham, E.T.; Gresshoff, P. Ontogeny and ultrastructure of spontaneous nodules in alfalfa (Medicago sativa). Protoplasma 1991, 162, 1–11. [Google Scholar] [CrossRef]
- Caetano-Anolles, G.; Joshi, P.; Gresshoff, P. Nodule morphogenesis in the absence of Rhizobium. In New Horizons in Nitrogen Fixation; Springer: Dordrecht, The Netherlands, 1993; pp. 297–302. [Google Scholar]
- Truchet, G.; Camut, S.; De Billy, F.; Odorico, R.; Vasse, J. TheRhizobium-legume symbiosis Two methods to discriminate between nodules and other root-derived structures. Protoplasma 1989, 149, 82–88. [Google Scholar] [CrossRef]
- Peralta, H.; Mora, Y.; Salazar, E.; Encarnación, S.; Palacios, R.; Mora, J. Engineering the nifH promoter region and abolishing poly-β-hydroxybutyrate accumulation in Rhizobium etli enhance nitrogen fixation in symbiosis with Phaseolus vulgaris. Appl. Environ. Microbiol. 2004, 70, 3272–3281. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, W.C.; Kadam, S.V.; Denison, R.F. Poly-3-hydroxybutyrate (PHB) supports survival and reproduction in starving rhizobia. FEMS Microbiol. Ecol. 2008, 65, 391–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, P.; Doharey, R.; Singh, S.; Singh, R.K.; Kumar, M.; Kumar, A. Communication and psychological behavior of the pigeon pea growers in Chitrakoot district, India. Int. J. Curr. Microbiol. App. Sci. 2017, 6, 2032–2037. [Google Scholar]
- Tilak, K.; Ranganayaki, N.; Manoharachari, C. Synergistic effects of plant-growth promoting rhizobacteria and Rhizobium on nodulation and nitrogen fixation by pigeonpea (Cajanus cajan). Eur. J. Soil Sci. 2006, 57, 67–71. [Google Scholar] [CrossRef]
- Schwinghamer, E.; Evans, H.; Dawson, M. Evaluation of effectiveness in mutant strains of Rhizobium by acetylene reduction relative to other criteria of N 2 fixation. Plant Soil 1970, 33, 192–212. [Google Scholar] [CrossRef]
- Krishnan, H.B. NolX of Sinorhizobium fredii USDA257, a type III-secreted protein involved in host range determination, is localized in the infection threads of cowpea (Vigna unguiculata [L.] Walp) and soybean (Glycine max [L.] Merr.) nodules. J. Bacteriol. 2002, 184, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Lorio, J.C.; Kim, W.S.; Krishnan, H.B. NopB, a soybean cultivar-specificity protein from Sinorhizobium fredii USDA257, is a type III secreted protein. Mol. Plant-Microbe Interact. 2004, 17, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, H.B.; Oehrle, N.W.; Natarajan, S.S. A rapid and simple procedure for the depletion of abundant storage proteins from legume seeds to advance proteome analysis: A case study using Glycine max. Proteomics 2009, 9, 3174–3188. [Google Scholar] [CrossRef] [PubMed]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alaswad, A.A.; Oehrle, N.W.; Krishnan, H.B. Classical Soybean (Glycine max (L.) Merr) Symbionts, Sinorhizobium fredii USDA191 and Bradyrhizobium diazoefficiens USDA110, Reveal Contrasting Symbiotic Phenotype on Pigeon Pea (Cajanus cajan (L.) Millsp). Int. J. Mol. Sci. 2019, 20, 1091. https://doi.org/10.3390/ijms20051091
Alaswad AA, Oehrle NW, Krishnan HB. Classical Soybean (Glycine max (L.) Merr) Symbionts, Sinorhizobium fredii USDA191 and Bradyrhizobium diazoefficiens USDA110, Reveal Contrasting Symbiotic Phenotype on Pigeon Pea (Cajanus cajan (L.) Millsp). International Journal of Molecular Sciences. 2019; 20(5):1091. https://doi.org/10.3390/ijms20051091
Chicago/Turabian StyleAlaswad, Alaa A., Nathan W. Oehrle, and Hari B. Krishnan. 2019. "Classical Soybean (Glycine max (L.) Merr) Symbionts, Sinorhizobium fredii USDA191 and Bradyrhizobium diazoefficiens USDA110, Reveal Contrasting Symbiotic Phenotype on Pigeon Pea (Cajanus cajan (L.) Millsp)" International Journal of Molecular Sciences 20, no. 5: 1091. https://doi.org/10.3390/ijms20051091
APA StyleAlaswad, A. A., Oehrle, N. W., & Krishnan, H. B. (2019). Classical Soybean (Glycine max (L.) Merr) Symbionts, Sinorhizobium fredii USDA191 and Bradyrhizobium diazoefficiens USDA110, Reveal Contrasting Symbiotic Phenotype on Pigeon Pea (Cajanus cajan (L.) Millsp). International Journal of Molecular Sciences, 20(5), 1091. https://doi.org/10.3390/ijms20051091